13.3
Impact Factor
Theranostics 2017; 7(4):1010-1025. doi:10.7150/thno.17736 This issue Cite
Review
Theranostic DNAzymes
1. Xiangya School of Pharmaceutical Sciences, Central South University, Changsha, Hunan, China, 410013.
2. Department of Chemistry, Waterloo Institute for Nanotechnology, University of Waterloo, Waterloo, Ontario, Canada, N2L 3G1.
Received 2016-9-27; Accepted 2016-10-31; Published 2017-2-23
Abstract
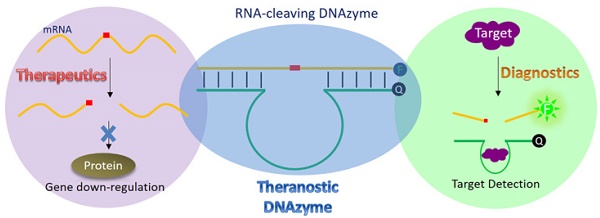
DNAzymes are catalytically active DNA molecules that are obtained via in vitro selection. RNA-cleaving DNAzymes have attracted significant attention for both therapeutic and diagnostic applications due to their excellent programmability, stability, and activity. They can be designed to cleave a specific mRNA to down-regulate gene expression. At the same time, DNAzymes can sense a broad range of analytes. By combining these two functions, theranostic DNAzymes are obtained. This review summarizes the progress of DNAzyme for theranostic applications. First, in vitro selection of DNAzymes is briefly introduced, and some representative DNAzymes related to biological applications are summarized. Then, the applications of DNAzyme for RNA cleaving are reviewed. DNAzymes have been used to cleave RNA for treating various diseases, such as viral infection, cancer, inflammation and atherosclerosis. Several formulations have entered clinical trials. Next, the use of DNAzymes for detecting metal ions, small molecules and nucleic acids related to disease diagnosis is summarized. Finally, the theranostic applications of DNAzyme are reviewed. The challenges to be addressed include poor DNAzyme activity under biological conditions, mRNA accessibility, delivery, and quantification of gene expression. Possible solutions to overcome these challenges are discussed, and future directions of the field are speculated.
Keywords: DNAzymes, RNA, biosensors, metal ions, delivery.
Introduction
Theranostics refers to combining disease diagnosis and therapy in one molecular package, which overcomes undesirable differences in bio-distribution when performed separately [1, 2]. The improved nanomaterials synthesis and bioconjugation has also boosted the development of this field. While many strategies have been explored to achieve this goal, artificial enzyme mimics have emerged as a promising direction for theranostics. Enzymes catalyze bio-related chemical transformations with catalytic turnovers, allowing signal amplification. If the reactions are designed to intervene disease-related processes and molecules, they can also achieve therapeutic effects. Using enzyme mimics instead of natural protein-based enzymes has a few important advantages. 1) Foreign proteins might elicit immune responses. 2) The production of protein enzymes usually requires fermentation with a high cost and the products are easily contaminated. 3) It is difficult to label proteins at specific sites, making in-situ production of signal quite challenging. Over the past few decades, many small molecules, polymers and nanomaterials have been studied to mimic a diverse range of enzymes [3-5]. Among them, DNAzymes (DNA-based catalysts) are particularly attractive [6-9].
The biggest advantage of DNAzymes for theranostics probably is amenability to combinatorial selection. Different DNAzymes can be evolved in vitro to fit specific needs, and this is more difficult to achieve for other catalytic molecules or nanomaterials. In addition, DNA is highly programmable and easy to modify and label with very low immunogenicity. Therefore, it is quite easy to rationally design DNAzyme sequences for theranostic applications. It is also straightforward to conjugate DNAzymes to various nanomaterials for signaling and delivery. Finally, compared to RNA and proteins, DNA is much more stable and cost-effective.
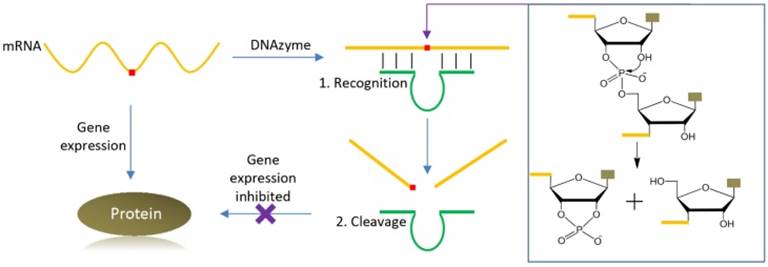
The first DNAzyme was reported in 1994 for RNA cleavage [10]. Since then, many different types of DNAzymes have been reported, catalyzing RNA/DNA cleavage, ligation, and phosphorylation, and other reactions [9, 11]. Among them, RNA-cleaving DNAzymes have been extensively used as biosensors [6-9], and they can also be used as therapeutic agents, thus fitting the need of theranostics. Figure 1 shows the cartoon of using such a DNAzyme for cleaving an mRNA, in which the 2′-hydroxyl group acts as internal nucleophile to attack the adjacent phosphodiester bond to initiate the cleavage reaction. The DNAzyme can be directed to the specific cleavage site by designing the substrate binding arms. Selective cleavage of viral RNA and oncogene related mRNA is quite useful for anti-viral and anti-cancer applications, respectively. It is also possible to use the same RNA molecule to produce fluorescence signal for its detection.
Reviews on RNA-cleaving DNAzymes have been published to cover various aspects such as for analytical applications [6, 7], nanotechnology [12], and chemical biology [8, 9]. Herein, the scope of this review is on their detection and therapeutic applications for in vitro and in vivo biochemical and biomedical studies. Therefore, we mainly cover those DNAzymes that can work in physiological conditions.
In vitro selection of DNAzymes
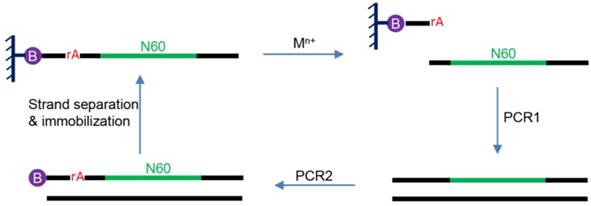
So far, no DNAzymes have been found in nature. This is not surprising since most DNAs in cells are double-stranded while the catalytic core of DNAzymes is typically single-stranded. Therefore, DNAzymes are enzyme mimics. All reported DNAzymes were isolated using a combinatorial biology technique called in vitro selection. To understand this process, an example of in vitro selection of RNA-cleaving DNAzymes is briefly introduced here (Figure 2) [10]. The initial selection library contains a 60-nucleotide random region, flanked by two constant regions for PCR primer binding. A typical library contains ~1014 random DNA sequences. The library also contains at least a ribonucleotide as the putative cleavage site. In this particular example, a ribo-adenine (rA) is used. A ribonucleotide is ~106-fold more liable to cleavage than deoxyribonucleotides [13]. At the 5′-end of this DNA, a biotin is labeled to immobilize the library on a streptavidin column. In the presence of a metal ion, a small fraction of the library that can fold into an active structure are cleaved at the rA site and released from the column. The cleaved products are amplified by two rounds of PCR to regenerate the library to seed next round of selection. PCR1 is used to produce the full-length of library and the PCR2 is for introducing the rA cleavage site and biotin tag. The iterative process is typically repeated for 5-10 rounds until the activity of the DNA pool reaches a plateau, and the library is then sequenced to identify the most active sequences.
Since the initial report of this protocol in 1994, significant advances have been made on DNAzyme selection, although the basic working principle remains the same: cleavage, separation and amplification. Separation methods that rely on denaturing polyacrylamide gel electrophoresis (dPAGE) have also been commonly used [14, 15]. Thanks to the development of DNA synthesis technique, fluorophore labels can be readily attached to the library to follow the progress of selection, avoiding the use of radio-isotopes. In addition, incorporation of a fluorophore/quencher pair right next cleavage site was reported to synchronize the cleavage reaction with fluorescence signaling [16].
A cartoon showing specific mRNA recognition and cleavage by a DNAzyme. The red square in the mRNA denotes for the targeted cleavage site. The DNAzyme can be directed to the cleavage site by designing the substrate binding arms using Watson-Crick base pairing, thus inhibiting gene expression. The general mechanism of the RNA cleavage reaction is also presented, where the 2′-OH group attacks the scissile phosphate to initiate the cleavage reaction.
A basic scheme of in vitro selection. The rA denotes ribo-adenine and the circled B denotes biotin moiety. The sequences that can cleave the rA bond in the presence of the added metal ions is harvested and amplified by PCR. The biotin is needed for library immobilization so that the negative strand after PCR can be washed away to generate the single-stranded DNA library. In addition, it allows the separation of the cleaved DNA from the rest of the library.
A key advantage of in vitro selection is that DNAzymes can be isolated under physiological conditions or even using biological fluids directly to ensure their activity for the intended applications [17]. On the other hand, extreme conditions such as high temperature or low pH can be used for other tailored applications [18, 19]. In addition, the selection library can be designed to fine-tune the function of DNAzymes. For instance, with the above single ribonucleotide bearing library, the DNAzymes are obtained for cleaving RNA/DNA chimera substrates, which is preferred for analytical applications. For mRNA cleavage, on the other hand, the library should contain a long stretch of RNA sequence, so that the resulting DNAzymes are able to cleave all-RNA substrates [20].
Some representative DNAzymes
Over the past 22 years, many RNA-cleaving DNAzymes have been isolated for various purposes. As such, different ways can be used to classify the reported DNAzymes. For example, DNAzymes can be classified based on their metal ion cofactors. Metal ions are an indispensable part of DNAzyme catalysis, especially divalent metal ions. For environmental monitoring, many selections were intentionally carried out in the presence of certain heavy metal ions. For biomedical applications, the metal ions of interest are Na+, K+, Mg2+, and Ca2+. Most other metal ions, although they can be effective cofactors, do not exist in a high concentration in blood or cells. Thus they are unlikely to be useful for this purpose. In addition, DNAzymes can be classified as cleaving all-RNA or a single RNA embedded in a chimeric DNA substrate. Finally, DNAzymes can be classified to contain only natural nucleotides or with modified ones. Modified nucleotides expand the chemical functionality of DNA and they may enhance the cleavage rate, but they complicate the selection process and make DNA synthesis much more expensive.
Here, we briefly review a few DNAzymes that might be of interest to the theranostic community with a historical perspective. The first DNAzyme was isolated in 1994 for cleaving a single RNA linkage in a chimeric substrate using Pb2+ [10]. It was later confirmed by Lu and co-workers that it is extremely specific for Pb2+ [21]. This DNAzyme is highly efficient with a rate >10 min-1 [22], but apparently it is not useful for cleaving cellular RNA since it is only active with Pb2+ and cannot cleave all-RNA substrate.
