13.3
Impact Factor
Theranostics 2017; 7(3):789-804. doi:10.7150/thno.18133 This issue Cite
Review
Progress in Exosome Isolation Techniques
1. Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore 639798.
2. Department of Chemistry, National University of Singapore, Singapore 117543.
3. Department of Pharmacy, National University of Singapore, Singapore 117543.
Received 2016-10-29; Accepted 2016-11-20; Published 2017-1-26
Abstract

Exosomes are one type of membrane vesicles secreted into extracellular space by most types of cells. In addition to performing many biological functions particularly in cell-cell communication, cumulative evidence has suggested that several biological entities in exosomes like proteins and microRNAs are closely associated with the pathogenesis of most human malignancies and they may serve as invaluable biomarkers for disease diagnosis, prognosis, and therapy. This provides a commanding impetus and growing demands for simple, efficient, and affordable techniques to isolate exosomes. Capitalizing on the physicochemical and biochemical properties of exosomes, a number of techniques have been developed for the isolation of exosomes. This article summarizes the advances in exosome isolation techniques with an emphasis on their isolation mechanism, performance, challenges, and prospects. We hope that this article will provide an overview of exosome isolation techniques, opening up new perspectives towards the development more innovative strategies and devices for more time saving, cost effective, and efficient isolations of exosomes from a wide range of biological matrices.
Keywords: Exosome, isolation
1. Introduction
The discovery of exosomes dates back to 1983 in two independent papers published respectively by Harding et al. 1 and Pan et al., 2 and later confirmed by Pan et al.3 In these papers, the authors cultured immature red blood cells - reticulocytes with labeled transferrin receptors to trace the movement of transferrin receptors from plasma membranes into the reticulocytes. Surprisingly, it was observed that the labeled transferrin receptors are internalized within the reticulocytes, and then repackaged into small (~50 nm) vesicles inside them. These vesicles, originally thought to be extracellular to be trafficked to lysosomes for destruction, are subsequently secreted out of the maturing blood reticulocytes into extracellular space.4 Later, in the year 1989, Johnstone et al. coined these vesicles “exosomes”.5 Exosomes belong to a large family of membrane vesicles referred to as extracellular vesicles, which generally include microvesicles (100-350 nm),6 apoptotic blebs (500-1000 nm),7 and exosomes (30-150 nm).8 These extracellular vesicles are believed to be involved in many biological processes and prominently in intercellular communication. Their pathophysiological roles are being decoded in various medical conditions and diseases including cancer. When examined under an electron microscope, exosomes show characteristic cup-shaped morphology, appearing as flattened spheres with diameters ranging from 30 to 150 nm (Figure 1).8 The cup-shaped morphology is most likely originated from the sample preparation process of conventional electron microscopy during which the exomes are extremely dehydrated, thus leading to the collapse of the exosomes. In contrast, the exosomes remain fully hydrated in cryo-electron microscopic examinations, round-shaped morphology is obsrved.9,10 A diagrammatic representation of a typical exosome is also shown in Figure 1.10
(A) An electron microscopic image of exosomes (Reproduced with permission from reference 8) and (B) a diagrammatic representation of a medium size exosome (Reproduced with permission from Reference 10).
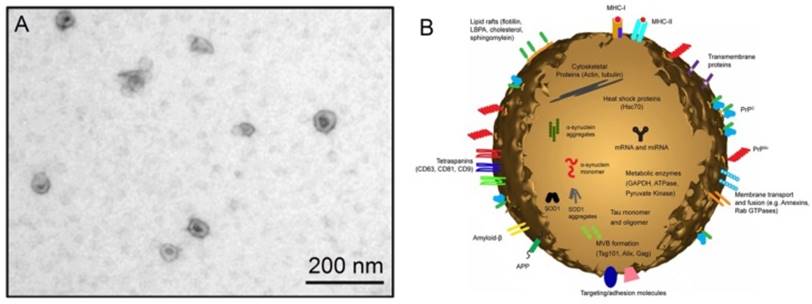
As seen in Figure 1, exosomes have a characteristic lipid bilayer which has an average thickness of ~5 nm.8 The lipid components of exosomes include ceramide (sometimes used to differentiate exosomes from lysosomes), cholesterol, sphingolipids, and phosphoglycerides with long and saturated fatty-acyl chains. The outer surface of exosomes is rich in saccharide chains, such as mannose, polylactosamine, alpha-2,6 sialic acid, and N-linked glycans.11
Of great interest is proteins found on the surface and in the core of exosomes. As one of the most important cargoes that exosomes carry, the proteins can provide invaluable information associated with the physiological states of the parental cells of exosomes. Many exosomes contain proteins that are common among all exosomes regardless of the types of cells which secrete them, whilst only a small fraction of proteins are cell-specific, reflecting the type and pathophysiological conditions of those secreting cells.12,13 Proteins typically found in exosomes include platelet derived growth factor receptor, lactadherin, transmembrane proteins and lysosome associated membrane protein-2B,14,15 membrane transport and fusion proteins like annexins, flotillins, GTPases, heat shock proteins, tetraspanins, proteins involved in multivesicular body biogenesis, as well as lipid-related proteins and phospholipases.16,17 These characteristic proteins therefore serve as good biomarkers for the isolation and quantification of exosomes. Another key cargo that exosomes carry is nucleic acids including deoxynucleic acids (DNA), coding and non-coding ribonucleic acid (RNA) like messenger RNA (mRNA) and microRNA (miRNA).18 These nucleic acids are often needed to be accurately quantified after exosomes have been isolated since their profiles meticulously reflect many kinds of medical conditions and diseases.19,20
Exosomes have received tremendous attention in recent years due to the fact that the biological fingerprints of exosomes practically mirror those of the parental cells they are originated.21 Although the exact biological functions of exosomes remain to be fully deciphered, increasing evidence has indicated that exosomes play a vital role in many cellular processes like cell-cell communication, coagulation, antigen presentation, waste management, as well as transfer of proteins and nucleic acids. Even though exosomes were discovered more than three decades ago,1,2 researchers are just starting to unravel the mystery of these extremely small extracellular vesicles. Originally considered as experimental artefacts, waste, or remains of deceased cells,2 exosomes are now believed to be an central part of a new form of cell-cell communication.22-24 Cell-cell communication is crucial in all multicellular organisms and were previously thought only to be possible via contact-dependent (direct contact between adjacent cells), paracrine (binding of short-ranged signaling molecules on receptors of nearby cells), endocrine (binding of long-ranged hormones on far away cells via bloodstream), or neuronal (electrical impulses travelling along neurons) signaling. Recent research has highlighted possible exosome-mediated cell-cell communication in many processes, for example immune signaling,25,26 angiogenesis,27,28 senescence,29,30 proliferation,31 differentiation,32 and implicated in many human diseases like neurodegenerative disorders, cancer, and AIDS.33-37 More importantly, growing evidence has also suggested that exosomes play a key role in facilitating tumorigenesis by regulating angiogenesis, immunity, and metastasis.38 Circulating exosomes in body fluids and blood in particular are potentially non-invasive or minimally invasive biomarkers for early diagnosis and prognosis of various types of cancer.39,40 As such, exosomes not only contribute to a greater understanding of cell physiology and pathology, but also have a great potential to be translated to various clinical applications, ranging from diagnosis and prognosis to nucleic acid delivery and cancer therapeutics.
The fast advances in the understanding of exosomes and their biological functions have been providing fundamentally new insights into the physiological roles that exosomes play and their prominent relevance to human health.41 Consequently, the isolation and quantification of exosomes have become a major initiative in both basic research and clinical applications.42,43 As the first step towards improving human health, exosomes have to be reliably and efficiently isolated from complex biological matrices like blood, urine, and cerebrospinal fluid since they are currently tested as next-generation biomarkers in those body fluids. To date, five groups of exosome isolation techniques have been developed. They are differential ultracentrifugation-based techniques, size-based techniques, immunoaffinity capture-based techniques, exosome precipitation, and microfluidics-based techniques.42 In the following sections, we will review all the five groups of techniques with typical examples illustrating their mechanisms and typical workflows of exosome isolation. A critical comparison of exosome isolation techniques is attempted and major challenges and prospects of the field are discussed before concluding remarks.
2. Exosome Isolation Techniques
In order to facilitate the study and application of these unique extracellular vesicles, it is crucial that exosomes are specifically isolated from a wide spectrum of cellular debris and interfering components. The techniques employed in the isolation of exosomes should exhibit high efficiency and are capable of isolating exosomes from various sample matrices. To examine the quality of isolated exosomes, several optical and non-optical techniques have been developed to gauge their size, size distribution, morphology, quantity, and biochemical composition.43 With the fast advances in science and technology, many techniques have been developed for the isolation of exosomes in appreciable quantity and purity. Each technique exploits a particular trait of exosomes, such as their density, shape, size, and surface proteins to aid their isolation. Variants within each group also bring about a unique set of advantages and disadvantages to exosome isolation. These exosome isolation techniques will be discussed in detail in the following sections.
