13.3
Impact Factor
Theranostics 2017; 7(2):482-492. doi:10.7150/thno.16816 This issue Cite
Research Paper
Functional Genomic mRNA Profiling of Colorectal Adenomas: Identification and in vivo Validation of CD44 and Splice Variant CD44v6 as Molecular Imaging Targets
1. Department of Gastroenterology and Hepatology, University of Groningen, University Medical Centre Groningen, Groningen, the Netherlands;
2. Institute of Toxicology and Genetics, Karlsruhe Institute of Technology, Karlsruhe, Germany;
3. Amcure GmbH, Eggenstein-Leopolshafen, Germany;
4. Department of Pathology, University of Groningen, University Medical Centre Groningen, Groningen, the Netherlands;
5. Department of Medical Oncology, University of Groningen, University Medical Centre Groningen, Groningen, the Netherlands;
6. Department of Surgery, Nuclear Medicine and Molecular Imaging and Intensive Care, University of Groningen, University Medical Centre Groningen, Groningen, the Netherlands.
* These authors contributed equally and jointly supervised the work.
Received 2016-7-12; Accepted 2016-11-7; Published 2017-1-6
Abstract
Colorectal cancer (CRC) is the third leading cause of cancer-related deaths worldwide. High adenoma miss rates, especially seen in high-risk patients, demand for better endoscopic detection. By fluorescently 'highlighting' specific molecular characteristics, endoscopic molecular imaging has great potential to fulfill this need. To implement this technique effectively, target proteins that distinguish adenomas from normal tissue must be identified. In this study we applied in silico Functional Genomic mRNA (FGmRNA) profiling, which is a recently developed method that results in an enhanced view on the downstream effects of genomic alterations occurring in adenomas on gene expression levels. FGmRNA profiles of sporadic adenomas were compared to normal colon tissue to identify overexpressed genes. We validated the protein expression of the top identified genes, AXIN2, CEMIP, CD44 and JUN, in sporadic adenoma patient samples via immunohistochemistry (IHC). CD44 was identified as the most attractive target protein for imaging purposes and we proved its relevance in high-risk patients by demonstrating CD44 protein overexpression in Lynch lesions. Subsequently, we show that the epithelial splice variant CD44V6 is highly overexpressed in our patient samples and we demonstrated the feasibility of visualizing adenomas in ApcMin/+ mice in vivo by using a fluorescently labeled CD44v6 targeting peptide. In conclusion, via in silico functional genomics and ex vivo protein validation, this study identified CD44 as an attractive molecular target for both sporadic and high-risk Lynch adenomas, and demonstrates the in vivo applicability of a small peptide drug directed against splice variant CD44v6 for adenoma imaging.
Keywords: Functional Genomics, Cancer Genetics, Colorectal adenomas, Molecular Targeted Imaging, CD44 / CD44v6.
Introduction
Despite its decreasing mortality, colorectal cancer (CRC) is still the third most common cause of cancer-related death worldwide (1). The development of CRC is a multistep process, known as the adenoma-carcinoma sequence: In approximately 10-20 years, normal colonic mucosa can slowly transform via premalignant adenomatous polyps into invasive cancer (2). The adenoma-carcinoma sequence is characterized by consecutive genetic alterations, such as common 'driver' mutations in the APC gene, the proto-oncogene K-RAS, the tumor suppressor gene TP53 and activation of the Wnt-signaling pathway (3).
Early endoscopic identification of adenomas and subsequent radical removal is the keystone in breaking the adenoma-carcinoma sequence, thereby preventing CRC development. Nevertheless, early detection and radical removal are still challenging tasks due to non-specific tissue morphology. When using standard white-light endoscopy, this results in detection miss rates of up to 25% for sporadic adenomas (4). In particular flat and small lesions, often seen in high-risk populations like Lynch patients, are notoriously difficult to detect, resulting in adenoma detection miss rates of up to 50% (5). Especially in these patients, missed lesions can rapidly progress to cancer, underscoring the necessity of improving endoscopic detection strategies. Therefore, endoscopic techniques based on molecular imaging are emerging (6). Molecular imaging can improve optical discrimination of aberrant tissue by labeling specific molecular characteristics with targeted fluorescent monoclonal antibodies or peptide probes. Molecular imaging preferably targets proteins that are selectively overexpressed in dysplastic or malignant cells. Protein overexpression can be the downstream effect of genomic alterations that occur frequently within the adenoma-carcinoma sequence. Thus, by fluorescently 'highlighting' the overexpressed target proteins within dysplastic lesions, an optical molecular method is created that can serve as a 'red-flag' strategy for distinguishing dysplastic tissue in real-time during endoscopic evaluation.
Effective implementation of this molecular-based technique in gastrointestinal endoscopy requires identification of target proteins that can detect dysplasia with a high specificity. To identify relevant target proteins, DNA changes that occur in adenomas can be addressed; the difference between adenomas and normal colorectal tissue is that adenomas exhibit genomic alterations that might lead to downstream changes in gene expression levels. These changed gene expression levels, in turn, influence the downstream cellular phenotype, and by doing so, they affect protein expression levels. Nevertheless, these genomically driven changes in gene expression levels are rather subtle and usually overshadowed by non-genetic processes, such as the circadian rhythm or prosprandial metabolic influences (7). To be able to identify target proteins that distinguish adenomas from normal colorectal tissue, we therefore applied a recently introduced method called functional genomic mRNA (FGmRNA) profiling. With FGmRNA-profiling we are able to correct gene expression data for major non-genetic factors, thereby enhancing our view on the downstream effect of genomic alteration occurring in adenomas on gene expression levels (7). Subsequently, to evaluate the protein expressions levels and validate our FGmRNA-profiling results, we performed immunohistochemistry (IHC) analyses for the top five genes that were identified. In addition, we evaluated CD44 expression in Lynch patient material. Finally, we performed ApcMin/+ mice experiments evaluating the applicability of a small peptide drug directed against the CD44 splice variant v6 for targeted adenoma imaging purposes.
Material and methods
In silico functional genomics: Functional Genomic mRNA (FGmRNA) Profiling
We used FGmRNA-profiling to capture the downstream effect of genomic alterations at gene expression levels. For a detailed description of this bioinformatics method we refer to Fehrmann et al.(7) In short, by performing a principal component analysis (PCA) on expression profiles of publicly available samples, Fehrmann et al. demonstrated that a limited number of 'Transcriptional Components' (TCs) capture the major regulators of the mRNA transcriptome. Subsequently, a subset of TCs that described non-genetic regulatory factors was identified. These non-genetic TCs were used as covariates to correct microarray expression data, with which they observed that the residual expression signal (i.e. FGM profile) captures the downstream consequences of genomic alterations on gene expression levels.