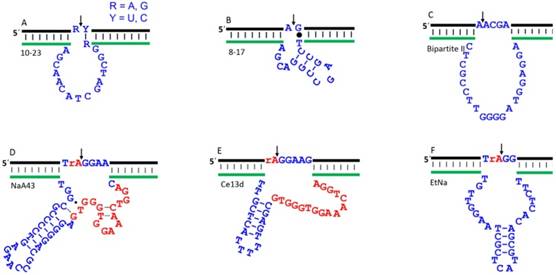
In 1997, the landmark work by Santoro and Joyce made the community very excited [20]. The 10-23 and 8-17 DNAzymes reported in this paper can cleave full-RNA substrates in the presence of Mg2+ (Figure 3A, B). At high Mg2+ concentrations, the 10-23 DNAzyme can rival protein enzymes in terms of catalytic efficiency defined by kcat/Km. One notable merit of the 10-23 is that it cleaves all purine-pyrimidine junctions, which offers excellent flexibility to target specific sites in an RNA sequence. Due to its high catalytic efficiency, general substrate cleavage junctions and small size, this DNAzyme have been extensively exploited for in vitro and in vivo therapeutic applications.
The substrate tolerance was reported to be less versatile for the 8-17 DNAzyme, which requires a G•T wobble pair next to the cleavage site (Figure 3B) [20]. Later, it was reported that it can cleave most of the dinucleotide junctions, although the efficiency can vary a few orders of magnitude [23]. Further studies suggested that the 10-23 DNAzyme is a variant of the 8-17 [24]. We reported that the 8-17 is even faster than the 10-23 for cleaving chimeric substrates in the presence of Mg2+ [25]. The 8-17 motif has been repeated selected by several labs using Zn2+ [26], Mg2+ [27, 28], Ca2+ [17, 28], Cd2+ [29], and other conditions with the best activity observed with Pb2+ [30]. Its recurrence has been attributed to the small size, high tolerance to mutations, and high activity [27]. Because of their importance, works on rational modification of these DNAzymes were reported to improve its activity [31, 32]. In 2001, a new DNAzyme named Bipartite II was reported by Sen to cleave full-RNA substrate, with a kcat of ~1.4 min-1 at 30 mM Mg2+ (Figure 3C) [33]. Since its activity is lower, its applications were not widely explored.
After these early attempts, the selection of DNAzyme for all-RNA substrate was rarely performed later (except some studies using modified nucleotides). Instead, most selections were performed to cleave RNA/DNA chimeric substrates for analytical or nanotechnology applications. A few Mg2+-dependent DNAzymes were isolated, but their activities were quite low [34]. Many interesting progresses were made by using transition metal ions [7, 11, 35]. Many DNAzymes were reported to use Zn2+ [26, 36], UO22+ [14], Cu2+ [37], Cd2+ [29, 38], Hg2+ [39], and Ag+ [40]. We also isolated a series of lanthanide-dependent DNAzymes [15, 41-43]. However, since most of these metals do not exist in high concentrations in biological fluids, their biomedical applications are unlikely to be fruitful and are not further discussed here.
Recently, a few new DNAzymes have been discovered to use physiologically relevant metals. For example, Lu and coworkers reported a highly specific Na+-dependent DNAzyme named NaA43 (Figure 3D) [44]. It is quite active under physiological concentration of Na+, and can reach a rate of 0.1 min-1 in presence of 400 mM Na+ alone with exceptional Na+ specificity. This DNAzyme was then used to design a biosensor for intracellular Na+ imaging. Interestingly, the NaA43 shared a stretch of 16 nucleotides in this catalytic core with a lanthanide-dependent DNAzyme we reported, named Ce13d (Figure 3E) [15]. Through detailed characterization, NaA43 and Ce13d contain the same Na+ binding aptamer in their structures, and their identical sequence is important for Na+ binding [45, 46]. The Na+ binding aptamer in the Ce13d was further dissected by various biological and spectroscopic techniques, by which the aptamer was found to have a Kd value of 20-40 mM Na+ [47, 48].
We also isolated another Na+-dependent DNAzyme called EtNa (Figure 3F), which however requires molar concentration of Na+ to achieve fast cleavage in water [49]. It can be accelerated by organic solvents. With 50% EtOH, its rate is ~0.04 min-1, but the involvement of organic solvents renders it unsuitable for in vivo applications. Interestingly, it is quite active with Ca2+ in water. With 2 mM Ca2+, it has a rate of 0.07 min-1.
By observing the six DNAzymes presented in Figure 3, they all have a very similar overall structure. The enzyme strands recognize their substrates by the two substrate binding arms, which can be arbitrary sequences as long as the base pairing interactions are maintained. This offers excellent flexibility for targeting specific regions in RNA. Another feature is that the cleavage activity of DNAzymes can be specifically tested by using inactive mutants. Most DNAzymes can be inactivated by making a single point mutation to a critical nucleotide [30, 50]. This does not change its structure or binding affinity to the substrate, but fully abolishes its activity. This property is valuable to rigorously test the mechanism of gene inhibition in cells (vide infra). Finally, the effect of DNAzymes can be inactivated in vivo by adding antidote, which is a simple sequence fully complementary to the DNAzyme sequence. This concept has been demonstrated in aptamer binding [51-53], and it should also be applicable to DNAzymes.
The secondary structures of a few representative RNA-cleaving DNAzymes. (A) 10-23; (B) 8-17; (C) Bipartite II. These three DNAzymes are able to cleave all-RNA substrates, which can be used as therapeutic agents for intracellular RNA cleavage. The (D) NaA43; (E) Ce13d; and (F) EtNa DNAzymes can only cleave a single RNA embedded DNA substrate, which is mainly for sensing applications. The arrowheads denote cleavage site. The “rA” denotes the single ribo-adenosine linkage. The identical nucleotides in NaA43 and Ce13d catalytic core are highlighted in red. These DNAzymes can all use or interact with physiologically important metal ions including Mg2+, Ca2+, and Na+.
Cleaving RNA for therapeutic applications
Numerous oligonucleotide-based biopharmaceuticals have been tested for gene therapy, such as anti-sense oligonucleotides, small interfering RNAs (siRNAs), ribozymes and DNAzymes. Their main targets are mRNA, and the key steps involve mRNA recognition and cleavage. Recognition of mRNA relies on hybridization. The size of the human genome is about 3 billion base pairs, which is roughly equal to 416. Therefore, in principle, it is possible to specifically recognize a particular segment in any RNA by designing a 16-nucleotide sequence. This length is quite comparable with all of above oligonucleotides-based technologies.
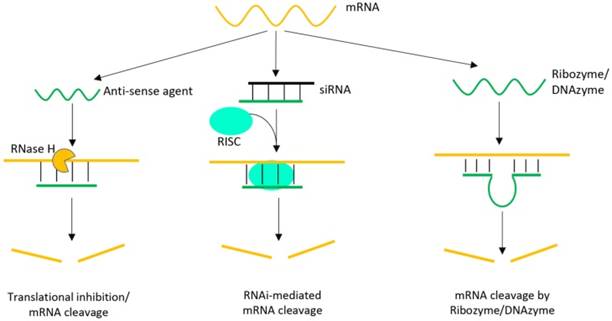
The mRNA cleavage mechanisms, on the other hand, are quite different among them, as summarized in Figure 4. For antisense oligonucleotides, Ribonuclease H (RNase H) is involved to bind and cleave mRNA after hybridization. While the concept is quite simple, the efficiency is quite moderate. The siRNA method also carries out mRNA cleavage but with a more complicated pathway. A siRNA is a class of double-stranded RNA with 2-3 nt overhangs at the 3′-end of each strand, in which one strand (the guide strand) is complementary to target mRNA. After entering cells, the double-stranded siRNA unwinds and the guide strand is incorporated into a nuclease-containing multi-protein complex (called RNA-induced silencing complex, RISC), which cleaves the mRNA. The efficiency of siRNA is much higher than anti-sense oligonucleotides. Due to the intrinsic catalytic activity, ribozymes and DNAzymes are capable of cleaving mRNA independent of endogenous nucleases. In this regard, they have the advantage to bypass cellular machineries. In addition, DNA is more cost-effective and stable compared to RNA-based reagents.
Schematic illustration of various oligonucleotide-directed gene therapeutic strategies, including anti-sense oligonucleotides, siRNA and ribozymes/DNAzymes [54, 55]. In each case, target specificity is achieved by hybridization, but the cleavage mechanisms are different.
The efficacy of ribozymes for mRNA cleaving therapeutics has been demonstrated in several cases, and a few vector-expressed or synthetic ribozymes have entered phase I/II trials, exhibiting good bioavailability and biocompatibility [56, 57]. Compared to ribozymes, DNAzymes are more cost-effective, easier to synthesis, more stable and can be easily labeled, presenting a better candidate for such applications. So far, DNAzymes have showed potential as therapeutic agents for various diseases, such as antiviral, antibacterial, anti-cancer and anti-inflammatory [58-60], as well as atherosclerosis [61]. DNAzyme was initially tested for the HIV RNA cleavage in vitro [20]. In one study, Tandom and coworkers designed a 10-23 DNAzyme against the HIV-1 integrase gene, which is responsible for the integration the HIV-1 genome into the host DNA [62]. The DNAzyme was tested with transiently transfected cell, which showed 70-80% of HIV integrase protein inhibition. Wu et al reported the first example of using DNAzymes for anti-cancer treatment by targeting the bcr-abl oncogene, which is associated with chronic myelogenous leukaemia [63]. In vitro experiments showed that the DNAzyme can efficiently cleave the target, while the inactive DNAzyme mutant (with a single critical nucleotide mutation) failed to produce activity. After delivery into cell, the DNAzyme inhibited the related protein expression by 40%, and inhibited cell growth by >50% after 6 days. The efficacy of the DNAzyme was also demonstrated using freshly isolated cancer cells from patients. DNAzyme was also designed to treat Epstein-Barr virus (EBV) infection by targeting the gene of the latent membrane protein (LMP1), a key protein in EBV-mediated carcinogenesis [64]. The DNAzyme down regulated the LMP1 expression and inhibited cellular signal transduction activated by LMP1. Combined with radiation treatment, the DNAzyme significantly induced apoptosis of the target cells, and the tumor size reduced by 60% in a disease-bearing mouse model. Given that more than >50 targets were demonstrated using the 10-23 DNAzyme alone, and the number is even greater than that with other DNAzymes [58, 65, 66], we do not intend to review them one-by-one. They all share the same strategy of gene silencing. While many promising applications have been reported, no FDA approved DNAzyme-based drugs have made their way to the market yet. This indicates a few challenges in this field. DNAzymes have not reached the same stage of development compared to other gene silencing techniques. Here, we try to provide some thought on this front that might be useful for future developments.