2.1 Ultracentrifugation-based isolation techniques
When a heterogeneous mixture (suspension) is subjected to a centrifugal force - centrifugation, particulate constituents in the suspension will be sedimented according to their density, size, and shape. More dense and/or bigger particles settle out first. Centrifugation is usually employed to separate and purify particulate materials as well as to analyze the hydrodynamic properties of polymeric materials including biopolymers like nucleic acids and proteins.44 Depending on the centrifugal force applied, particles in a suspension can be separated sequentially according to their physical properties and the density and viscosity of the solvent. Ultracentrifugation is a centrifugation process optimized for generating exceptionally high centrifugal forces up to 1,000,000 × g. There are two types of ultracentrifugation - analytical and preparative ultracentrifugation. Analytical ultracentrifugation is utilized to investigate the physicochemical properties of particulate materials and molecular interactions of polymeric materials. As for exosome isolation, preparative ultracentrifugation plays an important part since it is meant to fractionate small bioparticles such as viruses, bacteria, subcellular organelles, and extracellular vesicles. Ultracentrifugation-based exosome isolation is considered to be the gold standard and is one of the most commonly used and reported techniques in exosome isolation. It is estimated that ultracentrifugation accounts for 56% of all exosome isolation techniques employed by users in exosome research.45 This approach is often perceived by many as easy to use, requiring very little technical expertise, affordability over time (i.e. one ultracentrifuge machine for long term use), and moderately time-consuming with little or no sample pretreatments. For these reasons, ultracentrifugation-based techniques have become a rather popular option among researchers in exosome research. There are two types of preparative ultracentrifugation - differential ultracentrifugation and density gradient ultracentrifugation.
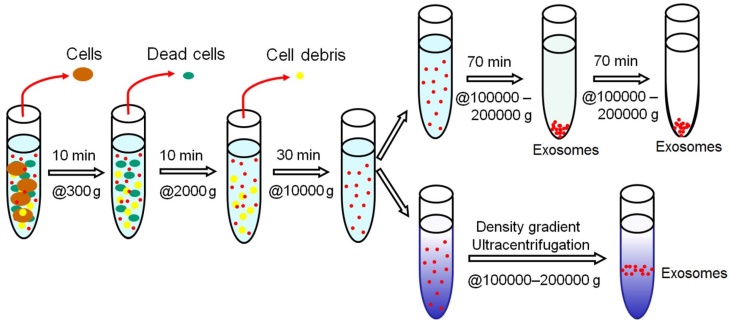
The isolation of exosomes by differential ultracentrifugation usually consists of a series of centrifugation cycles of different centrifugal force and duration to isolate exosomes based on their density and size differences from other components in a sample. For ultracentrifugation, the centrifugal force used typically ranges from ~100,000 to 120,000 × g. Before the start of isolation, a cleaning step is usually carried out for human plasma/serum to rid of large bioparticles in a sample and the sample is spiked with protease inhibitors to prevent the degradation of exosomal proteins.46 Between runs during exosome isolation, the supernatant is aspired and depending on the centrifugal force used, either the supernatant or the pellet is re-suspended in an appropriate medium such as phosphate buffered saline (PBS) and subjected to subsequent runs of centrifugation with increasing centrifugal force. Finally, the isolated exosomes are once again re-suspended and stored at -80⁰C until further analysis. This method of isolating exosomes is also known as the pelleting method or simple ultracentrifugation method.47 A schematic illustration of the workflow of differential ultracentrifugation is presented in Figure 2.48
Variations of ultracentrifugation also exist, such as density gradient ultracentrifugation. There are two types of density gradient ultracentrifugation, namely isopycnic ultracentrifugation and moving-zone ultracentrifugation. The use of density gradient ultracentrifugation has become increasingly popular in the isolation of extracellular vesicles like exosomes. In density gradient ultracentrifugation, separation of exosomes is accomplished based on their in size, mass, and density in a pre-constructed density gradient medium in a centrifuge tube with progressively decreased density from bottom to top. A sample is layered as a narrow band onto the top of the density gradient medium and subjected to an extended round of ultracentrifugation. Upon applying a centrifugal force, solutes including exosomes in the sample move as individual zones through the density gradient medium towards the bottom each at its specific sedimentation rate, thus leading to discrete solute zones. The separated exosomes can then be conveniently recovered by simple fraction collection. While a continuous gradient is used for analytical applications, a discontinuous gradient (stepped gradient) is more suited for preparative purposes in which the separated exosomes are located at the interface of the density gradient layers, thus greatly facilitating their harvesting. Unlike differential ultracentrifugation, a downside of density gradient ultracentrifugation is that its capacity is largely limited by the narrow load zone.
In isopycnic ultracentrifugation, a density gradient medium embracing the entire range of densities of solutes in a sample is loaded to a centrifuge tube. The separation of exosomes from other solutes into a discrete zone exclusively depends on their density difference from those of all other solutes provided that a sufficient period of centrifugation is engaged. During centrifugation, exosomes sediment along the density gradient medium to where they have the same density as the medium - isopycnic position. After the exosomes have reached their isopycnic position, the centrifugal force further focuses the exosomes into a sharp zone and upholds them there, implying that isopycnic ultracentrifugation is static. Alternatively, a sample containing exosomes can be uniformly mixed with a gradient medium in the case of self-generating gradient materials such as cesium chloride. During centrifugation, the exosomes move to their isopycnic position while a density gradient of cesium chloride is generated. Exosomes can then be extricated from the density region of interest between 1.10 and 1.21 g/ml, where they are concentrated.49 The aliquot obtained from the density region of interest is then subjected to a brief ultracentrifugation at ~100,000 × g to afford pure exosome pellets which are re-suspended in PBS for further study.
Schematic representation of isolating exosomes by differential ultracentrifugation. All centrifugations are carried out at 4⁰C.
In moving-zone ultracentrifugation, a sample containing exosomes is loaded as a thin zone on top of a gradient density medium of having a lower density than that of any of the solutes. Unlike isopycnic ultracentrifugation where separation of exosomes is solely dependent on their density difference from all other solutes, the exosomes in the sample are separated based on their size and mass instead of density. This allows the separation of extracellular vesicles with similar densities but different sizes. Because the densities of the solutes including exosomes are greater than the density of the gradient medium, moving-zone ultracentrifugation is dynamic rather than static. In other words, all solutes will eventually pellet at the bottom of the centrifuge tube when a prolonged period of centrifugation is executed. Therefore, the duration of centrifugation must be carefully optimized. In addition, to prevent exosomes from pelleting out, a high-density cushion is often layered at the bottom of the centrifuge tube. In contrast, exosomes will never sediment to the bottom of the centrifuge tube in isopycnic ultracentrifugation irrespective of the duration of centrifugation.
Due to the heterogeneity of exosomes and considerable overlap in size of extracellular vesicles, differential ultracentrifugation often suffers from contamination and exosome losses. Ultracentrifugation is often coupled to isopycnic or moving-zone techniques to allow the exosomes of relatively low densities to float and to further purify the exosomes. This technique has been known to be able to improve the quantity of exosomes isolated.50,51 After characterization, the fraction of interest containing the exosomes is diluted in PBS and subjected to another round of ultracentrifugation at ~100,000 × g to yield pure exosomes for further analysis.
2.2 Size-based isolation techniques
One of the popular size-based exosome isolation techniques is ultrafiltration. The fundamentals of ultrafiltration are no different from conventional membrane filtration in which the separation of suspended particles or polymers is primarily dependent on their size or molecular weight. Therefore, based on their size, exosomes can be isolated using membrane filters with defined molecular weight or size exclusion limits.52 Ultrafiltration is faster than ultracentrifugation and does not require special equipment.53 However, the use of force may result in the deformation and breaking up of large vesicles which may potentially skew the results of downstream analysis.54
Nanomembrane concentrators with short periods of centrifugations have been shown to be able to rapidly enrich urinary exosomes as effective as ultracentrifugation.55 It was shown that as little as 0.5 mL urine is sufficient for a successful isolation. To confirm the success of exosome isolation, Western blot was engaged to detect exosomal biomarkers and electron microscopy employed to examine the typical features of exosomes. The nanomembrane concentrators have the potential to aid the diagnosis of renal diseases. 55
For cell-free samples like urine, serum, cerebrospinal fluid, and cell culture medium, a commercial exosome isolation kit for exosome isolation and RNA extraction from isolated exosomes has been developed.56 The kit leverages on a syringe filter-based rapid fractionation process during which exosomes and other extracellular vesicles are captured. Two membranes are tandemly configured in the syringe filter so that exosomes are captured on the lower membrane whereas larger extracellular vesicles such as apoptotic bodies and microvesicles are retained on the upper membrane when a sample is passed through the two membranes. RNAs carried by the exosomes are then released when the captured exosomes are lysed by passing an RNA extraction cocktail through the lower membrane. Subsequently, miRNA detection and expression profiling are accomplished by quantitative real-time reverse transcriptase-polymerase chain reaction (qRT-PCR).57 Compared to ultracentrifugation and exosome precipitation, ultrafiltration has the highest exosomal RNA yield from urine as quantified by fluorescent staining.