In the present study, we generated FGmRNA profiles for adenomas and healthy control colorectal mucosa samples. We used microarray expression profiles from the Gene Expression Omnibus (GEO - www.ncbi.nlm.nih.gov/geo/), previously described by Heijink et al.(8) In short, we included data from existing Affymetrix intensity files (CEL files) of mRNA from adenomas and healthy control colorectal mucosa hybridized to HG-U133 plus 2.0 (Affymetrix, Santa Clara, CA, USA). Pre-processing and aggregation of CEL files was performed with Affymetrix Power Tools version 1.15.2, using apt-probe set-summarize and applying the robust multi-array average (RMA) algorithm, using the Affymetrix GeneChip Array CDF layout files (downloaded March 2014). We used the probe annotation file provided by Affymetrix (version 34). Quality control was performed using Principal Component Analysis (PCA) on the sample correlation matrix as described previously.(8)
Immunohistochemistry (IHC) details and methods.
| Target protein | Protein location | Antigen retrieval | 1st antibody | 2nd /3rd antibody |
|---|---|---|---|---|
| Axin-2 | Cytoplasm | Citrate buffer (10 mM, pH 6.0) | Abcam, ab32197 [0.9 mg/ml] (1) Rabbit-anti-human, polyclonal, Incubation: 1.8 ug/ml, 60min/RT | - GAR-HRP 1:100, - RAG-HRP 1:100, 30min/RT |
| CEMIP | Cytoplasm (ER membrane) | Citrate buffer (10 mM, pH 6.0) | Proteintech21129-1-AP [0.2 mg/ml] Rabbit-anti-human, polyclonal, Incubation: 2 μg/ml, 60min/RT | - GAR-HRP 1:100, - RAG-HRP 1:100, 30min/RT |
| CD44 | Cell membrane | Citrate buffer(10 mM, pH 6.0) | Biolegend, IM7 [0.5 mg/ml] (2)Rat-anti-human, monoclonalIncubation: 5 μg/ml, overnight/4 °C | - RARa-HRP 1:50, - GAR-HRP 1:100, 30min/RT |
| CD44v6 | Cell membrane | Citrate buffer (10 mM, pH 6.0) | Abcam ab30436 [1 mg/ml] (3) Mouse-anti-human, monoclonal Incubation: 10 μg/ml, 60min/RT | -RAM-HRP 1:50, -GAR-HRP 1:50, 30min/RT |
| Citrate buffer (10 mM, pH 6.0) | Thermofisther 9A4 [0.5mg/ml] Rat-anti-mouse, monoclonal Incubation: 2.5 μg/ml, 60min/RT | - RAR-HRP 1:100- GAR-HRP 1:10030 min/RT | ||
| C-Jun | Nucleus | Citrate buffer (10 mM, pH 6.0) | Santa Cruz (4)H-79, sc-1694 [0.2 mg/ml] Rabbit-anti-human, polyclonal Incubation: 0.8 μg/ml, 60min/RT | - GAR-HRP 1:100, - RAG-HRP,1:100, 30min/RT |
| CELF2 | NucleusCytoplasm | Varying buffers(pH6.0 - 9.0) | Abcam, ab50734 [1mg/ml]Rabbit-anti-human, polyclonal Incubation: 1 - 20 μg/mlVarying incubation times(up to 12 hours) | - GAR-HRP 1:100, - RAG-HRP,1:100, 30min/RT |
Abbreviations: RT: Room temperature; HRP: horseradish peroxidase; RARa: Rabbit-anti-Rat; GAR: goat-anti-rabbit; RAM: rabbit-anti-mouse; RAG, rabbit-anti-goat. References: (1) Winkler T et al. Wnt signaling activates Shh signaling in early postnatal intervertebral discs, and re-activates Shh signaling in old discs in the mouse. PLoS One 9:e9844 (2) NY Frank et al. ABCB5-Mediated Doxorubicin Transport and Chemoresistance in Human Malignant Melanoma. Cancer Res. 2005 May 15;65(10):4320-33. (3) T Okada et al. Coexpression of EpCAM, CD44 variant isoforms and claudin-7 in anaplastic thyroid carcinoma. PLoS One. 2014 Apr 11;9(4):e94487 (4) N Marqués et al. Regulation of protein translation and c-Jun expression by prostate tumor overexpressed 1. Oncogene. 2014 Feb 27;33(9):1124-34.
Immunohistochemistry (IHC)
IHC for FGmRNA validation-purposes was performed on formalin-fixed and paraffin-embedded (FFPE) tissue blocks containing sporadic high-grade dysplastic (HGD) adenomatous polyps (n=27) and normal adjacent colorectal mucosa (n=17). All tissue samples were derived from individuals who underwent colonoscopy between 2008 and 2013 at the University Medical Centre Groningen (UMCG, Groningen, the Netherlands). Furthermore, to evaluate CD44 expression in a high-risk CRC population, we performed a panCD44 staining on Lynch material. We collected FFPE tissue blocks derived from patients diagnosed with Lynch syndrome (LS) and who underwent surveillance colonoscopy or surgery between 1984 and 2011 at the UMCG. The LS tissue blocks contained LGD (low-grade dysplasia, n=63), HGD (n=58) or carcinoma tissue (n=32) with normal adjacent mucosa (n=52, n=42, n=21, respectively). In addition, next to the anti-panCD44 antibody, we also more selectively analyzed its epithelial splice variant CD44v6 in an extended sample-size of HGD sporadic adenomatous polyps (n=69) and normal adjacent mucosa (n=51).
From each tissue block four-micron sections were cut, mounted on amino-propylethyoxy-silan-coated glass slides, deparaffinized in xylene and rehydrated in a series of alcohol dilutions. Secondly, heat-induced antigen retrieval was conducted using a 10 mM citrate buffer (pH 6) followed by blocking of the endogenous peroxidase activity by incubation with a hydrogen-peroxide solution (3%), diluted with phosphate buffered saline (PBS). Subsequently, the primary antibodies against Axin-2, CEMIP, CELF2, CD44 (panCD44/CD44v6) and c-Jun were diluted in PBS and added as described in Table 1. Afterwards the second and third antibodies were diluted in PBS with 1% bovine serum albumin (BSA) and 1% human AB-serum and subsequently incubated, each for 30 minutes. Finally, specific staining was visualized using 3,3' diaminobenzidine (DAB) substrate chromogen solution and counterstaining of the nuclei with hematoxylin.