Concerns of DNAzyme in vivo activity. First, concerns on the activity of DNAzymes have been raised by the community. Most of the current DNAzymes achieve high activity with a metal concentration much higher than that available in cells. For example, most biochemical assays on the 10-23 DNAzyme involved 50 mM Mg2+, while the intracellular Mg2+ concentration is below 2 mM. Under this condition, its activity is quite low (essentially cannot be measured in some reports). One study even concluded that the effect of 10-23 DNAzyme on gene silence is simply due to antisense contribution instead of cleavage [67]. We measured both the anti-sense and cleavage effect of the 8-17 DNAzyme for an extracted mRNA, and found that the role of DNAzyme depends on its relative activity [25]. In the presence of Mg2+, only the antisense effect was observed, while upon addition of Zn2+, the cleavage activity of the 8-17 DNAzyme dominated, which is attributable to its preferential activation with Zn2+. This also indicates that one needs to be extremely careful when DNAzymes are used for gene silencing, and control experiments using inactive DNAzyme mutants are needed before drawing a conclusion. In addition, due to the complex intracellular conditions, such as molecular crowding and proteins that might bind DNAzymes [68-70], DNAzymes that work in buffer may turn out to be inactive in vivo, which may also contribute to inconsistent results.
Several methods can be used to achieve more active DNAzymes. The most standard method is to tune the selection pressure. As was mentioned above, in vitro selection condition can be tailored to push DNAzymes with desired properties. By decreasing metal concentration during selection step, novel DNAzymes that function optimally under physiologically relevant conditions might be obtained [71]. This idea however is complicated by the tyranny of the 8-17 motif, masking other DNAzymes [72]. We recently isolated the Ca2+-dependent EtNa DNAzyme accidentally in isopropanol [49]. Under this condition, the 8-17 DNAzyme is inactive. Therefore, if the 8-17 can be suppressed, it is still possible to obtain new and faster DNAzymes. Since the 8-17 DNAzyme is only active with divalent metal ions, chelators such as EDTA would be a simpler solution to mask this tyranny motif if the selection is performed for monovalent metal ions. Indeed, EDTA has been employed during the selection of the Na+-specific NaA43 DNAzyme [44].
Compared to the twenty amino acids, the four nucleotides have quite limited chemical diversity. To improve the functionality of DNAzymes, modified nucleotides have also been incorporated into the selection library. Perrin and coworkers initially modified the DNA library with imidazoles and cationic amines, and isolated an RNase A mimicking DNAzyme with a kcat of ~0.045 min-1 in its cis-form (in presence of 200 mM Na+) [73, 74]. Later, they introduced an additional cationic guanidine group and identified an even faster DNAzyme with a rate of 0.134 min-1 with 200 mM Na+ [75]. This DNAzyme is faster than the above NaA43, indicating an improvement over the unmodified catalysts. This method can also be used for selecting DNAzymes for cleaving all-RNA substrate. For example, Williams and coworkers reported a DNAzyme incorporating imidazolyl and amino groups that can recognize and cleave a 12-nt RNA substrate with rate of 0.07 min-1 in 200 mM Na+ [76]. More recently, Perrin group collectively used imidazole, ammonium, and guanidinium groups and isolated a DNAzyme with a comparable rate [77]. A few challenges have limited the use of such modified DNAzymes. First, most of the modified nucleotides are not commercially available right now, making it inaccessible to many other laboratories. In addition, these modified nucleotides cannot be incorporated into DNA through the standard PCR process, thus complicating the selection process. An alternative method for extending the chemical functionality of DNA is to use the artificially expanded genetic information system (AEGIS) developed by Benner and co-workers [78]. In the AEGIS system, other pairs of nucleotides have been added to the four standard nucleotides to form orthogonal base pairs, and they can all copied by certain polymerases using PCR. This idea has been successfully applied for aptamer selections, which demonstrates its power in the evolution of functional nucleic acid with better functionalities [79]. Since DNA might be degraded by various enzymes in biological fluids, increasing the stability of DNAzyme is also helpful for sustaining its efficacy.
In addition to searching for new DNAzymes with higher activity, it is also possible to co-deliver DNAzymes and metals to cell to increase the local concentration of certain metal ions and thus enhance DNAzyme activity. For example, Tan and coworkers reported a DNAzyme-MnO2 nanosystem for gene-silencing therapy [80]. MnO2 nanoparticle can adsorb the DNAzyme and the complex was efficiently internalized by the cells. After entering into cells, MnO2 was reduced to Mn2+ ions by intracellular reducing agents such as GSH. The resulting Mn2+ then served as a highly effective metal cofactor for DNAzyme activation.
Target RNA accessibility. Another issue that needs to be taken into consideration is the accessibility of the target site in mRNA. Sometimes an intended recognition region in mRNA may be unavailable for DNAzyme due to folding of the mRNA. Indeed, it is reported that only ~10% of possible sites in mRNA is readily cleaved by DNAzyme [81, 82]. In a typical assay in buffer, annealing is the first step for DNAzyme activity test to ensure the appropriate binding of the substrate, but this is unlikely to be possible in vivo. To address this problem, one feasible way is to screen the entire length of the target RNA to find the most efficient site [81]. However, this trial-and-error process is quite cumbersome, and each DNAzyme or mRNA has to be tested individually. For a given mRNA and DNAzyme, it is also possible to perform a combinatory selection to search for optimally accessible cleavage sites by fixing the catalytic core and randomizing the target binding regions [83]. Alternatively, a simpler solution is to modify the substrate binding regions of DNAzymes with 2′-O-methyl or locked nucleic acid (LNA) to enhance the base pair binding affinity, which can compete the internal structures for mRNA binding [84, 85].
Quantification of gene suppression. Finally, to quantify the therapeutic efficacy, it is often required to measure the fraction of mRNA cleavage. The most common way is based on real-time PCR, which is performed by reverse transcribing the mRNA into cDNA for PCR amplification. In most cases, the reverse transcription step is performed using a poly-T primer that can copy all mRNA with a poly-A tail. Thus, the target mRNA is mixed with thousands of other RNA. The specificity relies on the PCR primer. Therefore, one way to improve is to also design specific primers for reverse transcription. Even for the PCR quantification itself, quite a few potential pitfalls need to be noted. To account for the difference in cell density, a robust internal standard (usually a house keeping gene) needs to be measured at the same time. In addition, a suppression of 50% gene expression is reflected only in one cycle of difference in the PCR, which has really limited the sensitivity of this method. Therefore, complementary methods such as Western blotting should also be carried out. The situation becomes even more complicated in animal models, requiring carefully designed controls.
Involving nanomaterials. Despite these challenges, an important advantage of DNA is that it can be readily conjugated to various nanomaterials, much easier than RNA or proteins. Nanomaterials have been the main players in the theranostics field. DNA can be attached either via physisorption or by covalent conjugation. It has been demonstrated that a high density of DNA on a nanoparticle can form an interesting conjugate that can be internalized by many cells and even penetrate the skin barrier [86-88]. Nanomaterials can also retard DNA degradation by nucleases [89]. Using nanomaterials to deliver DNAzymes has been demonstrated in a few cases using gold nanoparticles (AuNPs) [90-92], graphene oxide (GO) [93], and MnO2 nanosheets [80].
DNAzymes for diagnosis
While the field of DNAzyme started with the goal of RNA cleavage, its analytical applications have been studied much more. Using RNA-cleaving DNAzymes alone has enabled the detection of many different analytes, especially metal ions. Such DNAzymes are mainly from direct combinatorial selection. In addition, by incorporating aptamers to make aptazymes, many more analytes can be detected. We summarize their biomedical relevance analytical applications below based on the type of analyte.
In vitro metal sensing
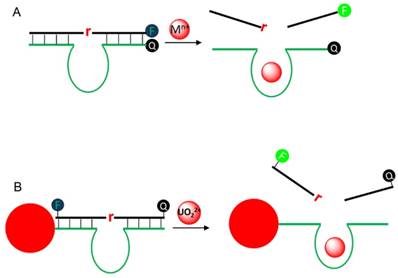
Metal ions are essential elements for human life, and their concentrations represent an important index for medical diagnostics. For example, a high blood Na+ concentration is associated with high blood pressure and water retention, while Ca2+ is highly important for biological signaling. Therefore, metal sensors have been long sought. While small molecule chelator and protein based sensors have been developed [94], RNA-cleaving DNAzymes have emerged as a new platform for this purpose. The initial idea was reported by Li and Lu using the 8-17 DNAzyme for Pb2+ detection [95]. Since then, we have witnessed a large growth of publication on metal sensing using DNAzymes [6, 96, 97]. The most common design principle is to label a fluorophore and a quencher at substrate and enzyme, respectively (Figure 5A). After hybridization, the fluorescence is quenched due to the close proximity to the quencher. In the presence of the target metal, the substrate is cleaved and the fluorophore bearing fragment releases from its quencher, resulting in fluorescence recovery. Typically, the fluorescence is monitored as a function of time, and the rate of fluorescence increase is proportional to metal concentration, allowing for quantification. Over the years, many variations of the design have been reported by changing the position and number of fluorophore/quencher [97].
For intracellular metal detection, attaching DNAzymes on the surface of AuNPs is quite attractive, due to its high fluorescence quenching efficiency and ability to enter cell. As a proof-of-concept, Lu and coworkers adsorbed a uranyl-specific DNAzyme on AuNPs for uranyl ion imaging in living cells [90]. The substrate was labelled with an additional quencher to ensure a very low background (Figure 5B). The sensor can detect low micro molar UO22+ in buffer and image 100 μM UO22+ in living cell, while the inactive mutant DNAzyme showed much less intracellular fluorescence. Tang and coworkers similarly adsorbed two DNAzymes on the same nanoparticle for simultaneous imaging Zn2+ and Cu2+ in living cells, which can detect metal concentration lower than 2 μM [98].
To avoid non-specific cleavage of the substrate strand for intracellular imaging, a breakthrough was made recently using the caged DNAzyme. A caged DNAzyme refers to capping and inactivating the DNAzyme with a photolabile molecule. Upon a brief irradiation at a certain wavelength, the caging group peels off, accompanied by recovery of the DNAzyme activity. With this strategy, the dynamic function of DNAzyme can be controlled in a temporal and spatial fashion through convenient light exposure. The method was developed by Deiters and coworkers to precisely control gene expression [67]. Later, it is found that this technique is useful for intracellular sensing. For example, the Lu group first reported a general and effective method to cage DNAzyme by modification of scissile 2′-OH group, which was applied for intracellular imaging of Zn2+ and Pb2+ [99]. More recently, Xiang and coworkers reported another facile strategy to synthesize caged DNAzymes, and this method was implemented to the 8-17 DNAzyme for intracellular Zn2+ sensing [100].
While the concept has been demonstrated, the physiological relevance of these studies is modest due to the low abundance of these transition metals in biology. Recently, the Lu group made an exciting advancement with the discovery of a highly sensitive Na+-dependent NaA43 DNAzyme for intracellular Na+ sensing by using the caged DNAzyme strategy [44]. The sensor showed minimal fluorescence after entering cell, indicating DNAzyme remained intact during delivery process. Upon irradiation to uncage the DNAzyme and the influx Na+ into cell, the fluorescence gradually increased over 30 min, while with the same treatment, an inactive DNAzyme showed little fluorescence change. The results demonstrated that the turn-on fluorescence is attributed to the decaging of DNAzyme and subsequent Na+-induced cleavage. Caging is critical in this work to avoid non-specific cleavage before the sensor enters the cell.