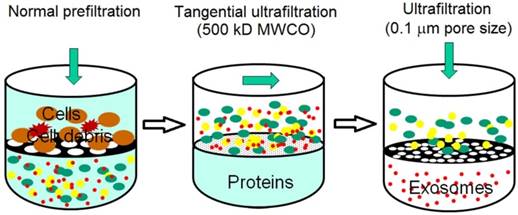
Sequential filtration is applied to isolate exosomes from cell culture supernatants, as summarized in Figure 3. Firstly, dead-end (normal) filtration depletes floating cells and large cell debris using a 100-nm membrane filter. Large and rigid components are eliminated, but large flexible components are able to pass through the filter, even if their diameter is larger than 100 nm. Secondly, the filtrate is subjected to tangential flow filtration with 500 kDa molecular weight cut-off (MWCO) hollow fibers. The concentrated retentate is then diafiltered to further deplete contaminants. Thirdly, the sample is filtered with a 100 nm track-etch filter. To maximize exosome recovery, the filters are washed at the end of each step and transmembrane pressure is monitored and maintained during the second and third steps. Finally, electron microscopy verifies the morphology and mass spectrometry confirms the presence of exosome-associated proteins. Sequential filtration allows the isolation of exosomes with high purity and functional integrity as a result of low manipulation forces. For example, tangential flow filtration combined with deuterium/sucrose-based density gradient ultracentrifugation was employed to isolate therapeutic exosomes for clinical trials.58,59 Its speed, automation, and scalability likely facilitate the translation of exosome research and applications from bench to bedside.60 In a phase I clinical trial, it was observed that exosomes from dendritic cell cultures promoted T-Cell response for tumor rejection.44
Another size-based separation technique applied to exosome isolation is size exclusion chromatography (SEC). In SEC, a porous stationary phase is utilized to sort macromolecules and particulate matters out according to their size. Components in a sample with small hydrodynamic radii are able to pass through the pores, thus resulting in late elution. Components with large hydrodynamic radii including exosomes, are excluded from entering the pores.61 For example, exosomes in mesenchymal stem cell-conditioned medium were isolated by size exclusion fractionation. Mesenchymal stem cell mediates cardioprotection during myocardial ischemia/reperfusion injury by secreting exosomes.62 It was observed that the isolated exosomes are structurally intact as examined by transmission electron microscopy. The size distribution of the isolated exosomes was measured by dynamic light scattering and nanoparticle tracking analysis where a laser illuminates the exosomes as individual point-scatters under Brownian motion and their size is computed from their velocity. Western blot was also employed to evaluate the quantity and purity of the isolated exosomes. In another report, using ascetic fluids from ovarian cancer patients, exosomes were isolated by SEC. Western blot was again performed to confirm the biomarkers associated with the isolated exosomes.63 SEC has also been used in combination with other techniques. For instance, ultracentrifugation followed by SEC greatly enriches urinary exosomes in comparison with the yields obtained solely by ultrafiltration or ultracentrifugation, thereby favorably impacting on the progress of the quest for biomarkers for renal diseases.64
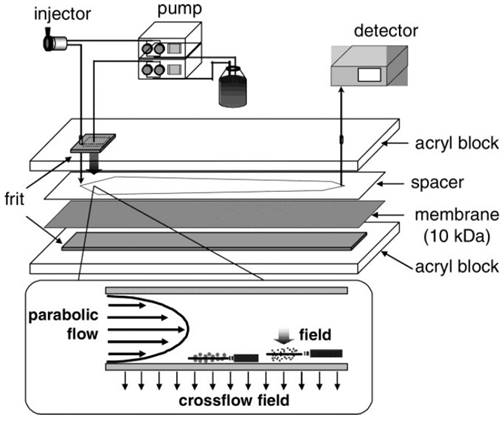
A relatively new member of the size-based exosome isolation techniques is flow field-flow fractionation (F4). As illustrated in Figure 4, F4 uses a porous rectangular channel. While a parabolic flow runs along the channel axis carries a sample towards the end of the channel, a crossflow across the channel controls the retention of the sample.65 The crossflow distributes particulate components against the channel wall based on the diffusivity of the components. Smaller particles diffuse further from the accumulation wall and are eluted earlier than larger ones. Successful attempts have been made using F4 to isolate exosomes from human neural stem cell culture. The fraction containing exosomes was confirmed by transmission electron microscopy and liquid chromatography-mass spectrometry.65 In another report, substantial improvement in the scalability of microfluidic F4 devices was achieved with an asymmetrical field-flow fraction system coupled with in-line ultraviolet and dynamic light scattering detectors.66 This integrated system is able to rapidly isolate and characterize exosomes, thus having the potential to greatly facilitate exosome research and application.
To provide a viable alternative to differential/gradient ultracentrifugation for processing large sample volumes, a simple and effective separation technique termed hydrostatic filtration dialysis (HFD) for enriching and isolating extracellular vesicles from urine was recently developed by Musante et al.67 Unlike conventional dialysis where the separation of solutes is achieved by diffusion across a dialysis membrane with a definite value of MWCO, a samples is forced through a dialysis tube by a low hydrostatic pressure. Solvent and small solutes pass through the dialysis tube freely while extracellular vesicles including exosomes are retained in the dialysis tube. It was demonstrated that exosomes in urine can be enriched by 100 times and further enrichment and purification can be realized by differential centrifugation. It was also demonstrated that HFD outperforms ultracentrifugation-based techniques with much reduced labor and cost.67
Schematic illustration of sequential filtration. Firstly, cells and cell debris are removed. Secondly, free protein is filtered out and the sample is concentrated. Finally, extracellular vesicles larger than 100 nm is removed. (Reproduced with permission from reference 60)
Schematic representation of F4. Sample components experience two opposite forces: crossflow field and diffusion. Parabolic flow and equilibrium positions of sample components are illustrated. (Reproduced with permission from reference 65)
2.3 Immunoaffinity capture-based techniques
The presence of plenty of proteins and receptors in the membrane of exosomes offers an excellent opportunity to develop highly specific techniques for the isolation of exosomes by tapping on immunoaffinitive interactions between those proteins (antigens) and their antibodies, and specific interactions between the receptors and ligands. Consequently, immunoaffinity capture-based techniques have been developed for the isolation of exosomes. Ideally, exosome biomarkers for immunoisolation are membrane-bound, lacking soluble counterparts, and solely expressed or highly concentrated on the surface of exosomes from specific biological sources.