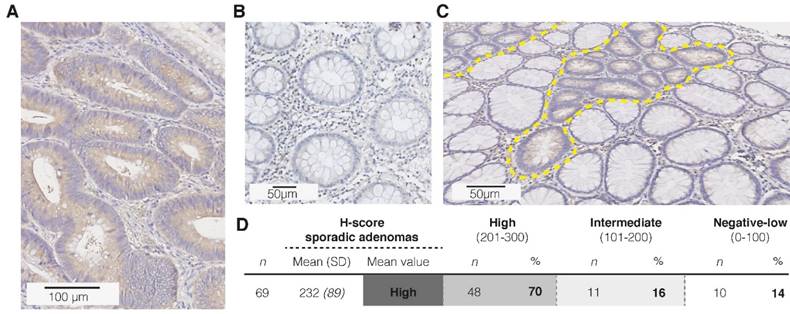
All human tissue samples were handled according to the guidelines of the University Medical Centre Groningen (UMCG) ethics board (www.ccmo.nl) and the code of conduct for responsible use of human tissue (www.federa.org). IHC staining protocols were optimized for their applicability in colorectal tissue with help of a gastrointestinal pathologist (AK). To ascertain specific binding of the antibody, positive tissue controls (colorectal cancer) and negative controls using an aspecific IgG monoclonal antibody were included in each stained sample batch. Staining intensities of the epithelial cells were independently scored visually by two separate observers (EH, JM) using a 0-3 scale, as was the fraction of cells stained per intensity. Subsequently, H-scores were generated (continuous scale: 0-300) by combining the evaluated intensity and frequency (formula used: 1x (percentage of cells weakly stained [1+] + 2x (percentage of cells moderately scored [2+]) + 3x (percentage of cells strongly stained [3+]), creating expression categories representative for overall protein expression for both adenoma and normal adjacent mucosa (0-100 = negative/low; 101-200 = intermediate; 201-300 = high) (9).
CD44v6-targeted fluorescence imaging of intestinal adenomas
To evaluate the potential of CD44v6-targeted adenoma imaging, we performed an imaging-experiment using APCMin/+ mice. This animal model is highly representative for human colorectal adenoma research as the mutation of the APC gene is a known early event in the adenoma-carcinoma sequence (10). Furthermore, this model mimics the high-risk CRC population with familial adenomatous polyposis (FAP), where the underlying germ line APC mutation causes excessive polyp formation. In order to demonstrate the usefulness of this animal model in CD44v6-targeted imaging experiments, we performed anti-CD44v6 IHC on APCMin/+ mice intestines (n = 6; see Table 1 for IHC details). All animal experiments were approved by the local Animal Welfare Committee of the Karlsruhe Institute of Technology (KIT; license number 35-9185.81/G-247/15) and carried out in accordance with the German Animal Welfare Act of 2006. We used an in-house strain of ApcMin/+ mice, which were bred and kept in a conventional animal facility, monitored three times a week and given access to standard diet and water ad libitum. Two weeks prior to the imaging experiment the animals were kept on a chlorophyll free diet.
To perform the imaging experiments, we made use of a CD44v6-targeting peptide, which previously demonstrated its applicability in a CD44v6-overexpressing preclinical cancer model (11); To accomplish fluorescence imaging, we labeled the 14-amino-acid peptides, directed against mouse CD44v6 (specific peptide; QETWFQNGWQGKN) and human CD44v6 (control peptide; KEQWFGNRWHEGYR), with a commercially available fluorophore DY-681 (Dyomics GmbH, Jena, Germany). CD44v6 peptides are species-specific and do not cross-react (11-13). By labeling the peptides with the fluorescent dye, we created tracer products (CD44v6*DY-681) that are fluorescent in the far red light spectrum (absorption max: 691nm / emission max: 708 nm).
During the experiment, 12-16 week-old ApcMin/+ mice received an intravenous (IV) tracer injection [CD44v6*DY-681, mouse-specific or control peptide (human-targeting); 1mg/kg] and were sacrificed 2 hours later. Ex vivo fluorescence imaging of the longitudinally opened small intestines was performed, using the fresh or formalin-fixed specimens. In total, six mice received the specific, mouse-targeting, tracer product. As control, six mice received the human-targeting tracer product and three mice were imaged at baseline (without tracer administration). Ex vivo white-light and fluorescence imaging was performed for the harvested organs for both 700 and 800 nm emission wavelengths, using a Pearl Impulse Small Animal Imaging System (LI-COR Biosciences, Bad Homburg, Germany). All images were acquired at a fixed distance, using the same channel settings (exposure times, laser intensities). Post-imaging processing of the files and visual fluorescence scaling, in order to accomplish adequate visual comparison, was performed using ImageJ software Vers. 1.48 (ImageJ, U.S. National Institutes of Health, Bethesda, MD, USA).
Statistical analyses
We performed a two-class comparison between FGmRNA profiles of adenomas and healthy control colorectal mucosa. The two-sample Student t-test was used to detect significant differences in FGmRNA expression. To control the false discovery rate, we performed a multivariate permutation (MVP) test with a false discovery rate (FDR) of 5% and a confidence interval (CI) of 80%. The identified FGmRNA overexpressing genes were ranked and listed according to the level of significance. For IHC analysis, the difference in staining intensities and distribution among the three LS disease stages (LGD, HGD and carcinoma) was analyzed using the non-parametric Kruskal-Wallis test (IBM SPSS 22.0.0; IBM Corporation, Armonk, NY, USA). For the statistical analyses of the IHC results P values < 0.05 were considered significant.
Results
Functional genomic mRNA profiling identifies differentially expressed genes in sporadic colorectal adenomas
FGmRNA-profiling was applied to publicly available expression profiles of 26 normal colon mucosa samples and 47 adenoma samples. After multivariate permutation testing, we identified 4 524 genes that show FGmRNA overexpression in adenoma samples compared to normal colon tissue (FDR 5%, CI 80% - Table S1).