In addition to intracellular sensing, blood and serum present another important biological matrix for diagnostic application. Serum mainly contains four types of metal, Ca2+, Mg2+, Na+ and K+. The concentrations of these metals are strictly controlled, and any deviation is a sign of physiological disorder. Therefore, their concentration monitoring is important in medicine. We recently studied the stability of a DNAzyme in undiluted serum and also performed an in vitro selection in serum [17, 101]. We found that DNAzymes can work stably in serum with minimal degradation and a few mutants of the 8-17 DNAzyme showed substantial activity due to the presence of Ca2+ and Mg2+. One major issue for fluorescent sensing in serum is the light adsorption and scattering by serum matrixes. In this regard, the lateral flow technology is a powerful tool, which usually uses gold nanoparticles for color production and separates the molecular recognition from detection spatially. The concept of developing DNAzyme-based lateral flow sensors has been demonstrated [102, 103].
(A) A typical sensor design for DNAzyme signaling. F and Q denote for fluorophore and quencher, respectively. (B) A UO22+ sensor design by adsorption the DNAzyme on AuNPs. AuNPs are potent fluorescence quenchers and also help cell uptake.
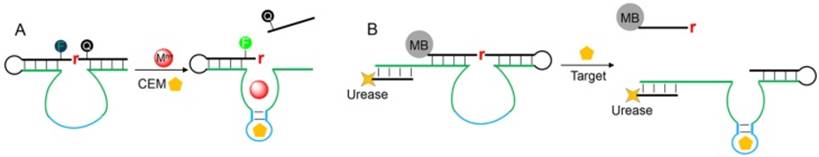
(A) A schematic illustration of the RNA-cleaving fluorescent DNAzyme for CEM detection. (B) Converting DNAzyme activity into pH change for colorimetric sensing by immobilizing DNAzyme on a magnetic bead (MB) and extending the enzyme strand to hybridize with a urease bearing DNA.
Bacterial and cancer cell sensing
Bacterial detection is an important diagnostic task, since bacterial infection is responsible for many infectious diseases and posts serious threats to public health. To use DNAzymes for bacterial detection, the Li group have performed in vitro selection to isolate RNA-cleaving fluorescent DNAzymes (RFDs) that respond to the crude extracellular mixture (CEM) produced by certain bacterial pathogens [104]. In their selection library, fluorophore and quencher are labeled just adjacent to the single RNA cleavage site, so that the obtained DNAzymes are coded with signaling capabilities without the need of further sensor design (Figure 6A). The resulting DNAzymes should contain two domains in their catalytic core: the CEM binding domain for allosteric control of the DNAzyme activity, and a catalytic domain for hydrolytic reaction. The RFD was then optimized to detect bacterial down to 1 colony-forming unit (CFU) after 4 h of culturing [105], and was also used for other detection platform [106]. To visualize the bacterial concentration, Li and coworkers extended the enzyme strand to hybridize with a urease tagged DNA and immobilized the DNAzyme on a magnetic bead (Figure 6B) [107]. The cleavage reaction released urease into solution, which subsequently catalyzed the hydrolysis of urea into carbon dioxide and ammonia to raise solution pH value. The pH change can be visualized by litmus dye to produce colorimetric signal. Since each urease can turnover ~1014 times, another layer of signal amplification was achieved, allowing a detection limit of ~103 E.coli cells (without cell culturing) with high selectivity. More recently, the same group further moved forward by reporting a novel DNAzyme that can recognize specific strain of bacterial with exquisite selectivity [108]. They also implemented this selection strategy to isolate DNAzyme for cancer cell detection [109]. To do so, the MDA-MB-231 cell lysates were used as the allosteric element during the selection process, and the resulting DNAzyme can detect cell lysate proteins down to 0.5 μg/mL (equivalent to ∼5000 cell/mL), with high selectivity against other subtypes of breast cancer cells.
Nucleic acids sensing
Another important class of analyte is nucleic acids, for which DNAzymes have also been used. Due to their catalytic nature, DNAzyme-based fluorescent biosensors for nucleic acids are also called catalytic molecular beacons. Compared to traditional molecular beacons, catalytic beacons are advantageous in several aspects. First, catalytic beacons have more choices available for positioning the fluorophore and quencher labels to suppress background signal. Second, after cleavage, the fluorophore can fully release from its quencher to produce a high signal. In addition, since a catalytic reaction is involved, signal amplification can be readily achieved. In an early study, Todd and coworkers developed a real-time DNA detection method by using a procedure called DzyNA-PCR, which allowed the detection of 10 copies of the target [110]. They designed two PCR primers for a target DNA. One primer was encoded with the complement of 10-23 DNAzyme sequence, so that the sense-strand DNAzyme can be generated by PCR amplification. Then, the substrate with fluorophore/quencher flanking the cleavage site was introduced as a reporter for signaling. The amount of fluorescence increase reflected PCR efficiency, which was in turn proportion to the target DNA concentration. To bypass the time-consuming PCR process, Suenaga and coworkers reported a gel electrophoresis method for a specific rRNA in a microbial community [111]. Since the DNAzyme cleaved the substrate in a sequence specific fashion, the relative abundance of a particular rRNA was quantified in a mixed sequence population.
Some elegant methods were also developed through the design of allosteric DNAzymes. It is reported that the DNAzyme activity can be regulated by a DNA effector through modulating the association between enzyme and substrate strand [112, 113]. To do so, one substrate binding arm was designed to have insufficient base pairs to inhibit DNAzyme activity. At the same time, both the substrate and enzyme strands were extended to be partially complementary to the DNA effector (Figure 7A). In the presence of the DNA effector, a three-way junction was formed to activate the enzyme. Inspired by this structure, Yasuhiro and coworkers constructed a target-assisted self-cleavage (TASC) probe for DNA detection [114]. Note that this probe used a cis-form DNAzyme to promote target binding, and the signal was generated by labeling fluorophore and quencher nearby the cleavage site. Another way of modulating DNAzyme activity by target DNA is the MNAzyme strategy reported by Todd group (Figure 7B) [115]. The MNAzyme was produced by splitting a DNAzyme at the catalytic core into two inactive halves. Each half contained a sensor arm that can hybridize with part of the target DNA to assemble an active DNAzyme. An obvious advantage of this design is to have catalytic signal amplification. Each target nucleic acid can activate a DNAzyme, which can cleave many substrates, achieving a detection limit down to 10 copies of the target gene. The design can be coupled with AuNPs to achieve point-of-care detection [116], and conjugated with cascade enzyme for signal amplification [117].
(A) A design of allosteric DNAzymes with activity modulated by the DNA effector. (B) A scheme showing the design of the MNAzyme to form an active catalytic core. (C) Combining the MNAzyme design with a strand displacement reaction to design enzyme-free signal amplification sensor for DNA detection. Reprinted with the permission from ref. [118]. Copyright 2011 American Chemical Society.
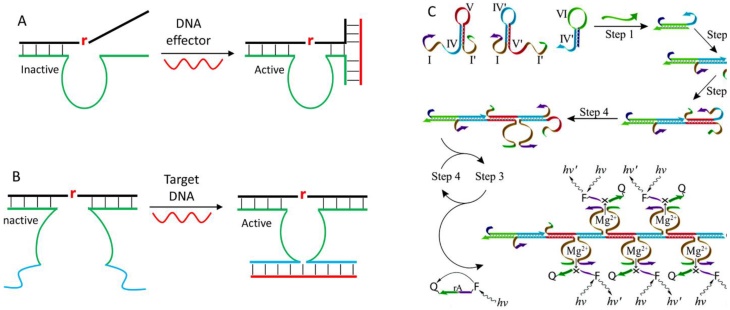
Another way of signal amplification for highly sensitive DNA detection is to exploit a strand displacement reaction combined with the MNAzyme concept. Willner and coworkers designed two split DNAzyme halves with two hairpin structures, and introduced a probe hairpin for target binding (Figure 7C) [118]. The presence of target DNA opened the probe hairpin, which subsequently triggered the strand displacement reaction to form tandem DNAzyme for multiple substrate cleavage. Such system can detect DNA down to 10-14 M, which rivals those of protein enzyme-based assays.
Sensing other analytes
Small molecules such as metabolites are an important class of analytes. While antibodies can be raised to bind small molecules, the affinity is usually low and competitive assays are often needed for their detection. In this regard, aptamers provide a useful alternative. Aptamers are single-stranded nucleic acids that can selectively bind target molecules. Compared with antibodies, aptamers have comparable or even better target binding affinity for small molecules. As such, aptamers have attracted significant attention for diagnostic applications [119].
Coupling DNAzyme with aptamers can produce sensors for a broader range of analytes while achieving highly specific signals. For example, aptazyme, which is a combination of aptamer and DNAzyme, has enabled the regulation of DNAzyme activity via aptamer binding. Many rational design methods have been developed to couple aptamers to DNAzymes. In one study, Li and coworkers designed an aptazyme sensor for ATP (Figure 8A) [120]. They extended one end of the 8-17 DNAzyme with the ATP aptamer, and a competitor DNA was used to block substrate binding. Addition of ATP removed the blocking DNA and facilitated substrate binding. Therefore, the ligand binding was translated to enzyme activation. Because of predictable and tailorable secondary structure of both DNAzyme and aptamer, aptazyme can be constructed with excellent versatility. Many aptamer sequences were integrated into DNAzymes for various target detection [121-123]. Interestingly, aptazymes for small molecule metabolites have also been found in Nature, and an excellent example is the glmS riboswitch ribozyme [124]. Typically, the riboswitch regulates gene expression by switching its conformation upon target binding, just like an aptamer. The glmS is catalytic riboswitch, in which the target binding activates the mRNA cleavage reaction to down regulate gene expression.
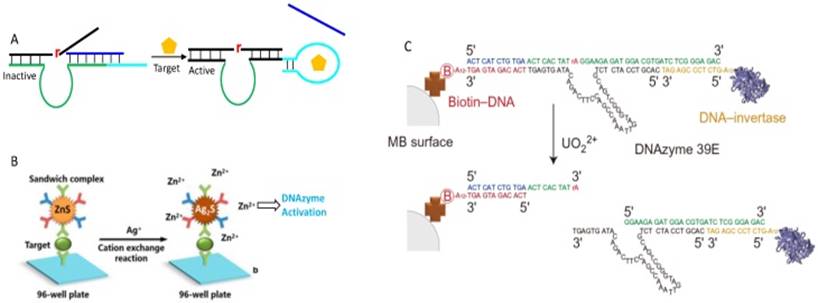
In addition to directly using DNAzymes for target recognition, DNAzymes can also be used only for signal generation. For example, the Tan group developed a concept of DLISA, which refers to the DNAzyme-based ELISA immunoassay system (Figure 8B) [125]. In their system, the first antibody was immobilized on a 96-well plate, and the secondary antibody was attached to the surface of zinc-sulfide nanocrystals (ZnS NCs). In the presence of the target, the ZnS NCs was captured on the 96-well plate through the formation of sandwich immunocomplex. The addition of Ag+ triggered the cation exchange reaction in nanocrystal to form Ag2S and release Zn2+ into solution, which subsequently activated the DNAzyme for substrate cleavage. Such a design was applied for the human lgG detection, with a detection limit of 2 fg/mL (3×10-17 M). In another study, DNAzyme was coupled with glucose oxidase (GOx) for sub-mM glucose detection in biological samples [126]. This sensor was designed based on the fact that the DNAzyme required both Cu2+ and H2O2 for activity [127, 128]. The GOx can quantitatively catalyze glucose forming gluconic acid and H2O2, and the product was exploited by DNAzyme for substrate cleavage in presence of sufficient Cu2+.