For example, a microplate-based enzyme-linked immunosorbent assay (ELISA) was developed for capturing and quantifying exosomes from plasma, serum, and urine. ELISA results were expressed as absorbance values to quickly compare the expression of known surface biomarkers and as a form of instantaneous readouts of the yield and specificity of exosomes. The absorbance values can also be extrapolated to quantify the captured exosomes through calibration using standards with known exosome counts. The specificity and yield of exosomes by means of the microplate-based immunoaffinity approach were assessed with respect to ultracentrifugation. Plasma samples were pre-cleaned by a brief round of low-speed centrifugation to eliminate cellular debris and large bioparticles and to concentrate extracellular vesicles. This microplate-based immunoaffinity capture approach produced comparable results to those obtained by ultracentrifugation with much less sample volumes, demonstrating the superiority of immunoaffinity capture over ultracentrifugation. Moreover, the RNA yield achieved from the captured exosomes using the microplate was higher than that obtained by ultracentrifugation. Exosomal RNA was extracted from as little as 100 μL of plasma. The amount of RNA extracted from 400 μL of plasma by the microplate was comparable to that obtained by ultracentrifugation of 2.5 mL of the same sample.45
To add value to immunoaffinity capture, Zarovni et al. developed submicron-size magnetic particles for uses in immunoaffinity capture - magneto-immunocapture.45 With as little as 1.0 mL of cell culture supernatant, it was found that the capture efficiency of antibody-coated magnetic particles is close to that of ultracentrifugation. Even though with a comparable yield to that of ultracentrifugation, immunoaffinity capture still has the edges of being rapid, easy to use, and compatible with routine bench equipment. When applied to plasma samples, the yield achieved by the magneto-immunocapture capture was 10 to 15 times higher than that obtained by ultracentrifugation, evidenced by Western blot and fluorescence-activated cell sorting.45 Being the first characterized member of the tetraspanin family, CD63 membrane protein are abundantly expressed on most human exosomes.68,69, As such, CD63 on exosome surface presents an attractive strategy for selective isolation of exosomes from complex sample matrices. For example, a commercial exosome isolation kit has been developed based on the concept of magneto-immunocapture.70 Exosomes of high purity have been successfully isolated from pre-enrich exosomes. On the other hand, this kit cannot be used to directly fish out exosomes from pristine samples and the success of exosome isolation largely depends on the quality the pre-enriched exosomes. In a similar attempt, exosome isolation from plasma of acute myeloid leukemia (AML) patients was achieved by using magnetic microbeads coated with antibody for CD34 which is a unique biomarker of AML blasts. The isolated exosomes had the typical exosomal morphology when examined by transmission electron microscopy and their molecular profiles were comparable to their parental blasts as inspected by Western blot. The isolated exosomes also retained their biological activity to mediate immune suppression. These findings suggested that the blast-derived exosomes might be useful in diagnosis and prognosis of AML in future.71 In a most recent report, Nakai are co-workers described an immunoaffinity capture procedure for isolating extracellular vesicles and exosomes in particular by leveraging on an exosome binding molecule - Tim4 protein.72 It has been demonstrated that Tim4 exhibits Ca2+-dependent binding to phosphatidylserine. The specific interaction between Tim4 protein molecules immobilized on magnetic beads and phosphatidylserine molecules on the surface of exosomes efficiently isolates the exosomes from sample matrices. Moreover, the release of the captured exosomes from the magnetic beads can be conveniently accomplished by removing Ca2+ with the addition of a complexing agent. It was reported that better quality and purity of exosomes are successfully isolated from cell culture media and biofluids than those obtained using other techniques.72 Furthermore, a detailed investigation suggested that immunoaffinity capture is the superior strategy to isolate exosomes from colon cancer cell culture media as compared to ultracentrifugation and density gradient ultracentrifugation.73 Again, magnetic beads coated with antibodies targeting the epithelial cell adhesion molecule (CD326), which is over expressed on tumor exosomes, were utilized. Electron microscopic and Western blot examinations were positive for exosomal morphology and biomarkers, and mass spectrometry was employed to reaffirm the success of the isolation of exosomes.65 Moreover, the application of immunoaffinity capture in cancer diagnosis was attempted with magnetically activated cell sorting.56 Serum samples from normal controls, patients with benign diseases, and patients with ovarian cancer were incubated with magnetic beads coated with anti-epithelial cell adhesion molecules, previously shown to be expressed by exosomes from epithelial tumors. Transmission electron microscopic tests revealed vesicular structures characteristic of exosomes. The exosomal nature was further confirmed by Western blot. More importantly, exosomal miRNAs from ovarian cancer patients exhibited distinctly different profiles from those observed in patients with benign diseases, while exosomal miRNAs could not be detected in the normal controls. These data suggested that exosomal miRNA profiles could potentially be used as diagnostic biomarkers for ovarian cancer.56 Compared to the microplate-based approach, immunoaffinity capture using magnetic beads has a higher capture efficiency and better sensitivity by virtue of the larger surface area and a near homogeneous capturing process. Additionally, the magnetic bead-based approaches do not impose the upper limit to the starting sample volume and can be scaled up or down without any difficulty.
To significantly enhance its capacity, immunoaffinity capture was hyphenated to mass spectrometry - mass spectrometric immunoassay.74 To capture exosomes, antibodies were immobilized onto highly porous monolithic silica micropipette tips. CD9 was chosen as antigen because it is abundantly expressed on the surface of exosomes derived from diverse origins. An automated multichannel pipette system allowed the isolation of exosomes from 12 serum samples simultaneously within 10 min. This technique was applied to proteome-wide mass spectrometric profiling of lung cancer and identified CD91 as a biomarker.
To maximize its high specificity, exosomes isolated by other techniques can be further purified by immunoaffinity capture. For example, magnetically activated cell sorting using anti-epithelial cell adhesion molecules was employed to purify tumor exosomes isolated from plasma samples of lung cancer patients by SEC. RNA was extracted from the isolated exosomes and used for miRNA profiling by microarrays. The results indicated that exosomal miRNAs might be useful for screening lung cancer.75 Another example combined differential ultracentrifugation, ultrafiltration, and immunoaffinity capture to isolate exosomes from colorectal cancer cell culture media.76 After differential ultracentrifugation and ultrafiltration isolation, immuno-electron microscopy and Western blot confirmed the identity of the isolated exosomes. Then, the exosomes were purified by immunoaffinity capture using colon epithelial cell-specific A33 antibody-coated magnetic beads. Proteomic analysis identified various proteins in colon cancer as potential diagnostic biomarkers. Likewise, exosomes from human B cell culture media were first purified by differential ultracentrifugation and sucrose density gradient ultracentrifugation. As the final purification step, exosomes were captured onto magnetic beads coated with anti-major histocompatibility complex class II antibody.77
2.4 Exosome precipitation
By altering their solubility or dispersibility, exosomes can be settled out of biological fluids. For this purpose, water-excluding polymers such as polyethylene glycol (PEG) is engaged.53 The water-excluding polymers tie up water molecules and force less soluble components out of solution.53,68 Generally, samples are incubated with a precipitation solution containing PEG with a molecular weight of 8000 Da.54 After incubation at 4 °C overnight, the precipitate containing exosomes is isolated by means of either low speed centrifugation or filtration.53 Exosome precipitation is easy to use and does not require any specialized equipment.54 This allows easy integration into clinical usage by exploiting existing technologies, and is scalable for large sample sizes.
Currently, several exosome precipitation kits are commercially available and some of the kits are compatible with body fluids including serum, plasma, ascites, urine, cerebrospinal fluid, and culture medium.78 Before carrying out precipitation, samples need to be first pre-cleaned from cells and cellular debris. It was shown that urinary exosome precipitation with those kits achieves the highest yield compared to differential ultracentrifugation and the nanomembrane concentrators. And the highest quantities of miRNAs and mRNAs were extracted for their subsequent profiling analysis.79 The polymeric networks formed by Tamm-Horsfall protein in urine are first reduced by dithiothreitol to alleviate possible trapping of exosomes during the initial urine pre-processing by low speed centrifugation, Afterwards, the exosomes can be quantified by CD9 ELISA. The purity of exosomal proteins can be evaluated by Western blot and RNA quantified by qRT-PCR.80,81 Some total exosome isolation kits developed offer reagents for cell culture medium, urine, serum, and other body fluids like cerebrospinal fluid, ascitic fluid, amniotic fluid, milk, and saliva, as well as a kit for plasma.82 Total RNA and protein can then be purified using a total exosomal RNA and protein isolation kit. The total exosomal RNA and protein isolation kit has enabled the identification of three potential exosomal miRNA biomarkers for prostate cancer.83 Exosomes were sized and quantified by means of nanoparticle tracking analysis. The isolated exosomal miRNA was further analyzed by qRT-PCR. A big disadvantage of the polymer-based exosome precipitation is the co-precipitation of other non-exosome contaminants, such as proteins and polymeric materials.45 These issues bring about pre- and post-isolation steps. The pre-isolation step removes subcellular particles such as lipoproteins, while the post-isolation step removes the polymeric materials using a Sephadex G-25 column.
Lectins are sugar-binding and cell-agglutinating proteins. Several lectins with high affinity towards saccharide residues on the surface of urinary exosomes have been identified. Hence, Lectin-induced exosome agglutination was explored for urinary exosome isolation.84 Atomic force microscopic examinations successfully identified surface-attached conglomerates of exosomes. The size distribution of the isolated exosomes was analyzed by dynamic light scattering. Proteins released from the isolated exosomes were evaluated by Western blot. Exosomal RNAs were extracted and miRNAs in the extract were quantified by means of qRT-PCR. It was found that several miRNAs are significantly expressed in patients with prostate cancer, thus suggesting that miRNA profiles of urinary exosomes isolated by the lectin induced agglutination may be potentially valuable for prostate cancer diagnosis.50
2.5 Microfluidics-based isolation techniques
The fast advances of microfabrication technology have offered a rare opportunity for the fabrication of microfluidics-based devices to rapidly and efficiently isolate exosomes, tapping on both the physical and biochemical properties of exosomes at microscales. In addition to the usual approaches like size, density, and immunoaffinity, innovative sorting mechanisms such as acoustic,85 electrophoretic and electromagnetic manipulations86 can be implemented. With the use of such devices, significant reductions in sample volume, reagent consumption, and isolation time are expected.