Ex vivo validation of protein expression: AXIN2, CEMIP, CD44 and JUN
To determine whether the FGmRNA-overexpressing genes could be translated into differences at the protein level, we analyzed the downstream protein expression for the top five genes in sporadic adenomas via immunohistochemistry (IHC). Since the FGmRNA-profiling results are ranked according to their level of significance, at least the top FGmRNA-overexpressing genes should show an observable difference at the protein level. In addition, validation of the top five FGmRNA-overexpressing genes provided a complete cellular coverage, as we were able to evaluate protein expression at each cellular location (e.g. cytoplasm, ER membrane, cell surface and nucleus).
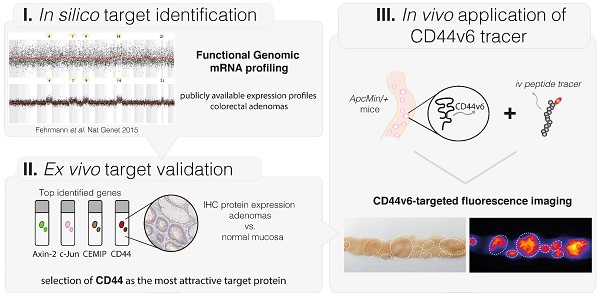
Figure 1A contains the IHC results and the representative images of the IHC stainings, showing a cytoplasmic and/or membranous staining pattern with an overall high staining intensity for the proteins Axin-2, CEMIP and CD44. For these three proteins, the majority of samples showed a positive staining of the dysplastic epithelial cells (78-96%; incl. intermediate and high staining intensities). For CD44, 85% of the sporadic adenoma tissue samples were positive (22/26), of which the vast majority (86%; 19/22) expressed a high staining intensity. It should be noted that CD44 is present on both the epithelial cells and in the stromal extracellular regions, including lymphocytes and macrophages (14). We observed that the panCD44-antibody, recognizing CD44s as well as CD44v antigens, also strongly stained the surrounding stromal tissue (Figure S1). The anti c-Jun staining shows a nuclear staining pattern, with an overall intermediate intensity score. Unfortunately, we were not able to optimize the staining of CELF2, so protein expression for this target could not be evaluated. Figure 1B demonstrates that for all four evaluated targets, Axin-2, CEMIP, CD44 and c-Jun, a significant difference in staining intensity between adenoma tissue and the adjacent normal mucosa could be observed (P < 0.001).
Target selection in sporadic adenomas: 18 possible imaging targets identified
The localization of the protein products determines whether they could be suitable as an imaging target. Cell membrane receptors and specific extracellularly secreted proteins are presumed to be the most attractive targets. This is because proteins on the cell surface or extracellular space are more easily accessible for targeting monoclonal antibodies or peptide probes, following both systemic and local administration. Furthermore, imaging targets should preferably be restricted to the adenomatous tissue, for example by anchoring or binding to membrane receptors. This enables 'highlighting' of the aberrant area and fluorescent visualization during endoscopy. Therefore, we categorized the protein products of the top fifty FGmRNA-overexpressing genes based on their subcellular localizations (Figure 2 - based on http/www.proteinatlas.org, http://www.genecards.org).
Figure 2 depicts the first 50 genes encoding proteins and their subcellular localization. By categorizing the proteins based on their localization, thus identifying cell membrane receptors and specific extracellularly secreted proteins, 18 proteins were selected that could serve as potential imaging targets, including CD44.
CD44 as a potential target in sporadic and high-risk CRC populations
Based on the degree of FGmRNA overexpression, the downstream protein upregulation and cellular membrane localization, we identified CD44 as an attractive target in sporadic adenomas. To determine whether CD44 would be of similar interest in high-risk CRC populations, we performed IHC on adenomatous and carcinomatous Lynch lesions. Figure 3A-C depicts representative images of both dysplastic and carcinomatous polyps, demonstrating the presence of CD44 throughout the complete adenoma-carcinoma sequence. The majority of the LGD, HGD and carcinoma samples showed positive epithelial staining results (Figure 3D, 46-73%; incl. intermediate and high staining intensities), with a reversed intensity-trend within the adenoma-carcinoma sequence (P = 0.006). All the adjacent normal mucosa, independent of the adenoma-carcinoma disease stage, showed weak to negative CD44 expression.
CD44v6-targeted fluorescence imaging of intestinal adenomas
It was previously established that the CD44 glycoproteins are potentially important components in CRC development (15), demonstrating tumor-initiating properties (16). In contrast to the standard CD44 isoform, expression in normal tissues is highly restricted for its v6 splice variant (17). CD44v6 has shown to be a prognostic factor in various solid cancer types, including colorectal carcinoma (18,19), and recent research demonstrated that CD44v6 expression is superior to CD44 expression, at least in respect to assessing the prognosis and clinical usefulness in sporadic CRC. Since CD44v6 is known to be functionally involved in intestinal tumorigenesis (20), and overexpression of this splice variant has been described to occur already early in the adenoma-carcinoma sequence (21), we wanted to investigated its imaging potential in precancerous intestinal lesions.
First of all, to confirm the hypothesized CD44v6 overexpression in sporadic adenomatous polyps, we performed an extended IHC analysis (n=69; Figure 4A-C). Figure 4D depicts that 86% of the samples showed indeed a positive epithelial staining (incl. intermediate and high staining intensities), of which the majority was of high intensity. Both the adjacent normal mucosa, as well as the surrounding stromal tissue, showed weak to negative results (n=52). Secondly, the membranous overexpression of CD44v6 observed in the adenomatous APCMin/+ mice crypts confirms the suitability of this specific animal model for CD44v6-targeted imaging experiments (Figure 5A).