Beyond fluorescence signaling
Most of the above examples used fluorescence for signaling. While fluorescence is sensitive, a fluorometer is needed for quantitative detection. Therefore, developments have also been made to use other signaling mechanisms. The most successful commercial biosensors are for glucose detection using electrochemistry. The glucose meters have already been engineered to a highly accurate and user-friendly stage at a low cost. Bearing this in mind, the Lu group reported a general design of using personal glucose meter (PGM) to quantify aptamer binding information and DNAzyme activity [129]. To do so, the DNAzyme substrate was elongated to hybridize with an invertase bearing DNA, and the DNAzyme was immobilized onto magnetic bead for separation (Figure 8C). The cleavage reaction released the invertase-labeled DNA from bead into solution to catalyze the hydrolysis of sucrose into glucose, which is then quantified by a glucose meter. This method was then adapted to other DNAzymes for various metal detection [130].
Finally, magnetic signal is quite desirable for theranostics signaling since it can achieve deep tissue penetration for non-invasive three-dimensional in vivo imaging. The Lu group has attached DNA to both Gd3+-based contrast agents and superparamagnetic iron oxide nanoparticles to produce smart MRI contrast agents responsive to various analytes [131-133]. In one example, they conjugated a Gd-DOTA contrast agent on the substrate of the UO22+-dependent DNAzyme, and streptavidin was labeled at the corresponding enzyme strand. Upon cleavage reaction, the Gd-DOTA label moved away from streptavidin, resulting in a change of its relaxivity for UO22+ quantification, with a detection range from 0.2 to 2 μM [134].
(A) A design of aptazyme by incorporating an aptamer sequence in DNAzyme substrate binding arm region. The target binding releases the competitor DNA to facilitate substrate hybridization. (B) Schematic illustration of the DLISA strategy. Reprinted with permission from ref. [125]. Copyright 2015 American Chemical Society. (C) Schematic of method using PGM to quantify DNAzyme activity. The DNAzyme was attached to a streptavidin-modified magnetic bead, and the substrate was extended to hybridize with DNA-invertase conjugates. Reproduced with permission from ref. [129]. Copyright 2011, Nature Publishing Group.
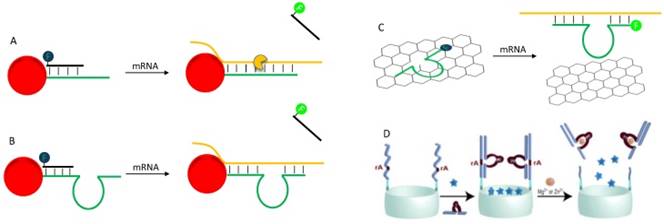
(A) A schematic illustration of AuNP-based nano-flares for mRNA depletion and detection. (B) A direct adaption of nano-flare concept for fabrication of theranostic DNAzyme. (C) A strategy of detection and knockdown mRNA by adsorption fluorophore-labeled DNAzyme on GO. (D) Schematic illustration of SiO2 mesoporous encapsulated with fluorophore and caped with DNAzyme for stimuli-responsive content release. Reprinted with the permission from ref. [137]. Copyright 2013 American Chemical Society.
Theranostic DNAzymes
So far, we have reviewed some representative applications of DNAzyme in therapeutics and diagnostics in separate windows. Examples of combining these two aspects are emerging. An excellent example of using oligonucleotides to develop theranostics is reported by Mirkin and co-workers, who constructed a nanoflare for simultaneously mRNA silencing and detection (Figure 9A) [135]. An anti-sense DNA was densely functionalized on AuNPs surface, and a fluorophore-labeled “flare” oligonucleotide was hybridized with part of the antisense DNA with fluorescence quenching. After entering cell, the antisense DNA hybrids with the mRNA to block its activity. At the same time, the flare oligonucleotide is released, resulting in a dose-dependent fluorescence increase. While the use of DNAzyme for this purpose is still to be demonstrated, in principle it should be feasible. For example, although not yet demonstrated, the above nanoflare concept can be readily adapted to DNAzyme by replacing the antisense DNA with DNAzyme (Figure 9B). Such a construct has already been used for delivery of DNAzymes [92], and for intracellular sensing of metal ions [90]. It is quite straightforward to achieve theranostic goal with this design.
In addition to covalent linking, physisorption of DNA on nanomaterials can also achieve theranostic applications. For example, Min and co-workers adsorbed a fluorophore labeled DNAzyme on graphene oxide (GO) to construct a theranostic DNAzyme (Figure 9C) [93]. In the presence of the target mRNA, the DNAzyme hybridizes with the mRNA and is released from GO, with fluorescence increase for mRNA detection. Then the formation of DNAzyme complex cleaves the mRNA. Other nanomaterials can allow more sophisticated designs. For example, Willner and coworkers encapsulated a series of fluorophores into mesoporous SiO2 nanoparticle, and then caped the pore by DNAzymes (Figure 9D) [136, 137]. In the presence of the target metal, the DNAzymes were activated to cleave the substrate, which broke the cap to allow contents release. Such system holds the promise for the design of theranostic platforms, in which the analyte activates DNAzyme to produce detectable signal, and at the same time drugs are released for therapeutic functions.
Delivery of DNAzymes
Just like other nucleic acid based therapeutics and diagnostics, DNAzymes also face considerable challenges related to its in vivo delivery. Two main issues need to be addressed for their successful in vivo applications: stability and intracellular penetration. DNAzymes are nucleic acids, which are liable to digestion by endo- and exo-nucleases. In addition, the negative charged phosphate backbone presents a primary barrier for cellular uptake due to electrostatic repulsion from cell surface [138]. Numerous advances have been made in developing strategies for DNAzyme in vivo delivery, which can either protect it from degradation or facilitate it cellular uptake.
While DNAzyme is preferred over ribozyme for in vivo applications, the lifetime of unmodified DNAzyme is still relatively short [139], rendering its function only within the relevant time period. To increase DNAzyme stability, a potent solution is chemical modification [140]. The phosphorothiolate DNA, 2′-O-Me-RNA modification and 3′-3′ inverted nucleotides have been demonstrated to remarkably enhance DNAzyme resistance to exonucleolytic degradation without compromising its catalytic activity [141-143]. Conjugate DNAzymes with polyamines and peptides, terminal modification of N3′-P5′ phosphoramidate bonds were also reported to enhance stability [144, 145]. It is also known that the L-formed DNA is more stable to resist nuclease digestion than naturally occurring D-form DNA [146]. Based on this fact, Tan and coworkers constructed L-DNAzyme using L-DNA [147]. This DNAzyme maintained its activity with L-DNA substrate, while the biostability was significantly improved. Therefore, its application is less affected by biological matrixes from cell or serum [148].
To deliver DNAzymes into cell, several methods have been developed. The standard method relies on cationic polymers for transfection [149]. In this case DNAzymes are merely regarded as a general nucleic acid without considering its characteristics. While this method can achieve high delivery efficiency, a subsequent release step is required before DNAzyme can perform its functions. In addition, cationic polymers or liposomes are quite toxic to cells.
By attaching a high density of DNA to AuNPs, the Mirkin group discovered three important features: 1) such AuNPs can enter cells without the need of cationic transfection agents [87]; 2) AuNP can enhance DNA stability against nuclease cleavage [89]; and 3) the DNA binding affinity to its target DNA is also improved [150]. Motivated by these observations, several ribozymes and DNAzymes have been delivered into cell using AuNPs [90-92]. Additional advantages of AuNPs are large DNA loading capacity and strong fluorescence quenching efficiency, which make them attractive for DNAzyme theranostic applications [135]. Non-thiolated DNA with a poly-A block can also be attached to AuNPs [151-153]. This method was used for immobilizing DNAzymes, and the spacing between each DNAzyme can be precisely regulated by controlling the length of the poly-A sequence, which allows for tunable and enhanced DNAzyme activity [154]. As other delivery vehicles, the internalized AuNPs have to escape from endosomes/lysosomes to exert their function. Other materials, such as cationic polypeptide [44], iron oxide nanoparticles (IONPs) [155], and GO [93], have also been used for intracellular delivery. With the development of DNA nanotechnology, various DNA nanostructures with well-defined dimensions and shapes have been reported as potential carriers for DNAzyme in vivo delivery. For example, DNA tetrahedron nanostructures were found to readily enter mammalian cells without the use of transfection agents [156]. Based on this, a DNAzyme was grafted to a DNA tetrahedron for intracellular imaging of metal ions [157]. Tan and coworkers reported a DNA dendrimer scaffold that can efficiently deliver DNAzymes into cells with sufficient stability, excellent biocompatibility and retained DNAzyme catalytic activity [158].
Conclusions and future perspectives
Theranostics is a relatively young field. The concept of theranostics was coined in 2002 [159]. Most work on this topic has been focused on designing sophisticated inorganic nanomaterials for imaging and therapy at the same time. In this article, we reviewed the progress based on DNAzymes. Although nanomaterials are also involved, they do not play a major role. The field of DNAzyme is 22 years old. Since its inception, DNAzymes were proposed for viral RNA cleavage. Using DNAzymes as biosensors has been practiced for at least 16 years. Although developing DNAzyme-centered theranostic modalities has not received much attention, as outlined here, DNAzymes have a huge potential for such applications. We have reviewed biomedical related work of using DNAzymes.
Based on the current status of the field, we also postulate some future directions that can facilitate the development of DNAzyme-based theranostics. 1) In vitro selection. To make a real biomedical impact, highly active DNAzymes working in biological fluids are needed. For DNAzyme activity, the physiological metal ions availability is a limitation currently. The two main sample matrixes are blood and cytoplasma. Blood has a high Ca2+ and Na+ concentration, while cells have more Mg2+ and K+. Currently, the best DNAzymes are only ~0.1 min-1 in such conditions even with extensively modified nucleotides. This is however not the chemical limit of in vivo RNA cleavage. Protein nucleases and even ribozymes can be much faster. Therefore, we believe there is still room for selecting new DNAzymes. Exciting developments have already been made by the recently reported Na+ and Ca2+ dependent DNAzymes. A key is to perform selections under conditions to avoid the 8-17 motif. So far, most work used RNA/DNA chimeric substrate containing only a single RNA linkage for analytical applications. The isolated DNAzymes are typically inactive if the substrate is replaced by the full-RNA analog. For practical RNA cleavage, it is likely that new selections using full-RNA substrates are required. 2) Theranostic systems and delivery. Developing theranostic systems based on DNAzyme is a relatively new concept, and only a few examples can be found in the literature at this moment. Most of the studies are still at the cell culture stage. To move to real animal studies, other design considerations need to be made. For example, the fluorescence-based assays will have to use near IR dyes, and magnetic materials might be introduced for MRI based imaging. All these are exciting new opportunities. We already outlined the challenges of intracellular delivery, and the challenges are even larger for delivery in animal models. Developing materials that can reach target organs remains a challenge for the whole nanomedicine field [160]. 3) Involving DNA nanotechnology. A beauty of DNA is its highly predictable structure. By designing DNA sequences, it is possible to form nanoparticles made of purely DNA. This may allow delivery, imaging, and therapy to be achieved in a single material, while avoiding inorganic nanomaterials that may have difficulty to obtain FDA approval. 4) Beyond RNA cleavage. The current discussion has been focused on RNA-cleaving DNAzymes, but other systems can be easily envisioned. For example, DNA-cleaving DNAzymes were also implemented for metal sensing [161-163], and their therapeutic potential was demonstrated for DNA excision repair [164]. In addition, aptazymes might be developed to respond to glucose. Upon cleavage, its associated nanocarriers can release insulin. Such a concept has already been demonstrated for metal ions [136, 137]. But to reach a clinical impact, much more work has to be done both fundamentally and to engineer such molecular devices.