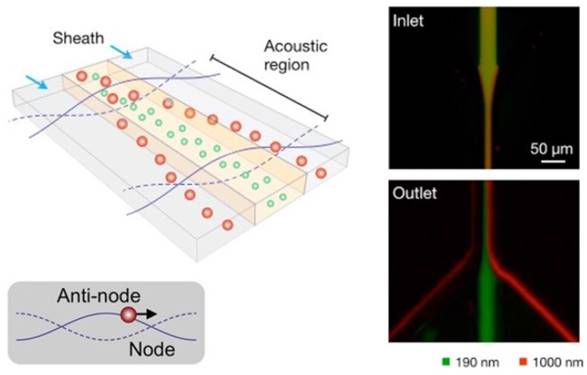
One such example was reported by Lee et al.85 As depicted in Figure 5, an acoustic nanofilter uses ultrasound standing waves to separate particulate constituents in a sample according to their size and density. Larger particles experience stronger radiation forces and migrate faster towards the pressure nodes. This in-flow size-fractionation technique was applied to cell culture media and red blood cell products.85
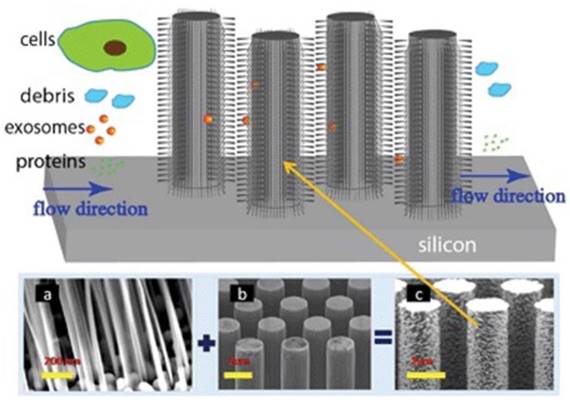
To effectively utilize the size difference between exosomes and all other extracellular vesicles and cellular debris, Wang and colleagues fabricated a porous silicon nanowire-on-micropillar structure which is made up of ciliated micropillars (Figure 6).87 This fabricated microfluidic device preferentially traps exosomes with diameters between 40 and 100 nm, while filters out proteins, other extracellular vesicles, and cellular debris. A retention rate of 60% was obtained when tested with 83-nm lipid vesicles in a sample volume up to 30 μL, whereas the retention rate of larger vesicles (500 nm) was only 10%. In addition, the trapped exosomes were recovered intact by dissolving the porous silicon nanowires in PBS buffer.87 To work directly with whole blood, Davies and co-workers tried to collect exosomes by sieving all other constituents in a whole blood sample through a nanoporous membrane in a microfluidic device. The whole blood sample is driven through the membrane by pressure or electrophoresis.86 Although the collection time was relatively short, its low collection efficiency of ~2% demands substantially more efforts.
Schematic depiction of the acoustic nanofilter (left). Acoustic radiation transported microvesicles to nodes. Larger ones move faster and are removed by sheath flow at the node region, while smaller ones are retained by the center flow. Smaller (green) and larger (red) fluorescent particles exit to the center and the side outlets, respectively (Right). (Reproduced with permission from reference 85)
Ciliated nanowire-on-micropillar. Before getting into the micropillars, cells are removed. The nanowires trap exosomes. (a) ciliated nanowires + (b) micropillars = (c) ciliated nanowire-on-micropillars. (Reproduced with permission from reference 87).
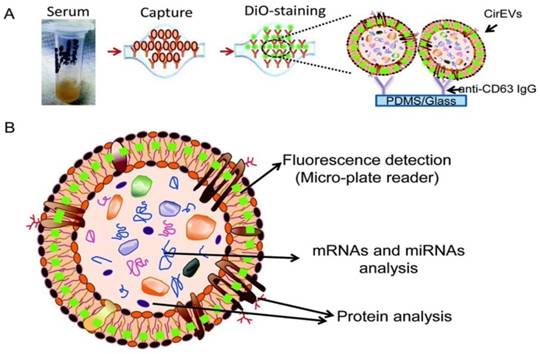
To substantially enhance the specificity and introduce exosome subtyping capability, Chen and co-workers first attempted to integrate immunoaffinity capture with a microfluidic chip for the isolation of specific exosomes.88 Similar to the conventional immunoaffinity capture approaches discussed earlier, at the heart of the exosome isolation lies the specific interaction between membrane-bound proteins (antigens) and their antibodies immobilized on the chip, thus allowing the isolation of specific exosomes from all other extracellular vesicles and constituents of a sample. Additional functionalities built on the chip enabled in situ characterization and nucleic acid extraction. As little as 400 μL of serum was needed with a run time of 60 min to complete one test. Along this direction, a commercial product - ExoChip is now available. ExoChip is an immunochip functionalized with anti-CD63, a commonly overexpressed antigen in exosomes. A fluorescent carbocyanine dye (DiO) specifically stains exosomes. Quantification was performed using a plate reader. ExoChip isolates exosomes with intact RNA for exosomal miRNA profiling. The experimental strategy using ExoChip is summarized in Figure 7.89 It was observed that significantly more exosomes from serum of pancreatic cancer patients are captured.89 Previous reports also showed increased secretion of exosomes in cancer patients. The exosomal identity was confirmed by means of Western blot.
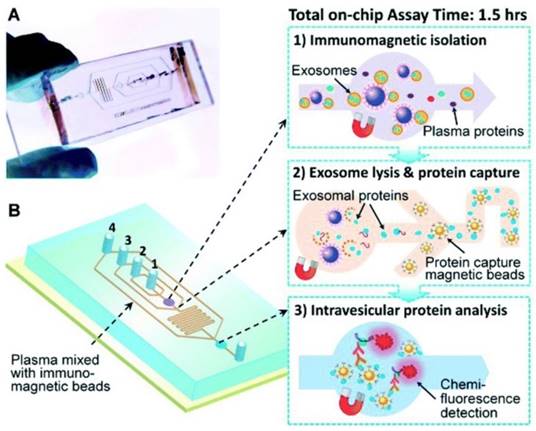
Another immunochip has integrated exosome isolation with enrichment, chemical lysis, protein immunoprecipitation, and chemiluminescent sandwich immunoassay with a much reduced sample volume of 30 μL of plasma and a run time under 100 min.90 As illustrated in Figure 8, in the first chamber, a plasma sample is mixed with antibody-labeled magnetic beads. Afterwards, a lysis buffer is added. In the second chamber, antibody-labeled magnetic beads are added to the lysate to capture intravesicular proteins. Detection antibodies and chemiluminescent reagents are then introduced.90
3. Comparison and Challenges of Exosome Isolation Techniques
Over the past decade, impressive progress has been made in the development of exosome isolation techniques and encouraging results have been obtained in decoding the mystery of exosomes. Meanwhile, it has also been proven that it is challenging to rapidly and efficiently isolate exosomes owing largely to the complexity of biological samples, complications from other extracellular vesicles due to considerable overlap of their physicochemical and biochemical properties, and the heterogeneity of exosome themselves. For example, although differential ultracentrifugation is currently considered as the gold standard of exosome isolation, in addition to its high workloads, exosomes isolated by using differential ultracentrifugation often contain proteins and lipoproteins. While improvements are achievable by hyphenating different isolation techniques like the hyphenation of ultracentrifugation and immunoaffinity capture tapping on the assets offered by both physical and biochemical worlds, one has to takes into account the additional workloads and cost.
Experimental strategy using ExoChip. (A) Serum is flowed through a CD63-Ab coated ExoChip. The captured exosomes are stained with membrane specific dye (DiO). (B) Fluorescently stained exosomes are measured using a microplate reader and exosomal contents are analyzed using Western blot for protein and RT-PCR or a microarray for RNA. (Reproduced with permission from reference 89)
Integrated microfluidic exosome analysis. (A) Chip with microchannel and (B) workflow: immunomagnetic isolation, chemical lysis and intravesicular protein analysis. #1-4 are the inlets for exosome capture beads, washing/lysis buffer, protein capture beads, and ELISA reagents, respectively. (Reproduced with permission from reference 90)
Likewise, ultrafiltration is not without its limitations despite the fact that it is a rather popular exosome isolation technique. For instance, ultrafiltration can suffer from clogging and vesicle trapping,91 thus leading to reduce lifetime of the membranes and low isolation efficiency. Exosomes can also attach themselves onto the membranes and become unavailable for downstream analyses,92 thereby resulting in low yields and sometimes erroneous interpretations of testing results. In addition, the complexity shared by size-based isolation of exosomes is the presence of a large number of nanoparticles (some non-vesicular) with the same size as exosomes.53 SEC could potentially yield highly purified exosomes, but dedicated equipment is required and it is not trivial to scale up. Because SEC is typically performed using gravity flow, vesicle structure and integrity largely remain intact and the biological activity of exosomes is preserved. Moreover, SEC has excellent reproducibility. However, its long run time limits its scalability for high throughput applications.53
Immunoaffinity capture is an excellent platform for selectively isolating exosomes of a specific origin or even subpopulations of exosomes. However, as a young field, the best exosomal tags are still to be established.53 Because only a subset of exosomes expressing the antibody-recognized protein is captured, lower yields are usually obtained but with much higher purity than those isolated based on the physical properties of exosomes.54 Additionally, underestimations and false negatives may arise from tumor heterogeneity in antigen expression and antigen modulations as tumor progresses. Furthermore, the antigenic epitope may be blocked or masked.52
Although exosome precipitation is straightforward and the isolation of exosomes can be accomplished in one step, apart from varied yields, tedious sample preparation and cleanup procedures and the lack of a proper selective isolation mechanism inevitably compromise the purity of the isolated exosomes, thereby impairing downstream analysis. For instance, co-precipitations of exosomes with other cell constituents, particularly other extracellular vesicles, protein aggregates, or even highly abundant proteins have been observed in several biofluids like plasma and serum.93,94 Moreover, the varied viscosity and sample matrix demand different stringency criteria of exosome precipitation, thus seriously jeopardizing the standardization of the precipitation protocols.