Immunohistochemistry results of sporadic adenomas. (A) Table presenting IHC results of the four targets; the mean intensities and distribution within the different staining categories are presented using the H-score (Cohen's Kappa coefficient for inter-observer agreement: 0.485) (B) Representative images of strong staining intensities per target (brown), where the first two show a cytoplasmic staining, CD44 a membrane staining and c-Jun a staining of the nucleus. (C) Immunohistochemistry results of sporadic adenomas and their surrounding normal tissue, illustrating the significant difference in staining intensities between dysplasia and normal colon crypts per marker. Bar graphs demonstrating the differences observed in staining intensities (H-scores) between dysplasia and adjacent normal colon mucosal crypts per marker. ** P value < 0.001
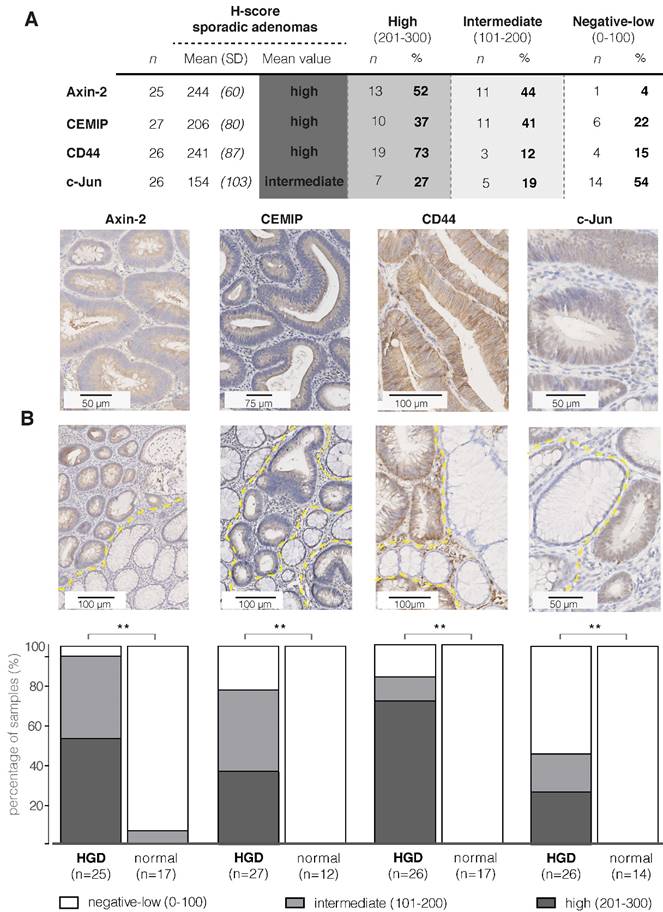
Cellular localization of protein products of the first 50 FGmRNA-overexpressing genes. The cell membrane and extracellular proteins that are restricted to the tumor site (e.g. by anchoring or binding to membrane receptors) are especially suitable as imaging targets (black circles). This results in already 18 potential imaging targets, including CD44. The top-five genes are represented in bold; genes with products at multiple locations are italicized. ER, endoplasmic reticulum.
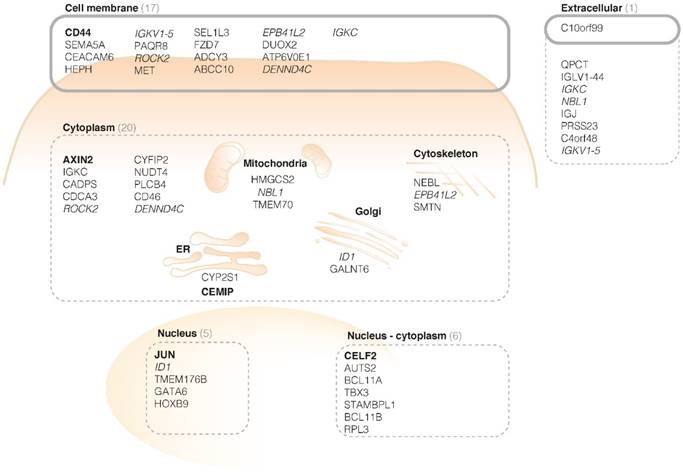
CD44 immunohistochemistry (IHC) results in high-risk Lynch lesions. Representative images of strong CD44 epithelial staining intensities (brown) in respectively LGD (A), HGD (B) and carcinoma samples (C). The high staining intensity in the aberrant crypts is in contrast to the CD44-negative surrounding normal colon tissue. (D) Bar graph presenting IHC results (H-score) for CD44 in Lynch lesions, illustrating a significant difference in staining intensity within the adenoma- carcinoma sequence. * P value < 0.05
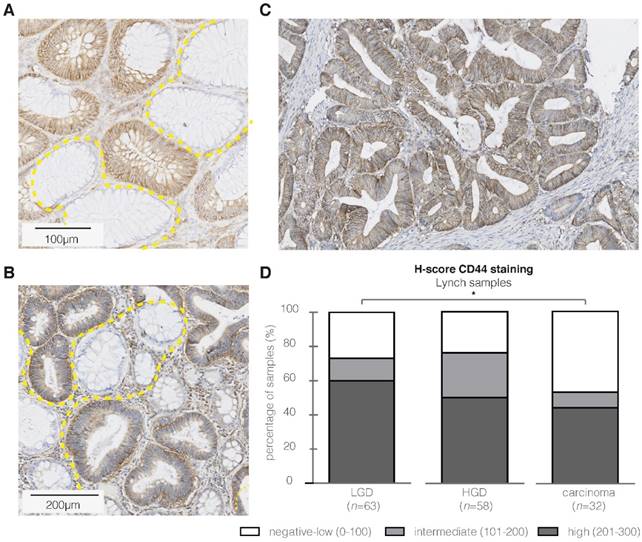
Immunohistochemistry (IHC) results for CD44v6 in sporadic adenomas. (A) Representative image of a strong staining intensity in high-grade adenomatous tissue, (B) and negative staining results in complete healthy colon crypts. (C) Representative image of the difference in CD44v6 staining intensity between normal and dysplastic colon crypts (dashed yellow lines). (D) H-score results for the extended sample-size (n = 69), resulting in an overall high CD44v6 staining intensity.
Results of CD44v6-targeted fluorescent imaging in ApcMin/+ mice. (A) CD44v6 overexpression can be observed in the cell membranes of the adenomatous crypts of the APCMin/+ mice, which confirms the suitability of this specific animal model for CD44v6-targeted imaging experiments. (B) Representative images of fresh resected and formalin-fixed murine intestines, previously injected with the specific (mouse-targeting) CD44v6*DY-681 fluorescent peptide tracer. The images illustrate the positive imaging results, as the fluorescent peaks correspond to the polyps identified on the white-light images (arrow heads) and moreover demonstrate the consistency of the observed signals among the different animals (n = 4). (C) Representative images of fresh and formalin-fixed murine intestines, previously injected with either the control CD44v6*DY681 tracer (human-targeting, n = 4) or no tracer (n = 2), demonstrating low to negative background signals in both control conditions.