Acknowledgements
We thank financial support from the Natural Sciences and Engineering Research Council of Canada (NSERC), Foundation for Shenghua Scholar of Central South University and the National Natural Science Foundation of China (Grant No. 21301195). Wenhu Zhou is supported by the Fellowship from the China Scholarship Council (CSC, Grant No. 201406370116).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kelkar SS, Reineke TM. Theranostics: combining imaging and therapy. Bioconj Chem. 2011;22:1879-903
2. Lammers T, Aime S, Hennink WE, Storm G, Kiessling F. Theranostic nanomedicine. Acc Chem Res. 2011;44:1029-38
3. Wang X, Hu Y, Wei H. Nanozymes in bionanotechnology: from sensing to therapeutics and beyond. Inorg Chem Front. 2016;3:41-60
4. Wei H, Wang E. Nanomaterials with enzyme-like characteristics (nanozymes): next-generation artificial enzymes. Chem Soc Rev. 2013;42:6060-93
5. Lin Y, Ren J, Qu X. Catalytically active nanomaterials: a promising candidate for artificial enzymes. Acc Chem Res. 2014;47:1097-105
6. Liu J, Cao Z, Lu Y. Functional Nucleic Acid Sensors. Chem Rev. 2009;109:1948-98
7. Xiang Y, Lu Y. DNA as sensors and imaging agents for metal ions. Inorg Chem. 2014;53:1925-42
8. Schlosser K, Li Y. Biologically inspired synthetic enzymes made from DNA. Chem Biol. 2009;16:311-22
9. Silverman SK. Catalytic DNA: scope, applications, and biochemistry of deoxyribozymes. Trends Biochem Sci. 2016;41:595-609
10. Breaker RR, Joyce GF. A DNA enzyme that cleaves RNA. Chem Biol. 1994;1:223-9
11. Hwang K, Hosseinzadeh P, Lu Y. Biochemical and biophysical understanding of metal ion selectivity of DNAzymes. Inorganica Chim Acta. 2016;452:12-24
12. Lu Y, Liu J. Functional DNA nanotechnology: emerging applications of DNAzymes and aptamers. Curr Opin Biotechnol. 2006;17:580-8
13. Li Y, Breaker RR. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2'-hydroxyl group. J Am Chem Soc. 1999;121:5364-72
14. Liu J, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB. et al. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc Natl Acad Sci U S A. 2007;104:2056-61
15. Huang P-JJ, Lin J, Cao J, Vazin M, Liu J. Ultrasensitive DNAzyme beacon for lanthanides and metal speciation. Anal Chem. 2014;86:1816-21
16. Mei SH, Liu Z, Brennan JD, Li Y. An efficient RNA-cleaving DNA enzyme that synchronizes catalysis with fluorescence signaling. J Am Chem Soc. 2003;125:412-20
17. Zhou W, Zhang Y, Ding J, Liu J. In Vitro Selection in Serum: RNA-Cleaving DNAzymes for Measuring Ca2+ and Mg2+. ACS Sensors. 2016;1:600-6
18. Liu Z, Mei SH, Brennan JD, Li Y. Assemblage of signaling DNA enzymes with intriguing metal-ion specificities and pH dependences. J Am Chem Soc. 2003;125:7539-45
19. Nelson KE, Bruesehoff PJ, Lu Y. In vitro selection of high temperature Zn2+-dependent DNAzymes. J Mol Evol. 2005;61:216-25
20. Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci U S A. 1997;94:4262-6
21. Lan T, Furuya K, Lu Y. A highly selective lead sensor based on a classic lead DNAzyme. Chem Commun. 2010;46:3896-8
22. Saran R, Liu J. A comparison of two classic Pb2+-dependent RNA-cleaving DNAzymes. Inorg Chem Front. 2016;3:494-501
23. Schlosser K, Gu J, Sule L, Li Y. Sequence-function relationships provide new insight into the cleavage site selectivity of the 8-17 RNA-cleaving deoxyribozyme. Nucleic Acids Res. 2008;36:1472-81
24. Wang B, Cao L, Chiuman W, Li Y, Xi Z. Probing the function of nucleotides in the catalytic cores of the 8-17 and 10-23 DNAzymes by abasic nucleotide and C3 spacer substitutions. Biochemistry. 2010;49:7553-62
25. Wang F, Saran R, Liu J. Tandem DNAzymes for mRNA cleavage: choice of enzyme, metal ions and the antisense effect. Biorg Med Chem Lett. 2015;25:1460-3
26. Li J, Zheng W, Kwon AH, Lu Y. In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme. Nucleic Acids Res. 2000;28:481-8
27. Cruz RP, Withers JB, Li Y. Dinucleotide junction cleavage versatility of 8-17 deoxyribozyme. Chem Biol. 2004;11:57-67
28. Faulhammer D, Famulok M. The Ca2+ Ion as a Cofactor for a Novel RNA-Cleaving Deoxyribozyme. Angew Chem Int Ed. 1996;35:2837-41
29. Kasprowicz A, Stokowa-Sołtys K, Wrzesiński J, Jeżowska-Bojczuk M, Ciesiołka J. In vitro selection of deoxyribozymes active with Cd2+ ions resulting in variants of DNAzyme 8-17. Dalton Trans. 2015;44:8138-49
30. Brown AK, Li J, Pavot CM, Lu Y. A lead-dependent DNAzyme with a two-step mechanism. Biochemistry. 2003;42:7152-61
31. Rong W, Xu L, Liu Y, Yu J, Zhou Y, Liu K. et al. 8-17 DNAzyme modified with purine analogs in its catalytic core: The conservation of the five-membered moieties of purine residues. Biorg Med Chem Lett. 2012;22:4238-41
32. Zhu J, Li Z, Wang Q, Liu Y, He J. The contribution of adenines in the catalytic core of 10-23 DNAzyme improved by the 6-amino group modifications. Biorg Med Chem Lett. 2016;26:4462-5
33. Feldman AR, Sen D. A new and efficient DNA enzyme for the sequence-specific cleavage of RNA. J Mol Biol. 2001;313:283-94
34. Breaker RR, Joyce GF. A DNA enzyme with Mg2+-dependent RNA phosphoesterase activity. Chem Biol. 1995;2:655-60
35. Lu Y. New Transition-metal-dependent DNAzymes as efficient endonucleases and as selective metal biosensors. Chem Eur J. 2002;8:4588-96
36. Santoro SW, Joyce GF, Sakthivel K, Gramatikova S, Barbas CF 3rd. RNA cleavage by a DNA enzyme with extended chemical functionality. J Am Chem Soc. 2000;122:2433-9
37. Huang P-JJ, Liu J. An ultrasensitive light-up Cu2+ biosensor using a new DNAzyme cleaving a phosphorothioate-modified substrate. Anal Chem. 2016;88:3341-7
38. Huang P-JJ, Liu J. Rational evolution of Cd2+-specific DNAzymes with phosphorothioate modified cleavage junction and Cd2+ sensing. Nucleic Acids Res. 2015;43:6125-33
39. Hollenstein M, Hipolito C, Lam C, Dietrich D, Perrin DM. A highly selective DNAzyme sensor for mercuric ions. Angew Chem Int Ed. 2008;47:4346-50
40. Saran R, Liu J. A silver DNAzyme. Anal Chem. 2016;88:4014-20
41. Huang P-JJ, Vazin M, Liu J. In vitro selection of a DNAzyme cooperatively binding two lanthanide ions for RNA cleavage. Biochemistry. 2016;55:2518-25
42. Huang PJ, Vazin M, Matuszek Z, Liu J. A new heavy lanthanide-dependent DNAzyme displaying strong metal cooperativity and unrescuable phosphorothioate effect. Nucleic Acids Res. 2015;43:461-9
43. Huang P-JJ, Vazin M, Liu J. In vitro selection of a new lanthanide-dependent DNAzyme for ratiometric sensing lanthanides. Anal Chem. 2014;86:9993-9
44. Torabi S-F, Wu P, McGhee CE, Chen L, Hwang K, Zheng N. et al. In vitro selection of a sodium-specific DNAzyme and its application in intracellular sensing. Proc Natl Acad Sci USA. 2015;112:5903-8
45. Zhou W, Zhang Y, Huang P-JJ, Ding J, Liu J. A DNAzyme requiring two different metal ions at two distinct sites. Nucleic Acids Res. 2016;44:354-63
46. Torabi S-F, Lu Y. Identification of the same Na+-specific DNAzyme motif from two in vitro selections under different conditions. J Mol Evol. 2015;81:225-34
47. Zhou W, Ding J, Liu J. A Selective Na+ Aptamer Dissected by Sensitized Tb3+ Luminescence. ChemBioChem. 2016;17:1563-70
48. Zhou W, Ding J, Liu J. A highly specific sodium aptamer probed by 2-aminopurine for robust Na+ sensing. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw845
49. Zhou W, Saran R, Chen Q, Ding J, Liu J. A New Na+-Dependent RNA-Cleaving DNAzyme with over 1000-fold Rate Acceleration by Ethanol. ChemBioChem. 2016;17:159-63
50. Peracchi A, Bonaccio M, Clerici M. A mutational analysis of the 8-17 deoxyribozyme core. J Mol Biol. 2005;352:783-94
51. Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D. et al. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature. 2002;419:90-4
52. Nimjee SM, Keys J, Pitoc G, Quick G, Rusconi C, Sullenger BA. A novel antidote-controlled anticoagulant reduces thrombin generation and inflammation and improves cardiac function in cardiopulmonary bypass surgery. Mol Ther. 2006;14:408-15
53. Cao Z, Tong R, Mishra A, Xu W, Wong GC, Cheng J. et al. Reversible cell-specific drug delivery with aptamer-functionalized liposomes. Angew Chem Int Ed. 2009;48:6494-8
54. Crooke ST. Progress in antisense technology. Annu Rev Med. 2004;55:61-95
55. Isaka Y, Imai E, Takahara S, Rakugi H. Oligonucleotidic therapeutics. Expert Opin Drug Discovery. 2008;3:991-6
56. Usman N, Blatt LM. Nuclease-resistant synthetic ribozymes: developing a new class of therapeutics. J Clin Invest. 2000;106:1197-202
57. Weng DE, Masci PA, Radka SF, Jackson TE, Weiss PA, Ganapathi R. et al. A phase I clinical trial of a ribozyme-based angiogenesis inhibitor targeting vascular endothelial growth factor receptor-1 for patients with refractory solid tumors. Mol Cancer Ther. 2005;4:948-55
58. Fokina AA, Stetsenko DA, François J-C. DNA enzymes as potential therapeutics: towards clinical application of 10-23 DNAzymes. Expert Opin Biol Ther. 2015;15:689-711