Despite the impressive progress made, none of the first generation microfluidic devices is ready to be translated to clinical trials since they are obstructed by issues such as scalability, validation, and standardization. Additionally, some of the devices require time-consuming sample pretreatments and others designed to work with pristine samples have very low isolation efficiency. So far, all microfluidic devices developed employ a single exosome sorting mechanism, thereby causing low yield or specificity. Also, their low processing capacity may impede downstream analysis because of insufficient amounts of proteins and nucleic acids in the isolated exosomes.
As such, existing exosome isolation techniques, albeit with many improvements from the past decade, have presented a new set of challenges to researchers in this field. A list of advantages and disadvantages of the exosome isolation techniques is summarized in Table 1.
Existing techniques to isolate exosomes do not have a one-size-fits-all model. Hyphenation of different isolation techniques has been raised in 2012, but a common consensus has not yet achieved.96 Since each isolate technique is based on a unique property/characteristic of exosome themselves like density, size, or immunoaffinity, low to moderate purities of the isolated exosomes are often observed. On the other hand, hyphenation of these techniques alludes to higher costs, more man hours spent on technical training for complicated techniques, and more separation steps, thus potentially resulting in greater error rates and lower recoveries in both pre- and post-analysis. The lower recoveries may adversely affect down-stream proteomic and genomic analysis and more seriously lead to erroneous diagnosis in clinical applications. Similarly, this is also the case if there is a need to isolate a unique fraction from a crude exosome extract for research/clinical treatment of specific diseases. Heterogeneity among exosomes isolated from the same parental cell type has been reported.97,98 This demands for a multiplex system for screening multiple exosome types isolated from the same origin/parent cell type, which could be a plausible direction in identifying subpopulations within the isolated exosome pool.
Another crucial challenge that should be tackled is to integrate the exosome isolation techniques to downstream analysis to alleviate the need to deal with exosome isolation and analysis separately. With integration, less time and fewer steps are required to run exosome assays, hence greatly improving the efficiency and the reliability of exosome isolation and analysis.
Researchers also face additional resistance to changes which stem from increasing complexity due to a lack of knowledge in handling new exosome isolation techniques. One example of existing techniques is the immunoaffinity capture-based isolation technique, which requires multiple and elaborate steps in sample preparation. This incurs time-consuming and tedious procedures, making the isolation process more prone to errors - both human and technical in nature. Additional barriers include proprietary rights of new exosome isolation techniques, which imply that the knowledge behind the techniques is not fully disclosed to the general scientific community. This may prevent the growth of knowledge and the understanding of existing and future exosome isolation techniques.
Comparison of exosome isolation techniques.
| Isolation Technique | Isolation principle | Potential Advantage | Potential Disadvantage |
|---|---|---|---|
| Ultracentrifugation- based techniques | Density, size, and shape based sequential separations of particulate constituents and solutes | Reduced cost and contamination risks with separation reagents, Large sample capacity and yields large amounts of exosomes | High equipment cost, cumbersome,, long run time, and labor intensive low portability - not available at point-of-care, high speed centrifugation may damage exosomes thus impeding downstream analysis.95 |
| Size-based techniques | Exosome isolation is exclusively based on the size difference between exosomes and other particulate constituents | Ultrafiltration: Fast, does not require special equipment, good portability, direct RNA extraction possible. SEC: high-purity exosomes, gravity flow preserves the integrity and biological activity; superior reproducibility, moderate sample capacity. | Ultrafiltration: low equipment cost, moderate purity of isolated exosomes, shear stress induced deterioration, possibility of clogging and vesicle trapping, exosomes loss due to attaching to the membranes. SEC: Moderate equipment cost, requires dedicated equipment, not trivial to scale up, long run time. |
| Exosome Precipitation | Altering the solubility or dispersibility or exosomes by the use of water-excluding polymers | Easy to use, does not require specialized equipment, large and scalable sample capacity | Co-precipitation of other non-exosomal contaminants like proteins and polymeric materials. Long run time, Requires pre-and post-cleanup. |
| Immunoaffinity capture-based techniques | Exosome fishing based on specific interaction between membrane-bound antigens (receptors) of exosomes and immobilized antibodies (ligands) | Excellent for the isolation of specific exosomes, Highly purified exosomes - much better than those isolated by other techniques, high possibility of subtyping. | High reagent cost, exosome tags need to be established, low capacity and low yields, only works with cell-free samples, tumor heterogeneity hampers immune recognition, antigenic epitope may be blocked or masked.54 |
| Microfluidics- based techniques | Microscale isolation based on a variety of properties of exosomes like immunoaffinity, size, and density. | Fast, low cost, portable, easy automation and integration, high portability. | Lack of standardization and large scale tests on clinical samples, lack of method validation, moderate to low sample capacity. |
Recent technological progress in exosome isolation techniques has increased the efficiency of exosome isolation, but users should be reminded of inherent advantages and disadvantages of each technique. Co-isolation of contaminating extracellular vesicles along with exosomes can result in false conclusions on proteomic or genomic interpretation of isolated exosomes. A selective, reproducible, robust, and high-throughput isolation technique is critical to meet the demands of the explosive activities in exosome research. The choice of eventual technique largely depends on biological samples to be tested, purity required, speed and cost, as well as integration with downstream proteomic or sequencing analysis. With existing exosome isolation techniques, a large percentage is deployed for the purpose of basic research. This is because translational technology from bench to bedside usually requires reasonable scalability, throughput and thorough validation in screening a large number of clinical samples. Future direction of exosome research should also take a more translational approach towards applications from bench to bedside and home diagnostics and the success of which will definitely be a great leap towards improving human health.
4. Conclusions and Outlooks
The explosive activities in exosome research have slowly evolved to meet the rapid developments in molecular medicine. For instance, the discovery of genetic materials such as miRNAs in exosomes has helped to shape the techniques for miRNA analysis. It has also helped to shape the field of therapeutics where exosomes are not only themselves therapeutic agents, but are also vehicles for targeted drug delivery and therapy. Nonetheless, significant and impactful discovery in the field of exosome research will only be possible with the development of highly efficient exosome isolation techniques. A careful selection of isolation techniques catered to specific biological samples and types of cargoes to be screened will improve the quality of the exosomes isolated and the validity of the results. Moreover, by hyphenating the developed exosome isolation techniques, exosome isolation techniques have the potential of going wide, allowing for the isolation of exosomes from many types of matrices by a single technique. By coupling to various exosome quantification techniques, they also have the potential of going deep, allowing for a multiplexed system that is capable of selective isolation of a specific exosome subtype from a heterogeneous exosome pool and performing exosome characterization and analysis.
To meaningfully assist the understanding exosomes and their functionalities, research efforts should be focused on the development of exosome isolation and detection techniques and devices that are capable of not only isolating, subtyping, and quantifying the total population of exosomes, but also analyzing the contents of exosomes to facilitate both basic research and clinical applications. The developed techniques and devices should also be thoroughly evaluated with a sufficiently large population of clinical samples for their robustness, sensitivity, and selectivity in order to be clinical relevant. In addition, a critical role that exosomes play is in cell-cell communication. Knowledge of the molecular specificity of exosomes will be instrumental in study of their pathogenic roles and to their potential use as biomarkers for early detection of cancer since early detection is vital in improving the survival rate of cancer patients. Usually, diagnosis and monitoring of solid tumors require biopsies. In addition to their invasive and cumbersome procedures, biopsies have potentially detrimental effect on stimulating cancer progression/metastases.99 It has been concurred that direct isolation of circulating nucleic acids (miRNA and DNA) and circulating tumor cells from blood or serum for cancer diagnosis may not be as feasible as sought owning to their low abundance.60 Therefore, exosomes signify an exciting new avenue of diagnosis and treatment. Abnormal levels of exosomal biomarkers likely indicate the presence of cancer or even cancer of a more advanced stage.100 As a group of promising biomarkers, the analysis of exosomes in blood may eventually offer a minimally invasive route for cancer diagnosis, prognosis, and therapy. However, much work needs to be done to identify and validate exosomal biomarkers to be utilized in cancer diagnosis and therapy.