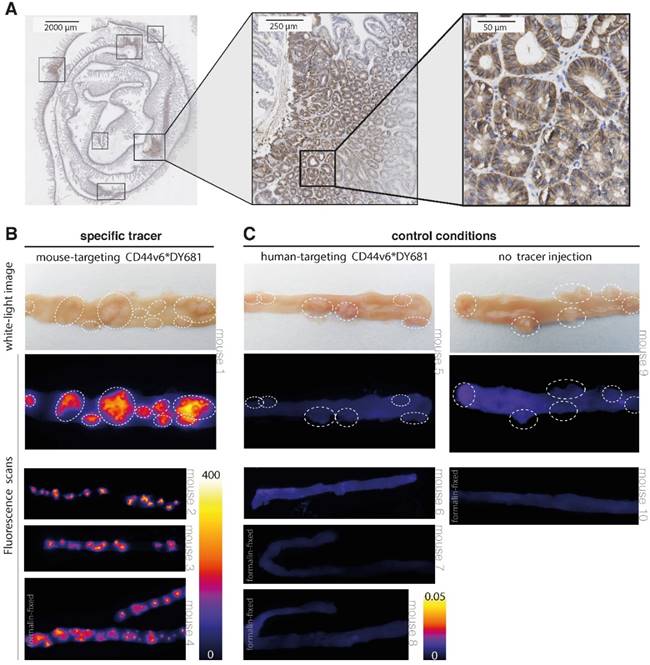
Our results show that a clear difference in fluorescent signals could be observed between the specific CD44v6-targeting tracer and the control conditions (human-targeting peptide and no peptide tracer). When using the specific (mouse-targeting) tracer product, high fluorescent signals can be visualized in the - dye specific - 700 nm channel (Figure 5B). Positive fluorescent results could be observed for the adenoma-containing intestines, urine-containing bladder and in a limited amount for the kidneys; the other harvested organs did not display any fluorescence (Figure S2 A). In contrast, low to negative fluorescent signals can be observed at baseline (no peptide) and following aspecific (human-targeting) tracer administration (Figure 5C and Figure S2 B). At 800 nm wavelengths, low fluorescent signals are present, fitting the emission spectrum of the fluorophore (Figure S3). The negative control results and the absence of non-specific emission signals together indicate the authenticity of the specific (mouse-targeting) CD44v6*DY-681 fluorescent results. Therefore, these imaging results confirm what we expected based on our FGmRNA-profiling results: CD44v6 is indeed an attractive target for intestinal adenoma imaging. In addition, our results show that wide-field, 'red-flag' molecular imaging of intestinal adenomas is feasible when using an intravenously administered, fluorescently labeled small peptide drug.
Discussion
CRC is a highly lethal condition and early endoscopic identification and removal of its precursor lesions, adenomatous polyps, could prevent this disease from developing. Molecular guided endoscopy has the potential to improve adenoma detection. This study presents an extensive list of FGmRNA-overexpressing genes that may serve as potential targets for molecular colorectal adenoma imaging. Based on protein location of the first 50 FGmRNA-overexpressing genes, i.e. membrane bound or extracellular, we were able to identify 18 potential targets for molecular imaging. We validated our FGmRNA results by demonstrating the differences in downstream protein expression levels between adenoma and normal colon tissue for the top FGmRNA-overexpressing genes. Based on the degree of FGmRNA-overexpression, the downstream protein membrane localization and cellular protein expression level, we identified CD44 as an attractive adenoma target in both sporadic adenomas and high-risk Lynch patients. In addition, we demonstrated the feasibility of CD44v6-targeted adenoma imaging, as we were able to fluorescently visualize adenomas in ApcMin/+ mice.
We are the first to perform molecular characterization of adenomas based on FGmRNA expression profiles. Genomic alterations can have downstream effects by changing gene and protein expression levels. Therefore, exploration of genomic alterations and the molecular downstream effects, specifically occurring in adenomas, might result in the identification of new imaging targets. However, although genomic alterations affect the expression levels of genes, they are in general overshadowed by non-genetic factors. Our FGmRNA-profiling method is capable of correcting this gene expression data for influencing non-genetic factors, allowing an enhanced view on gene expression levels (7). This enhanced view allows us to reanalyze and reinterpret publicly available adenoma expression profiles. The fact that the genes that show the most FGmRNA upregulation differ from the previously published results derived with use of the same expression profiles (8), at the time assessed without the FGmRNA-profiling method, strengthen the believe that we are able to provide new insights in pre-existing data. Interestingly, our top identified and validated genes, AXIN2, CEMIP, CD44 and JUN, are all known target-genes of the Wnt-signaling pathway (Figure S4). The Wnt-pathway is a well-known initiator of the adenoma-carcinoma sequence, illustrating the biological relevance of the identified genes (2,22). Thus, in addition to selecting potential targets for molecular imaging strategies, the genes we identified via FGmRNA-profiling also reflect the biology of the adenoma-carcinoma sequence. In this report we present an extensive, publicly accessible list of additional FGmRNA-overexpressing genes, which may contain other promising targets and could serve as a source to select new targets for molecular adenoma imaging (Table S1). However, it should be noted that, although FGmRNA-profiling can serve as a tool for identifying cancer associated genetic alterations with consecutive protein upregulation, it is unable to distinguish different gene splice variants.
Since molecular imaging could facilitate early lesion detection, this advanced imaging technique could be of high value; especially when considering high-risk CRC populations, such as patients with Lynch syndrome (LS). The role of CD44 in the adenoma-carcinoma sequence has been previously established (15,16,20,23-30). No results have yet been published on the role of CD44 in LS. In this study we demonstrate that CD44 is indeed overexpressed in LS adenomas and carcinomas. Its previously established biological role, combined with its demonstrated epithelial overexpression in sporadic and high-risk Lynch adenomas, makes CD44 an interesting imaging target; especially when targeting one of its epithelial splice variants and therefore limiting surrounding stromal involvement. Our results evidence the suggested adenoma-imaging potential of CD44v6: first of all, we have shown that the majority of sporadic adenomas indeed overexpress this particular epithelial splice protein, which is in concordance with literature (21,31,32). Even more importantly, our ex vivo imaging results clearly demonstrate that, with use of a fluorescently labeled CD44v6-targeting peptide, wide-field 'red flag' adenoma detection can be accomplished. We are the first to describe preclinical wide-field fluorescent imaging of CD44v6 using an IV injected fluorescent CD44v6 targeted peptide. Compared to other cancer types (11,33), no data has yet been published regarding CD44v6-targeting peptides in the context of colorectal cancer or adenomas. In addition, our approach makes for a 'red-flag' adenoma identification strategy, suitable for future clinical translation. Firstly, it is based on a peptide tracer product, which is an attractive molecular targeting probe due to its relative small-sized structure, reduced immunogenicity and low large-scale production costs (34). And secondly, recent technical advances have let to the availability of endoscopic wide-field molecular imaging systems, suitable for clinical use (35-37).