59. Dass CR, Choong PF, Khachigian LM. DNAzyme technology and cancer therapy: cleave and let die. Mol Cancer Ther. 2008;7:243-51
60. Karnati HK, Yalagala RS, Undi R, Pasupuleti SR, Gutti RK. Therapeutic potential of siRNA and DNAzymes in cancer. Tumor Biol. 2014;35:9505-21
61. Khachigian LM, Fahmy RG, Zhang G, Bobryshev YV, Kaniaros A. c-Jun regulates vascular smooth muscle cell growth and neointima formation after arterial injury Inhibition by a novel DNA enzyme targeting c-Jun. J Biol Chem. 2002;277:22985-91
62. Singh N, Ranjan A, Sur S, Chandra R, Tandon V. Inhibition of HIV-1 Integrase gene expression by 10-23 DNAzyme. J Biosci. 2012;37:493-502
63. Wu Y, Yu L, McMahon R, Rossi JJ, Forman SJ, Snyder DS. Inhibition of bcr-abl oncogene expression by novel deoxyribozymes (DNAzymes). Hum Gene Ther. 1999;10:2847-57
64. Lu Z, Ma X, Yang L, Wang Z, Zeng L, Li ZJ. et al. DNAzymes targeted to EBV-encoded latent membrane protein-1 induce apoptosis and enhance radiosensitivity in nasopharyngeal carcinoma. Cancer Lett. 2008;265:226-38
65. Fu S, Sun L. DNAzyme-based therapeutics for cancer treatment. Future Med Chem. 2015;7:1701-7
66. Baum D, Silverman S. Deoxyribozymes: useful DNA catalysts in vitro and in vivo. Cell Mol Life Sci. 2008;65:2156-74
67. Young DD, Lively MO, Deiters A. Activation and deactivation of DNAzyme and antisense function with light for the photochemical regulation of gene expression in mammalian cells. J Am Chem Soc. 2010;132:6183-93
68. Nakano S, Miyoshi D, Sugimoto N. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem Rev. 2014;114:2733-58
69. Miyoshi D, Sugimoto N. Molecular crowding effects on structure and stability of DNA. Biochimie. 2008;90:1040-51
70. Chinen AB, Guan CM, Mirkin CA. Spherical nucleic acid nanoparticle conjugates enhance G-quadruplex formation and increase serum protein interactions. Angew Chem Int Ed. 2015;54:527-31
71. Zillmann M, Limauro SE, Goodchild J. In vitro optimization of truncated stem-loop II variants of the hammerhead ribozyme for cleavage in low concentrations of magnesium under non-turnover conditions. RNA. 1997;3:734-47
72. Schlosser K, Li Y. A versatile endoribonuclease mimic made of DNA: characteristics and applications of the 8-17 RNA-cleaving DNAzyme. ChemBioChem. 2010;11:866-79
73. Perrin DM, Garestier T, Hélène C. Bridging the gap between proteins and nucleic acids: a metal-independent RNAseA mimic with two protein-like functionalities. J Am Chem Soc. 2001;123:1556-63
74. Lermer L, Roupioz Y, Ting R, Perrin DM. Toward an RNaseA mimic: a DNAzyme with imidazoles and cationic amines. J Am Chem Soc. 2002;124:9960-1
75. Hollenstein M, Hipolito CJ, Lam CH, Perrin DM. A self-cleaving DNA enzyme modified with amines, guanidines and imidazoles operates independently of divalent metal cations (M2+). Nucleic Acids Res. 2009;37:1638-49
76. Sidorov AV, Grasby JA, Williams DM. Sequence-specific cleavage of RNA in the absence of divalent metal ions by a DNAzyme incorporating imidazolyl and amino functionalities. Nucleic Acids Res. 2004;32:1591-601
77. Hollenstein M, Hipolito CJ, Lam CH, Perrin DM. Toward the combinatorial selection of chemically modified DNAzyme RNase A mimics active against all-RNA substrates. ACS Comb Sci. 2013;15:174-82
78. Benner SA. Understanding nucleic acids using synthetic chemistry. Acc Chem Res. 2004;37:784-97
79. Sefah K, Yang Z, Bradley KM, Hoshika S, Jiménez E, Zhang L. et al. In vitro selection with artificial expanded genetic information systems. Proc Natl Acad Sci USA. 2014;111:1449-54
80. Fan H, Zhao Z, Yan G, Zhang X, Yang C, Meng H. et al. A smart DNAzyme-MnO2 nanosystem for efficient gene silencing. Angew Chem Int Ed. 2015;127:4883-7
81. Cairns MJ, Hopkins TM, Witherington C, Wang L, Sun L. Target site selection for an RNA-cleaving catalytic DNA. Nat Biotechnol. 1999;17:480-6
82. Kurreck J, Bieber B, Jahnel R, Erdmann VA. Comparative study of DNA enzymes and ribozymes against the same full-length messenger RNA of the vanilloid receptor subtype I. J Biol Chem. 2002;277:7099-107
83. Sriram B, Banerjea AC. In vitro-selected RNA cleaving DNA enzymes from a combinatorial library are potent inhibitors of HIV-1 gene expression. Biochem J. 2000;352:667-73
84. Schubert S, Fürste JP, Werk D, Grunert H-P, Zeichhardt H, Erdmann VA. et al. Gaining target access for deoxyribozymes. J Mol Biol. 2004;339:355-63
85. Vester B, Lundberg LB, Sørensen MD, Babu BR, Douthwaite S, Wengel J. LNAzymes: incorporation of LNA-type monomers into DNAzymes markedly increases RNA cleavage. J Am Chem Soc. 2002;124:13682-3
86. Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AK, Han MS, Mirkin CA. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027-30
87. Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Oligonucleotide loading determines cellular uptake of DNA-modified gold nanoparticles. Nano Lett. 2007;7:3818-21
88. Ding Y, Jiang Z, Saha K, Kim CS, Kim ST, Landis RF. et al. Gold nanoparticles for nucleic acid delivery. Mol Ther. 2014;22:1075-83
89. Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Polyvalent DNA nanoparticle conjugates stabilize nucleic acids. Nano Lett. 2008;9:308-11
90. Wu P, Hwang K, Lan T, Lu Y. A DNAzyme-gold nanoparticle probe for uranyl ion in living cells. J Am Chem Soc. 2013;135:5254-7
91. Rouge JL, Sita TL, Hao L, Kouri FM, Briley WE, Stegh AH. et al. Ribozyme-spherical nucleic acids. J Am Chem Soc. 2015;137:10528-31
92. Yehl K, Joshi JP, Greene BL, Dyer RB, Nahta R, Salaita K. Catalytic deoxyribozyme-modified nanoparticles for RNAi-independent gene regulation. ACS Nano. 2012;6:9150-7
93. Kim S, Ryoo S, Na H, Kim Y, Choi B, Lee Y. et al. Deoxyribozyme-loaded nano-graphene oxide for simultaneous sensing and silencing of the hepatitis C virus gene in liver cells. Chem Commun. 2013;49:8241-3
94. Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440-50
95. Li J, Lu Y. A highly sensitive and selective catalytic DNA biosensor for lead ions. J Am Chem Soc. 2000;122:10466-7
96. Gong L, Zhao Z, Lv Y, Huan S, Fu T, Zhang X. et al. DNAzyme-based biosensors and nanodevices. Chem Commun. 2015;51:979-95
97. Zhang X, Kong R, Lu Y. Metal ion sensors based on DNAzymes and related DNA molecules. Annu Rev Anal Chem. 2011;4:105
98. Li L, Feng J, Fan Y, Tang B. Simultaneous imaging of Zn2+ and Cu2+ in living cells based on DNAzyme modified gold nanoparticle. Anal Chem. 2015;87:4829-35
99. Hwang K, Wu P, Kim T, Lei L, Tian S, Wang Y. et al. Photocaged DNAzymes as a general method for sensing metal ions in living cells. Angew Chem Int Ed. 2014;126:14018-22
100. Wang X, Feng M, Xiao L, Tong A, Xiang Y. Postsynthetic modification of DNA phosphodiester backbone for photocaged dnazyme. ACS Chem Biol. 2015;11:444-51
101. Zhou W, Chen Q, Huang P-JJ, Ding J, Liu J. DNAzyme hybridization, cleavage, degradation, and sensing in undiluted human blood serum. Anal Chem. 2015;87:4001-7
102. Torabi SF, Lu Y. Small-molecule diagnostics based on functional DNA nanotechnology: a dipstick test for mercury. Faraday Discuss. 2011;149:125-35
103. Mazumdar D, Liu J, Lu G, Zhou J, Lu Y. Easy-to-use dipstick tests for detection of lead in paints using non-cross-linked gold nanoparticle-DNAzyme conjugates. Chem Commun. 2010;46:1416-8
104. Ali MM, Aguirre SD, Lazim H, Li Y. Fluorogenic DNAzyme probes as bacterial indicators. Angew Chem Int Ed. 2011;50:3751-4
105. Aguirre SD, Ali MM, Salena BJ, Li Y. A sensitive DNA enzyme-based fluorescent assay for bacterial detection. Biomolecules. 2013;3:563-77
106. Kang D-K, Ali MM, Zhang K, Huang SS, Peterson E, Digman MA. et al. Rapid detection of single bacteria in unprocessed blood using integrated comprehensive droplet digital detection. Nat Commun. 2014:5
107. Tram K, Kanda P, Salena BJ, Huan S, Li Y. Translating bacterial detection by DNAzymes into a litmus test. Angew Chem Int Ed. 2014;53:12799-802
108. Shen Z, Wu Z, Chang D, Zhang W, Tram K, Lee C. et al. A catalytic DNA activated by a specific strain of bacterial pathogen. Angew Chem Int Ed. 2016;55:2431-4
109. He S, Qu L, Shen Z, Tan Y, Zeng M, Liu F. et al. Highly specific recognition of breast tumors by an RNA-cleaving fluorogenic DNAzyme probe. Anal Chem. 2014;87:569-77
110. Todd AV, Fuery CJ, Impey HL, Applegate TL, Haughton MA. DzyNA-PCR: use of DNAzymes to detect and quantify nucleic acid sequences in a real-time fluorescent format. Clin Chem. 2000;46:625-30
111. Suenaga H, Liu R, Shiramasa Y, Kanagawa T. Novel approach to quantitative detection of specific rRNA in a microbial community, using catalytic DNA. Appl Environ Microbiol. 2005;71:4879-84
112. Wang DY, Sen D. A novel mode of regulation of an RNA-cleaving DNAzyme by effectors that bind to both enzyme and substrate. J Mol Biol. 2001;310:723-34