The demands of clinical applications involving diagnostics and therapeutics such as low cost, reliability, and speed can eventually be met with modifications to existing technologies for improved scalability. Adaptations can be accomplished with necessary modifications of current exosome isolation techniques, such as ultrafiltration, which has exhibited great potential among the exosome isolation techniques for handling and analyzing large batches of exosome isolation from human blood and plasma. More importantly, the occurrence of exosomes in easily available body fluids such as urine and saliva will pave the way for non-invasive modes of exosome isolation and analysis in clinical settings. With adequate research, exosome isolation and analysis may be promising in replacing conventional invasive modes of body fluid collection such as blood and cerebrospinal fluid. The discovery of exosomes has opened a huge and untapped market for home-based diagnostics using exosomes. Along with the complete decoding of exosomes and their functionalities, more exciting applications of exosomes especially in clinical practice are expected in the near future.
Acknowledgements
This work is supported by the Ministry of Education under the grant MOE-2014-T2-081.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983;97:329-339
2. Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967-978
3. Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 1985;101:942-948
4. Harding CV, Heuser JE, Stahl PD. Exosomes: looking back three decades and into the future. J. Cell Biol. 2013;200:367-371
5. Johnstone RM, Bianchini A, Teng K. Reticulocyte maturation and exosome release: transferrin receptor containing exosomes shows multiple plasma membrane functions. Blood. 1989;74:1844-1851
6. Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43-51
7. Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25:364-372
8. Lobb RJ, Becker M, Wen SW, Wong CSF, Wiegmans AP, Leimgruber A, Möller A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles. 2015;4:27031
9. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013;200:373-383
10. Bellingham SA, Guo BB, Coleman BM, Hill AF. Exosomes: vehicles for the transfer of toxic proteins associated with neurodegenerative diseases? Front. Physiol. 2012;3:124
11. Laulagnier K, Motta C, Hamdi S, Roy S, Fauvelle F, Pageaux JF, Record M. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem. J. 2004;380:161-171
12. Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol. Lett. 2006;107:102-108
13. Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J. Proteomics. 2010;73:1907-1920
14. Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29:341-345
15. Cooper JM, Wiklander PB, Nordin JZ, Al-Shawi R, Wood MJ, Vithlani M, Alvarez-Erviti L. Systemic exosome siRNA delivery reduced alpha-synuclein aggregates in brains of transgenic Mmice. Movement Disord. 2014;29:1476-1485
16. Conde-Vancells J, Rodriguez-Suarez E, Embade N, Gil D, Matthiesen R, Valle M, Falcon-Perez JM. Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J. Proteome Res. 2008;7:5157-5166
17. Subra C, Grand D, Laulagnier K, Stella A, Lambeau G, Paillasse M, Poirot M. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J. Lipid Res. 2010;51:2105-2120
18. Jenjaroenpun P, Kremenska Y, Nair VM, Kremenskoy M, Joseph B, Kurochkin IV. Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. Peer J. 2013;1:e201
19. Lasser C, Eldh M, Lotvall J. Isolation and characterization of RNA-containing exosomes. JoVE. 2012;59:e3037
20. Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Kohli M. Characterization of human plasma-derived exosome RNAs by deep sequencing. BMC genomics. 2013;14:319
21. Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function, Nat. Rev. Immunol. 2002;2:569-579
22. Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996;183:1161-1172
23. Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza D, Amigorena S. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat. Med. 1998;4:594-600
24. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654-659
25. Cereghetti DM, Lee PP. Tumor-derived exosomes contain microRNAs with immunological function: Implications for a novel immunosuppression mechanism. Microrna. 2014;2:194-204
26. Bobrie A, Colombo M, Raposo G, Thery C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011;12:1659-1668
27. Janowska-Wieczorek A, Wysoczynski M, Kijowski J, Marquez-Curtis L, Machalinski B, Ratajczak J, Ratajczak M. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int. J. Cancer. 2005;113:752-760
28. Li W, Mu D, Tian F, Hu Y, Jiang T, Han Y, Li X. Exosomes derived from Rab27a overexpressing tumor cells elicit efficient induction of antitumor immunity. Mol. Med. Rep. 2013;8:1876-1882
29. van Balkom BW, de Jong OG, Smits M, Brummelman J, den Ouden K, de Bree PM, Verhaar MC. Endothelial cells require miR-214 to secrete exosomes that suppress senescence and induce angiogenesis in human and mouse endothelial cells. Blood. 2013;121:3997-4006
30. Xu D, Tahara H. The role of exosomes and microRNAs in senescence and aging. Adv. Drug Deliv. Rev. 2013;65:368-375
31. Gutzeit C, Nagy N, Gentile M, Lyberg K, Gumz J, Vallhov H, Scheynius A. Exosomes derived from Burkitt's lymphoma cell lines induce proliferation, differentiation, and class-switch recombination in B cells. J. Immunol. 2014;192:5852-5862
32. Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70:9621-9630
33. Denzer K, Kleijmeer MJ, Heijnen HF, Stoorvogel W, Geuze HJ. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J. Cell Sci. 2000;113:3365-3374
34. Stahl PD, Barbieri MA. Multivesicular bodies and multivesicular endosomes: the "ins and outs" of endosomal traffic. Sci. STKE. 2002;2002:32
35. Ogorevc E, Kralj-Iglic V, Veranic P. The role of extracellular vesicles in phenotypic cancer transformation. Radiol. Oncol. 2013;47:197-205
36. Schorey JS, Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008;9:871-881
37. Konadu KA, Huang MB, Roth W, Armstrong W, Powell M, Villinger F, Bond V. Isolation of exosomes from the plasma of HIV-1 positive individuals. JoVE. 2016;107:e53495
38. Milane L, Singh A, Mattheolabakis G, Suresh M, Amiji MM. Exosome mediated communication within the tumor microenvironment. J. Control Release. 2015;219:278-294
39. Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Curry WT, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470-1476
40. Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, Reissfelder C. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177-182
41. Corrado C, Raimondo S, Chiesi A, Ciccia F, de Leo G, Alessandro R. Exosomes as intercellular signaling organelles involved in health and disease: Basic science and clinical applications. Int. J. Mol. Sci. 2013;14:5338-5366
42. Sharma A, Khatun Z, Shiras A. Tumor exosomes: cellular postmen of cancer diagnosis and personalized therapy. Nanomedicine. 2016;11:421-437
43. Khatun Z, Bhat A, Sharma S, Sharma A. Elucidating diversity of exosomes: biophysical and molecular characterization methods. Nanomedicine. 2016;11:2359-2377
44. Garrett HR, Grisham CM. Biochemistry, 5th ed. Belmont, CA: Brooks/Cole, Cengage Learning. 2013:111
45. Zarovni N, Corrado A, Guazzi P, Zocco D, Lari E, Radano G, Chiesi A. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods. 2015;87:46-58
46. Rechavi O, Erlich Y, Amram H, Flomenblit L, Karginov FV, Goldstein I, Kloog Y. Cell contact-dependent acquisition of cellular and viral nonautonomously encoded small RNAs. Genes Dev. 2009;23:1971-1979
47. Yamashita T, Takahashi Y, Nishikawa M, Takakura Y. Effect of exosome isolation methods on physicochemical properties of exosomes and clearance of exosomes from the blood circulation. Eur. J. Pharm. 2016;98:1-8
48. Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006;3:3-22
49. Miranda KC, Bond DT, Levin JZ, Adiconis X, Sivachenko A, Russ C, Russo LM. Massively parallel sequencing of human urinary exosome/microvesicle RNA reveals a predominance of non-coding RNA. PloS ONE. 2014;9:e96094
50. Grapp M, Wrede A, Schweizer M, Hüwel S, Galla HJ, Snaidero N, Gärtner J. Choroid plexus transcytosis and exosome shuttling deliver folate into brain parenchyma. Nat. Commun. 2013;4:2123
51. Booth AM, Fang Y. Fallon JK, Yang JM, Hildreth JE, Gould SJ. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J. Cell Biol. 2006;172:932-935
52. Quintana JF, Makepeace BL, Babayan SA, Ivens A, Pfarr KM, Blaxter M, Tanya VN. Extracellular Onchocerca-derived small RNAs in host nodules and blood. Parasit. Vectors. 2015;8:1
