13.3
Impact Factor
Theranostics 2017; 7(5):1088-1099. doi:10.7150/thno.18551 This issue Cite
Research Paper
Exome Sequencing of Oral Squamous Cell Carcinoma Reveals Molecular Subgroups and Novel Therapeutic Opportunities
1. Whole-Genome Research Core Laboratory of Human Diseases, Chang Gung Memorial Hospital, Keelung, Taiwan;
2. Department of Dermatology, Drug Hypersensitivity Clinical and Research Center, Chang Gung Memorial Hospital, Linkou, Taiwan;
3. Institute of Oral Sciences, Chung Shan Medical University, Taichung, Taiwan;
4. Department of Dentistry, Chung Shan Medical University Hospital, Taichung, Taiwan;
5. Department of Biomedical Sciences, Chung Shan Medical University, Taichung, Taiwan;
6. Department of Otorhinolaryngology-Head and Neck Surgery, Changhua Christian Hospital, Changhua, Taiwan;
7. Biodiversity Research Center, Academia Sinica, Taipei, Taiwan;
8. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan;
9. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan;
10. School of Medicine, Chung Shan Medical University, Taichung, Taiwan;
11. Department of Otolaryngology, Chung Shan Medical University Hospital, Taichung, Taiwan;
12. School of Medicine, College of Medicine, Chang Gung University, Taoyuan, Taiwan.
Abstract
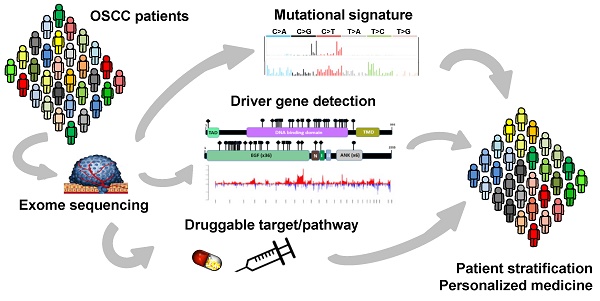
Oral squamous cell carcinoma (OSCC), an epithelial malignancy affecting a variety of subsites in the oral cavity, is prevalent in Asia. The survival rate of OSCC patients has not improved over the past decades due to its heterogeneous etiology, genetic aberrations, and treatment outcomes. Improvement in therapeutic strategies and tailored treatment options is an unmet need. To unveil the mutational spectrum, whole-exome sequencing of 120 OSCC from male individuals in Taiwan was conducted. Analyzing the contributions of the five mutational signatures extracted from the dataset of somatic variations identified four groups of tumors that were significantly associated with demographic and clinical features. In addition, known (TP53, FAT1, EPHA2, CDKN2A, NOTCH1, CASP8, HRAS, RASA1, and PIK3CA) and novel (CHUK and ELAVL1) genes that were significantly and frequently mutated in OSCC were discovered. Further analyses of gene alteration status with clinical parameters revealed that the tumors of the tongue were enriched with copy-number alterations in several gene clusters containing CCND1 and MAP4K2. Through defining the catalog of targetable genomic alterations, 58% of the tumors were found to carry at least one aberrant event potentially targeted by US Food and Drug Administration (FDA)-approved agents. Strikingly, if targeting the p53-cell cycle pathway (TP53 and CCND1) by the drugs studied in phase I-III clinical trials, those possibly actionable tumors are predominantly located in the tongue, suggesting a better prediction of sensitivity to current targeted therapies. Our work revealed molecular OSCC subgroups that reflect etiological and prognostic correlation as well as defined the landscape of major altered events in the coding regions of OSCC genomes. These findings provide clues for the design of clinical trials for targeted therapies and stratification of OSCC patients with differential therapeutic efficacy.
Keywords: Oral squamous cell carcinoma, exome sequencing, mutational signature, driver gene, targeted therapy.
 Global reach, higher impact
Global reach, higher impact