113. Wang DY, Lai BH, Feldman AR, Sen D. A general approach for the use of oligonucleotide effectors to regulate the catalysis of RNA-cleaving ribozymes and DNAzymes. Nucleic Acids Res. 2002;30:1735-42
114. Sando S, Sasaki T, Kanatani K, Aoyama Y. Amplified nucleic acid sensing using programmed self-cleaving DNAzyme. J Am Chem Soc. 2003;125:15720-1
115. Mokany E, Bone SM, Young PE, Doan TB, Todd AV. MNAzymes, a versatile new class of nucleic acid enzymes that can function as biosensors and molecular switches. J Am Chem Soc. 2009;132:1051-9
116. Zagorovsky K, Chan WC. A plasmonic DNAzyme strategy for point-of-care genetic detection of infectious pathogens. Angew Chem Int Ed. 2013;52:3168-71
117. Elbaz J, Moshe M, Shlyahovsky B, Willner I. Cooperative multicomponent self-assembly of nucleic acid structures for the activation of DNAzyme cascades: a paradigm for DNA sensors and aptasensors. Chem Eur J. 2009;15:3411-8
118. Wang F, Elbaz J, Orbach R, Magen N, Willner I. Amplified analysis of DNA by the autonomous assembly of polymers consisting of DNAzyme wires. J Am Chem Soc. 2011;133:17149-51
119. Famulok M, Hartig JS, Mayer G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem Rev. 2007;107:3715-43
120. Achenbach JC, Nutiu R, Li Y. Structure-switching allosteric deoxyribozymes. Anal Chim Acta. 2005;534:41-51
121. Tang J, Breaker RR. Rational design of allosteric ribozymes. Chem Biol. 1997;4:453-9
122. Liu J, Lu Y. Rational design of “turn-on” allosteric DNAzyme catalytic beacons for aqueous mercury ions with ultrahigh sensitivity and selectivity. Angew Chem Int Ed. 2007;119:7731-4
123. Huang J, He Y, Yang X, Wang K, Quan K, Lin X. Split aptazyme-based catalytic molecular beacons for amplified detection of adenosine. Analyst. 2014;139:2994-7
124. Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281-6
125. Hu R, Liu T, Zhang X-B, Yang Y, Chen T, Wu C. et al. DLISA: A DNAzyme-based ELISA for protein enzyme-free immunoassay of multiple analytes. Anal Chem. 2015;87:7746-53
126. Liu C, Sheng Y, Sun Y, Feng J, Wang S, Zhang J. et al. A glucose oxidase-coupled DNAzyme sensor for glucose detection in tears and saliva. Biosens Bioelectron. 2015;70:455-61
127. Carmi N, Balkhi SR, Breaker RR. Cleaving DNA with DNA. Proc Natl Acad Sci USA. 1998;95:2233-7
128. Carmi N, Breaker RR. Characterization of a DNA-cleaving deoxyribozyme. Biorg Med Chem. 2001;9:2589-600
129. Xiang Y, Lu Y. Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat Chem. 2011;3:697-703
130. Xiang Y, Lu Y. An invasive DNA approach toward a general method for portable quantification of metal ions using a personal glucose meter. Chem Commun. 2013;49:585-7
131. Yigit MV, Mazumdar D, Kim HK, Lee JH, Odintsov B, Lu Y. Smart “turn-on” magnetic resonance contrast agents based on aptamer-functionalized superparamagnetic iron oxide nanoparticles. ChemBioChem. 2007;8:1675-8
132. Yigit MV, Mazumdar D, Lu Y. MRI detection of thrombin with aptamer functionalized superparamagnetic iron oxide nanoparticles. Bioconj Chem. 2008;19:412-7
133. Xu W, Lu Y. A smart magnetic resonance imaging contrast agent responsive to adenosine based on a DNA aptamer-conjugated gadolinium complex. Chem Commun. 2011;47:4998-5000
134. Xu W, Xing H, Lu Y. A smart T 1-weighted MRI contrast agent for uranyl cations based on a DNAzyme-gadolinium conjugate. Analyst. 2013;138:6266-9
135. Prigodich AE, Seferos DS, Massich MD, Giljohann DA, Lane BC, Mirkin CA. Nano-flares for mRNA regulation and detection. ACS Nano. 2009;3:2147-52
136. Balogh D, Aleman Garcia MA, Albada HB, Willner I. Programmed synthesis by stimuli-responsive DNAzyme-modified mesoporous SiO2 nanoparticles. Angew Chem Int Ed. 2015;127:11818-22
137. Zhang Z, Balogh D, Wang F, Willner I. Smart mesoporous SiO2 nanoparticles for the DNAzyme-induced multiplexed release of substrates. J Am Chem Soc. 2013;135:1934-40
138. Patil SD, Rhodes DG, Burgess DJ. DNA-based therapeutics and DNA delivery systems: a comprehensive review. AAPS J. 2005;7:E61-E77
139. Dass CR, Saravolac EG, Li Y, Sun LQ. Cellular uptake, distribution, and stability of 10-23 deoxyribozymes. Antisense Nucleic Acid Drug Dev. 2002;12:289-99
140. Deleavey GF, Damha MJ. Designing chemically modified oligonucleotides for targeted gene silencing. Chem Biol. 2012;19:937-54
141. Cieslak M, Niewiarowska J, Nawrot M, Koziolkiewicz M, Stec WJ, Cierniewski CS. DNAzymes to β1 and β3 mRNA down-regulate expression of the targeted integrins and inhibit endothelial cell capillary tube formation in fibrin and matrigel. J Biol Chem. 2002;277:6779-87
142. Sioud M, Leirdal M. Design of nuclease resistant protein kinase Cα DNA enzymes with potential therapeutic application. J Mol Biol. 2000;296:937-47
143. Schubert S, GuÈl DC, Grunert HP, Zeichhardt H, Erdmann VA, Kurreck J. RNA cleaving '10-23' DNAzymes with enhanced stability and activity. Nucleic Acids Res. 2003;31:5982-92
144. Kubo T, Takamori K, Kanno K, Rumiana B, Ohba H, Matsukisono M. et al. Efficient cleavage of RNA, enhanced cellular uptake, and controlled intracellular localization of conjugate DNAzymes. Biorg Med Chem Lett. 2005;15:167-70
145. Takahashi H, Hamazaki H, Habu Y, Hayashi M, Abe T, Miyano-Kurosaki N. et al. A new modified DNA enzyme that targets influenza virus A mRNA inhibits viral infection in cultured cells. FEBS Lett. 2004;560:69-74
146. Ke G, Wang C, Ge Y, Zheng N, Zhu Z, Yang CJ. L-DNA molecular beacon: a safe, stable, and accurate intracellular nano-thermometer for temperature sensing in living cells. J Am Chem Soc. 2012;134:18908-11
147. Cui L, Peng R, Fu T, Zhang X, Wu C, Chen H. et al. Biostable L-DNAzyme for sensing of metal ions in biological systems. Anal Chem. 2016;88:1850-5
148. Wyszko E, Szymanski M, Zeichhardt H, Muller F, Barciszewski J, Erdmann VA. Spiegelzymes: sequence specific hydrolysis of L-RNA with mirror image hammerhead ribozymes and DNAzymes. PLoS One. 2013;8:e54741
149. Lin Tan M, Choong PF, Dass CR. DNAzyme delivery systems: getting past first base. Expert Opin Drug Deliv. 2009;6:127-38
150. Lytton-Jean AK, Mirkin CA. A thermodynamic investigation into the binding properties of DNA functionalized gold nanoparticle probes and molecular fluorophore probes. J Am Chem Soc. 2005;127:12754-5
151. Zhang X, Liu B, Servos MR, Liu J. Polarity control for nonthiolated DNA adsorption onto gold nanoparticles. Langmuir. 2013;29:6091-8
152. Pei H, Li F, Wan Y, Wei M, Liu H, Su Y. et al. Designed diblock oligonucleotide for the synthesis of spatially isolated and highly hybridizable functionalization of DNA-gold nanoparticle nanoconjugates. J Am Chem Soc. 2012;134:11876-9
153. Zhang X, Liu B, Dave N, Servos MR, Liu J. Instantaneous attachment of an ultrahigh density of nonthiolated DNA to gold nanoparticles and its applications. Langmuir. 2012;28:17053-60
154. Zhu D, Pei H, Chao J, Su S, Aldalbahi A, Rahaman M. et al. Poly-adenine-based programmable engineering of gold nanoparticles for highly regulated spherical DNAzymes. Nanoscale. 2015;7:18671-6
155. Li J, Hong C, Wu S, Liang H, Wang L, Huang G. et al. Facile phase transfer and surface biofunctionalization of hydrophobic nanoparticles using Janus DNA tetrahedron nanostructures. J Am Chem Soc. 2015;137:11210-3
156. Walsh AS, Yin H, Erben CM, Wood MJ, Turberfield AJ. DNA cage delivery to mammalian cells. ACS Nano. 2011;5:5427-32
157. Zhou W, Liang W, Li D, Yuan R, Xiang Y. Dual-color encoded DNAzyme nanostructures for multiplexed detection of intracellular metal ions in living cells. Biosens Bioelectron. 2016;85:573-9
158. Meng H, Zhang X, Lv Y, Zhao Z, Wang N, Fu T. et al. DNA dendrimer: an efficient nanocarrier of functional nucleic acids for intracellular molecular sensing. ACS Nano. 2014;8:6171-81
159. Funkhouser J. Reintroducing pharma: Theranostic revolution. Current Drug Discovery. 2002;2:17-9
160. Wilhelm S, Tavares AJ, Dai Q, Ohta S, Audet J, Dvorak HF. et al. Analysis of nanoparticle delivery to tumours. Nature Reviews Materials. 2016;1:16014
161. Xiao Y, Allen EC, Silverman SK. Merely two mutations switch a DNA-hydrolyzing deoxyribozyme from heterobimetallic (Zn2+/Mn2+) to monometallic (Zn2+-only) behavior. Chem Commun. 2011;47:1749-51
162. Gu H, Furukawa K, Weinberg Z, Berenson DF, Breaker RR. Small, highly active DNAs that hydrolyze DNA. J Am Chem Soc. 2013;135:9121-9
163. Liu N, Hou R, Gao P, Lou X, Xia F. Sensitive Zn2+ sensor based on biofunctionalized nanopores via combination of DNAzyme and DNA supersandwich structures. Analyst. 2016;141:3626-9
164. Wang M, Zhang H, Zhang W, Zhao Y, Yasmeen A, Zhou L. et al. In vitro selection of DNA-cleaving deoxyribozyme with site-specific thymidine excision activity. Nucleic Acids Res. 2014;42:9262-9
Author contact
![]() Corresponding author: Email: liujwca.
Corresponding author: Email: liujwca.
 Global reach, higher impact
Global reach, higher impact