53. Zeringer E, Barta T, Li M, Vlassov AV. Strategies for isolation of exosomes. Cold Spring Harb. Protoc. 2015:319-323
54. Batrakova EV, Kim MS. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J. Control Release. 2015;219:396-405
55. Cheruvanky A, Zhou H, Pisitkun T, Kopp JB, Knepper MA, Yuen PS, Star RA. Rapid isolation of urinary exosome biomarkers using a nanomembrane ultrafiltration concentrator. Am. J. Physiol. Renal Physiol. 2007;292:F1657-1661
56. Bioo Scientific Corporation, ExoMir™ Kit, 2016. http://www.biooscientific.com/exosome-analysis/exomir-kit
57. Channavajjhala SK, Rossato M, Morandini F, Castagna A, Pizzolo F, Bazzoni F, Olivieri O. Optimizing the purification and analysis of miRNAs from urinary exosomes. Clin. Chem. Lab Med. 2014;52:345-354
58. Escudier B, Dorval T, Chaput N, André F, Caby MP, Novault S, Boccaccio C. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of the first phase I clinical trial. J. Transl. Med. 2005;3:1-13
59. Morse MA, Garst J, Osada T, Khan S, Hobeika A, Clay TM, Hsu DH. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J. Transl. Med. 2005;3:1-8
60. Heinemann ML, Ilmer M, Silva LP, Hawke DH, Recio A, Vorontsova MA, Vykoukal J. Benchtop isolation and characterization of functional exosomes by sequential filtration. J. Chromatogr. A. 2014;1371:125-135
61. Feng Y, Huang W, Wani M, Yu X, Ashraf M. Ischemic preconditioning potentiates the protective effect of stem cells through secretion of exosomes by targeting Mecp2 via miR-22. PloS ONE. 2014;9:e88685
62. Lai RC, Arslan F, Lee MM, Sze NSK, Choo A, Chen TS, Pasterkamp G. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010;4:214-222
63. Taylor DD, Gercel-Taylor C. Tumour-derived exosomes and their role in cancer-associated T-cell signalling defects. Br. J. Cancer. 2005;92:305-311
64. Rood IM, Deegens JK, Merchant ML, Tamboer WP, Wilkey DW, Wetzels JF, Klein JB. Comparison of three methods for isolation of urinary microvesicles to identify biomarkers of nephrotic syndrome. Kidney Int. 2010;78:810-816
65. Kang D, Oh S, Ahn SM, Lee BH, Moon MH, Proteomic analysis of exosomes from human neural stem cells by flow field-flow fractionation, nanoflow liquid chromatography-tandem mass spectrometry. J. Proteome Res. 2008;7:3475-3480
66. Petersen KE, Manangon E, Hood JL, Wickline SA, Fernandez DP, Johnson WP, Gale BK. A review of exosome separation techniques and characterization of B16-F10 mouse melanoma exosomes with AF4-UV-MALS-DLS-TEM. Anal. Bioanal. Chem. 2014;406:7855-7866
67. Musante L, Tataruch D, Gu DF, Benito-Martin A, Calzaferri G, Aherne S, Holthofer H, Sci Rep. A Simplified Method to Recover Urinary Vesicles for Clinical Applications, and Sample Banking. Sci Rep. 2014;4:7532
68. Kim D, Nishida H, An SY, Shetty AK, Bartosh TJ, Prockop DJ. Chromatographically isolated CD63+CD81+ extracellular vesicles from mesenchymal stromal cells rescue cognitive impairments after TBI. Proc. Natl. Acad. Sci. USA. 2016;113:170-175
69. Yanez-Mo M, Barreiro O, Gordon-Alonso M, Sala-Valdes M, Sanchez-Madrid F. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 2009;19:435-446
70. https://www.thermofisher.com/order/catalog/product/10606D
71. Hong CS, Muller L, Boyiadzis M, Whiteside TL. Isolation and characterization of CD34+ blast-derived exosomes in acute myeloid leukemia. PloS ONE. 2014;9:e103310
72. Nakai W, Yoshida T, Diez D, Miyatake Y, Nishibu T, Imawaka N, Naruse K, Sadamura Y, Hanayama R. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci Rep. 2016;6:33935
73. Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, Simpson RJ. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293-304
74. Ueda K, Ishikawa N, Tatsuguchi A, Saichi N, Fuji R, Nakagawa H. Antibody-coupled monolithic silica microtips for highthroughput molecular profiling of circulating exosomes. Sci. Rep. 2014;4:6232
75. Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosome MicroRNA: A Diagnostic Marker for Lung Cancer. Clin. Lung Cancer. 2009;10:42-46
76. Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, Simpson RJ. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol. Cell Proteomics. 2010;9:197-208
77. Wubbolts R, Leckie RS, Veenhuizen PT, Schwarzmann G, Möbius W, Hoernschemeyer J, Stoorvogel W. Proteomic and biochemical analyses of human B cell-derived exosomes Potential implications for their function and multivesicular body formation. J. Biol. Chem. 2003;278:10963-10972
78. ExoQuick overview. System Biosciences Inc. https://www.systembio.com/microrna-research/exoquick-exosomes/overview
79. Alvarez ML, Khosroheidari M, Kanchi RR, DiStefano JK. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012;82:1024-1032
80. Gallo A. Alevizos I. Isolation of circulating microRNA in saliva. Methods Mol. Biol. 2013;1024:183-190
81. Ohno SI, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, Gotoh N. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol. Ther. 2013;21:185-191
82. Total exosome isolation. Thermo Fisher Scientific Inc. https://www.thermofisher.com/sg/en/home/life-science/cell-analysis/exosomes.html
83. Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MÁ, Sánchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011;2:282
84. Samsonov R, Shtam T, Burdakov V, Glotov A, Tsyrlina E, Berstein L, Malek A. Lectin-induced agglutination method of urinary exosomes isolation followed by mi-RNA analysis: Application for prostate cancer diagnostic. Prostate. 2016;76:68-79
85. Lee K, Shao H, Weissleder R, Lee H. Acoustic purification of extracellular microvesicles. ACS Nano. 2015;9:2321-2327
86. Davies RT, Kim J, Jang SC, Choi EJ, Gho YS, Park J. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab Chip. 2012;12:5202-5210
87. Wang Z, Wu HJ, Fine D, Schmulen J, Hu Y, Godin B, Liu X. Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles. Lab Chip. 2013;13:2879-2882
88. Chen C, Skog J, Hsu CH, Lessard RT, Balaj L, Wurdinger T, Carter BS, Breakefield XO, Toner M, Irimia D. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip. 2010;10:505-511
89. Kanwar SS, Dunlay CJ, Simeone DM, Nagrath S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip. 2014;14:1891-1900
90. He M, Crow J, Roth M, Zeng Y, Godwin AK. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip. 2014;14:3773-3780
91. Liga A, Vliegenthart AD, Oosthuyzen W, Dear JW, Kersaudy-Kerhoas M. Exosome isolation: a microfluidic road-map. Lab Chip. 2015;15:2388-2394
92. Xin H, Li Y, Buller B, Katakowski M, Zhang Y, Wang X, Chopp M. Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells. 2012;30:1556-1564
93. Gyorgy B, Modos K, Pallinger E, Paloczi K, Pasztoi M, Misjak P, Deli MA, Sipos A, Szalai A, Voszka I, Polgar A, Toth K, Csete M, Nagy G, Gay S, Falus A, Kittel A, Buzas EI. Detection and isolation of cell-derived microparticles are compromised by protein complexes resulting from shared biophysical parameters. Blood. 2011;117:e39-48
94. Kalra H, Adda CG, Liem M, Ang CS, Mechler A, Simpson RJ, Hulett MD, Mathivanan S. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics. 2013;13:3354-3364
95. Jeppesen DK, Hvam ML, Primdahl-Bengtson B, Boysen AT, Whitehead B, Dyrskjøt L, Ørntoft TF, Howard KA, Ostenfeld MS. Comparative analysis of discrete exosome fractions obtained by differential centrifugation J. Extracell. Vesicles. 2014;3:25011
96. Witwer KW, Buzas EI, Bemis LT, Bora A, Lässer C, Lötvall J, Thery C. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles. 2013;2:20360
97. Laulagnier K, Vincent-Schneider H, Hamdi S, Subra C, Lankar D, Record M. Characterization of exosome subpopulations from RBL-2H3 cells using fluorescent lipids. Blood Cells Mol. Dis. 2005;35:116-121
98. Smith.J Lee C, Rojalin T Carney RP, Hazari S Knudson A, Laaksonen T. Single exosome study reveals subpopulations distributed among cell lines with variability related to membrane content. J. Extracell. Vesicles. 2015;4:28533
99. Shyamala K, Girish HC, Sanjay Murgod S. Risk of tumor cell seeding through biopsy and aspiration cytology. J. Int. Soc. Prev. Community Dent. 2014;4:5-11
100. Alegre E, Zubiri L, Perez-Gracia JL, González-Cao M, Soria L, Martín-Algarra S, González A. Circulating melanoma exosomes as diagnostic and prognosis biomarkers. Clin. Chim. Acta. 2016;454:28-32
Author contact
![]() Corresponding authors: Tel: 6592-3991, 6516-3887, Fax: 6779-1691, e-mail: lipinedu.sg, chmgaozedu.sg.
Corresponding authors: Tel: 6592-3991, 6516-3887, Fax: 6779-1691, e-mail: lipinedu.sg, chmgaozedu.sg.
 Global reach, higher impact
Global reach, higher impact