Recently, the first in-human molecular targeted endoscopy results on the detection of colorectal polyps were published, using an intravenously administered peptide probe against c-Met (37); They reported that the adenoma identification increased with 19% when using c-Met targeted fluorescence endoscopy compared to white-light inspection. These results support our findings, since we identified MET to be among the 50 highly significant FGmRNA-overexpressing genes and denoted the c-Met protein as one of the 18 potential imaging targets (see Figure 2). In addition, the clinical potential of our molecular-targeted imaging approach is indicated by a recent clinical trial with a 89Zr-labeled CD44-targeting antibody, which showed the feasibility of tumor-targeted imaging by positron emission tomography (PET)(38), and a preceding case report that described microscopic crypt characterization on resected human colonic tissue with use of confocal laser endomicroscopy (CLE) after ex vivo topical application of fluorescein-isothiocyanate labeled CD44v6 antibodies (39).
Conclusion
We have shown that our bioinformatics and functional approach can identify biologically relevant targets that are potentially applicable for molecular imaging strategies. We have not only identified CD44 as an attractive colorectal adenoma target, but have also demonstrated the applicability of a small peptide, targeting its splice variant CD44v6, for molecular imaging purposes. Moreover, the availability of a human CD44v6-targeting peptide enables clinical translation of these results in de near future. Since the same need for diagnostic improvements exists for other - gastrointestinal - cancers, we envision a similar molecular target identification approach for a wide range of tumors, including esophageal, stomach and pancreatic cancer.
Abbreviations
CRC: colorectal cancer; FGmRNA: Functional Genomic mRNA; IHC: immunohistochemistry; LGD: low-grade dysplasia; HGD: high-grade dysplasia; GEO: Robust multi-array average: RMA; PCA: Principal Component analyses; FFPE: formalin-fixed paraffin embedded; LS: Lynch Syndrome; FAP: familial adenomatous polyposis.
Supplementary Material
Additional File 1Table S1. Complete list of identified FGmRNA-overexpressing genes.
Figures S1-S4.
Acknowledgements
This work was financially supported by grants from NWO-VENI (916-16025 to R.S.N.F), the Dutch Cancer Society / Alpe d'HuZes (RUG 2013-5960 to R.S.N.F and RUG 2012-5416 to W.B.N.) and the German Cancer Research Centre and Karlsruhe Institute of Toxicology (DKFZ-KIT; obtained by V.O.R.). In addition, we would like to thank research analyst W. Boersma-van Ek for her assistance in the laboratory and master student J. Meisner for contributing to the IHC staining and scoring process.
Contributions
E.H. contributed to the acquisition and analysis of the genetic, IHC and animal data and wrote the manuscript. V.O.R. and A.M.O. conceived and execution of the animal experiments. A.K. contributed to the optimization of the IHC protocols and assisted in assessment of the tissue samples. D.J.A.d.G., S.d.J. and G.M.v.D. contributed to data interpretation and revised the manuscript. R.S.N.F. and W.B.N. contributed equally; they conceived the study, contributed to the acquisition and interpretation of the data and critically revised drafts of the manuscript. All authors approved the final version of the manuscript.
Competing Interests
V.O.R. and A.M.O. are shareholders of the start-up company Amcure GmbH (Germany); Amcure employs A.M.O., but did not finance this work in any way.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7-30
2. Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M. et al. Genetic Alterations during Colorectal-Tumor Development. N Engl J Med. 1988;319:525-32
3. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546-58
4. van Rijn JC, Reitsma JB, Stoker J, Bossuyt PM, van Deventer SJ, Dekker E. Polyp miss rate determined by tandem colonoscopy: a systematic review. Am J Gastroenterol. 2006;101:343-50
5. Hazewinkel Y, Tytgat KMAJ, van Leerdam ME, Koornstra J-J, Bastiaansen BA, van Eeden S. et al. Narrow-band imaging for the detection of polyps in patients with serrated polyposis syndrome: a multicenter, randomized, back-to-back trial. Gastrointestinal Endoscopy. 2015;81:531-8
6. Goetz M, Wang TD. Molecular imaging in gastrointestinal endoscopy. Gastroenterology. 2010;138:828-33.e1
7. Fehrmann RSN, Karjalainen JM, Krajewska M, Westra H-J, Maloney D, Simeonov A. et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat Genet. 2015;47:115-25
8. Heijink DM, Fehrmann RSN, de Vries EGE, Koornstra JJ, Oosterhuis D, van der Zee AGJ. et al. A bioinformatical and functional approach to identify novel strategies for chemoprevention of colorectal cancer. Oncogene. 2011;30:2026-36
9. Hirsch FR. Epidermal Growth Factor Receptor in Non-Small-Cell Lung Carcinomas: Correlation Between Gene Copy Number and Protein Expression and Impact on Prognosis. J Clin Oncol. 2003;21:3798-807
10. Boivin GP, Washington K, Yang K, Ward JM, Pretlow TP, Russell R. et al. Pathology of mouse models of intestinal cancer: consensus report and recommendations. Gastroenterology. 2003;124:762-77
11. Tremmel M, Matzke A, Albrecht I, Laib AM, Olaku V, Ballmer-Hofer K. et al. A CD44v6 peptide reveals a role of CD44 in VEGFR-2 signaling and angiogenesis. Blood. 2009;114(25):5236-44
12. Matzke A, Herrlich P, Ponta H, Orian-Rousseau V. A five-amino-acid peptide blocks Met- and Ron-dependent cell migration. Cancer Res. 2005;65:6105-10
