13.3
Impact Factor
Theranostics 2017; 7(6):1437-1446. doi:10.7150/thno.16558 This issue Cite
Research Paper
Cross-Platform Comparison of Four Leading Technologies for Detecting EGFR Mutations in Circulating Tumor DNA from Non-Small Cell Lung Carcinoma Patient Plasma
1. Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Department of Clinical Laboratory, Peking University Cancer Hospital & Institute, Beijing, China;
2. Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Department of Thoracic Surgery I, Peking University Cancer Hospital & Institute, Beijing, China;
3. Shanghai AccuraGen Biotechnology Co. Ltd, China.
*Ting Xu and Xiaozheng Kang contributed equally to this study.
#Ke-Neng Chen and Guobing Xu jointly supervised this work.
Abstract
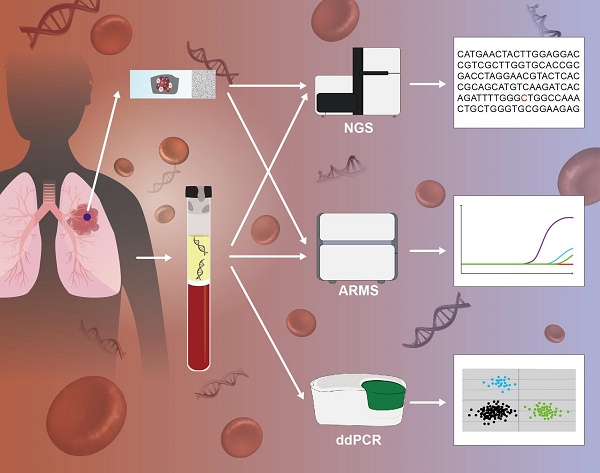
Analysis of circulating tumor DNA (ctDNA) is emerging as a powerful tool for guiding targeted therapy and monitoring tumor evolution in patients with non-small cell lung cancer (NSCLC), especially when representative tissue biopsies are not available. Here, we have compared the ability of four leading technology platforms to detect epidermal growth factor receptor (EGFR) mutations (L858R, exon 19 deletion, T790M and G719X) in ctDNA from NSCLC patients. Two amplification refractory mutation systems (cobas-ARMS and ADx-ARMS), a droplet digital polymerase chain reaction (ddPCR) and a next-generation sequencing (Firefly NGS) platform were included in the comparison. Fifteen EGFR mutations across twenty NSCLC patients were identified. Firefly NGS, cobas-ARMS and ddPCR all displayed superior sensitivity while ADx-ARMS was better suited for the qualitative detection of EGFR mutations with allele frequency higher than 1% in plasma and tissue samples. We observed high coincidence between the plasma and tissue EGFR mutational profiles for three driver mutations (L858R, exon 19 deletion and G719X) that are known targets of first generation EGFR-TKI therapies among patients who relapsed. Discrepancies between tissue and plasma EGFR mutational profiles were mainly attributable to spatial and temporal tumor heterogeneity, mutation inhibition due to therapy response and drug resistance (T790M). This study illustrates the challenges associated with selection of a technology platform for EGFR ctDNA analysis in the context of treatment evaluation and drug resistance detection.
Keywords: non-small cell lung cancer, epidermal growth factor receptor, quantitative real-time polymerase chain reaction, next-generation sequencing.
 Global reach, higher impact
Global reach, higher impact