13. Matzke-Ogi A, Jannasch K, Shatirishvili M, Fuchs B, Chiblak S, Morton J. et al. Inhibition of Tumor Growth and Metastasis in Pancreatic Cancer Models by Interference With CD44v6 Signaling. Gastroenterology. 2016;150:513-525.e10
14. Lesley J, Hyman R, Kincade PW. CD44 and its interaction with extracellular matrix. Adv Immunol. 1993;54:271-335
15. Wielenga VJ, van der Neut R, Offerhaus GJ, Pals ST. CD44 glycoproteins in colorectal cancer: expression, function, and prognostic value. Adv Cancer Res. 2000;77:169-87
16. Dalerba P, Dylla SJ, Park I-K, Liu R, Wang X, Cho RW. et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158-63
17. Li X-D, Ji M, Wu J, Jiang J-T, Wu C-P. Clinical significance of CD44 variants expression in colorectal cancer. Tumori. 2013;99:88-92
18. Heider K-H, Kuthan H, Stehle G, Munzert G. CD44v6: a target for antibody-based cancer therapy. Cancer Immunol Immunother. 2004;53:567-79
19. Wielenga VJ, Heider KH, Offerhaus GJ, Adolf GR, van den Berg FM, Ponta H. et al. Expression of CD44 variant proteins in human colorectal cancer is related to tumor progression. Cancer Res. 1993;53:4754-6
20. Zeilstra J, Joosten SPJ, van Andel H, Tolg C, Berns A, Snoek M. et al. Stem cell CD44v isoforms promote intestinal cancer formation in Apc(min) mice downstream of Wnt signaling. Oncogene. 2014;33:665-70
21. Afify A, Durbin-Johnson B, Virdi A, Jess H. The expression of CD44v6 in colon: from normal to malignant. Ann Diagn Pathol. 2016;20:19-23
22. Nusse R. Wnt signaling in disease and in development. Cell Res. 2005;15(1):28-32
23. Wittig BM, Goebel R, Weg-Remers S, Pistorius G, Feifel G, Zeitz M. et al. Stage-Specific Alternative Splicing of CD44 and α6β1 Integrin in Colorectal Tumorigenesis. Exp Mol Pathol. 2001;70:96-102
24. Weg-Remers S, Anders M, Lampe von B, Riecken EO, Schüder G, Feifel G. et al. Decreased expression of CD44 splicing variants in advanced colorectal carcinomas. Eur J Cancer. 1998;34:1607-11
25. Todaro M, Gaggianesi M, Catalano V, Benfante A, Iovino F, Biffoni M. et al. CD44v6 Is a Marker of Constitutive and Reprogrammed Cancer Stem Cells Driving Colon Cancer Metastasis. Stem Cell. 2014;14:342-56
26. Zeilstra J, Joosten SPJ, Dokter M, Verwiel E, Spaargaren M, Pals ST. Deletion of the WNT Target and Cancer Stem Cell Marker CD44 in Apc(Min/+) Mice Attenuates Intestinal Tumorigenesis. Cancer Res. 2008;68:3655-61
27. Misra S, Hascall VC, De Giovanni C, Markwald RR, Ghatak S. Delivery of CD44 shRNA/nanoparticles within cancer cells: perturbation of hyaluronan/CD44v6 interactions and reduction in adenoma growth in Apc Min/+ MICE. J Biol Chem. 2009;284:12432-46
28. Neumayer R, Rosen HR, Reiner A, Sebesta C, Schmid A, Tüchler H. et al. CD44 expression in benign and malignant colorectal polyps. Dis Colon Rectum. 1999;42:50-5
29. Heider KH, Hofmann M, Hors E, van den Berg F, Ponta H, Herrlich P. et al. A human homologue of the rat metastasis-associated variant of CD44 is expressed in colorectal carcinomas and adenomatous polyps. J Cell Biol. 1993;120:227-33
30. Hong I, Hong SW, Chang YG, Lee WY, Lee B, Kang YK. et al. Expression of the Cancer Stem Cell Markers CD44 and CD133 in Colorectal Cancer: An Immunohistochemical Staining Analysis. Ann Coloproctol. 2015;31:84-91
31. Coppola D, Hyacinthe M, Fu L, Cantor AB, Karl R, Marcet J. et al. CD44V6 expression in human colorectal carcinoma. Hum Pathol. 1998;29:627-35
32. Orzechowski HD, Beckenbach C, Herbst H, Stölzel U, Riecken EO, Stallmach A. Expression of CD44v6 is associated with cellular dysplasia in colorectal epithelial cells. Eur J Cancer. 1995;31A:2073-9
33. Zöller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11:254-67
34. Chen K, Chen X. Design and development of molecular imaging probes. Curr Top Med Chem. 2010;10:1227-36
35. Tjalma JJ, Garcia-Allende PB, Hartmans E, Terwisscha van Scheltinga AG, Boersma-van Ek W, Glatz J. et al. Molecular-Guided Endoscopy Targeting Vascular Endothelial Growth Factor A for Improved Colorectal Polyp Detection. J Nucl Med. 2016;57:480-5
36. Keller R, Winde G, Terpe HJ, Foerster EC, Domschke W. Fluorescence endoscopy using a fluorescein-labeled monoclonal antibody against carcinoembryonic antigen in patients with colorectal carcinoma and adenoma. Endoscopy. 2002;34:801-7
37. Burggraaf J, Kamerling IMC, Gordon PB, Schrier L, de Kam ML, Kales AJ. et al. Detection of colorectal polyps in humans using an intravenously administered fluorescent peptide targeted against c-Met. Nat Med. 2015;21:955-61
38. Houven van Oordt der CWM-V, Gomez-Roca C, Herpen CV, Coveler AL, Mahalingam D, Verheul HMW. et al. First-in-human phase I clinical trial of RG7356, an anti-CD44 humanized antibody, in patients with advanced, CD44-expressing solid tumors. Oncotarget. 2016:5
39. Neurath M, Kiesslich R. Molecular detection of CD44v6 on aberrant crypt foci by confocal laser endoscopy. Endoscopy. 2010;42(Suppl 02):E314-5
Author contact
![]() Corresponding authors: Wouter B Nagengast and Rudolf S N Fehrmann, University Medical Centre Groningen (UMCG), Hanzeplein 1, PO-box 30 001 9700 RB Groningen, The Netherlands, phone number: +3150 3612620 / Fax number: +3150 3619306, w.b.nagengastnl / r.s.n.fehrmannnl
Corresponding authors: Wouter B Nagengast and Rudolf S N Fehrmann, University Medical Centre Groningen (UMCG), Hanzeplein 1, PO-box 30 001 9700 RB Groningen, The Netherlands, phone number: +3150 3612620 / Fax number: +3150 3619306, w.b.nagengastnl / r.s.n.fehrmannnl
 Global reach, higher impact
Global reach, higher impact