13.3
Impact Factor
Theranostics 2016; 6(12):2183-2195. doi:10.7150/thno.17140 This issue Cite
Research Paper
Deficiency of ATP6V1H Causes Bone Loss by Inhibiting Bone Resorption and Bone Formation through the TGF-β1 Pathway
1. State Key Laboratory of Military Stomatology, National Clinical Research Center for Oral Diseases, Shaanxi Key Laboratory of Oral Diseases, Department of Oral Biology, Clinic of Oral Rare and Genetic Diseases, School of Stomatology, The Fourth Military Medical University, Xi'an, 710032, People's Republic of China.
2. Key Laboratory of Biomedical Information Engineering of Ministry of Education, and Institute of Molecular Genetics, School of Life Science and Technology, Xi'an Jiaotong University, Xi'an 710049, People's Republic of China.
3. School of Public Health and Tropical Medicine, Tulane University, New Orleans, LA 70112, USA.
*Contributed equally to this work.
Received 2016-8-8; Accepted 2016-8-15; Published 2016-9-13
Abstract
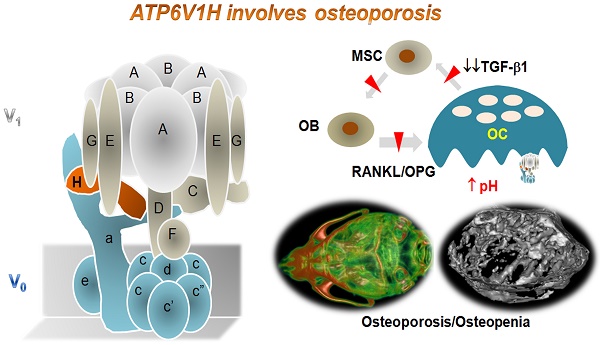
Vacuolar-type H +-ATPase (V-ATPase) is a highly conserved, ancient enzyme that couples the energy of ATP hydrolysis to proton transport across vesicular and plasma membranes of eukaryotic cells. Previously reported mutations of various V-ATPase subunits are associated with increased bone density. We now show that haploinsufficiency for the H subunit of the V1 domain (ATP6V1H) is associated with osteoporosis in humans and mice. A genome-wide SNP array analysis of 1625 Han Chinese found that 4 of 15 tag SNPs (26.7%) within ATP6V1H were significantly associated with low spine bone mineral density. Atp6v1h+/- knockout mice generated by the CRISPR/Cas9 technique had decreased bone remodeling and a net bone matrix loss. Atp6v1h+/- osteoclasts showed impaired bone formation and increased bone resorption. The increased intracellular pH of Atp6v1h+/- osteoclasts downregulated TGF-β1 activation, thereby reducing induction of osteoblast formation but the bone mineralization was not altered. However, bone formation was reduced more than bone resorption. Our data provide evidence that partial loss of ATP6V1H function results in osteoporosis/osteopenia. We propose that defective osteoclast formation triggers impaired bone formation by altering bone remodeling. In the future, ATP6V1H might, therefore, serve as a target for the therapy of osteoporosis.
Keywords: osteoporosis, V-ATPase, osteoclasts, ATP6V1H, CRISPR/Cas9, TGF-β1, pH, RANKL, OPG.
Introduction
Vacuolar-type H +-ATPase (V-ATPase), a highly conserved enzyme, mediates acidification in organelles by pumping protons and coupling with the energy of ATP hydrolysis. V-ATPase-dependent organelle acidification is necessary for intracellular processes such as protein sorting, zymogen activation, receptor-mediated endocytosis, and synaptic vesicle proton gradient generation. V-ATPase consists of a peripheral catalytic V1 domain and a membrane proton channel V0 domain [1]. Subunits of V-ATPase play important roles in the biological and physiological functions of osteoclasts [2] and the dysfunction of these subunits is usually associated with osteopetrosis and other bone abnormalities. T-cell immune regulator 1 (TCIRG1), also known as V-ATPase V0 subunit a3, resides in the ruffled border of osteoclasts and controls osteoclast-mediated extracellular acidification. Mutations in TCIRG1 gene cause autosomal recessive osteopetrosis in humans [3-6], and mice deficient in Tcirg1 (or Atp6i) have hypocalcemia and osteopetrorickets [7]. The V0D2 subunit of V-ATPase is required for efficient preosteoclast fusion. Atp6v0d2-deficient mice have increased bone mass and enhanced bone formation [8-10]. The C subunit of the V1 domain of V-ATPase regulates F-actin ring formation during osteoclast activation [11-13] and the reversible dissociation of V-ATPases[14]. Consequently, Atp6v1c1 deficiency severely impairs osteoclast acidification and bone resorption, whereas osteoclast differentiation is unaffected [11]. The V1B2 subunit of V-ATPase is associated with F-actin and facilitates the recruitment of V-ATPase complexes to the osteoclast ruffled border during polarization and bone resorption [15, 16]. Recent findings have shown that mutations in the ATP6V1B1 gene cause Zimmermann-Laband syndrome and dominant deafness-onychodystrophy syndrome, which might be related to impaired assembly of the V1 subcomplex of ATP6V 1B1 [17, 18].
Osteoporosis is a common metabolic bone disease characterized by reduced bone mineral density (BMD) and increased risk of osteoporotic fractures. Genetic factors have been closely associated with the risk of osteoporosis inciting research on the genes involved in osteoclasts functions in osteoporosis [19, 20]. One of our recent bivariate genome-wide association studies (GWAS) implicated ATP6V1G1 as a novel pleiotropic gene affecting BMD in osteoporosis patients [21]. This study suggested that other subunits of V-ATPase could also be associated with osteoclast function and bone disorders. ATP6V1H is a small subunit of V-ATPase that connects the V1 and V0 domains; its role in bone development and bone disorders remains unknown. Further GWAS screening of the osteoporosis population helped us identify ATP6V1H as an additional subunit of V-ATPase involved in osteoporosis. To define the function of ATP6V1H and its relationship with osteoporosis, we generated Atp6v1h knockout mice using the CRISPR/Cas9 technique and defined the bone phenotypes in this mouse model. We observed that the deficiency of ATP6V1H caused a lower bone turn over and bone loss by inhibiting osteoclasts formation and bone formation at the same time; bone formation was reduced more than bone resorption, resulting in a net bone loss. This imbalance in bone homeostasis occurred from altered interactions between osteoclasts and osteoblasts through the TGF-β1 pathway.
Results
ATP6V1H variants and low bone mass in a human population. 1625 Han Chinese were divided into three groups with different Z-scores (Table 1). We chose -1SD to -2SD of Z-score as a threshold to separate controls from osteopenia group with low bone mass and osteoporosis group. A Z-score below -2 was considered as a significant risk for osteoporosis. A GWAS analysis involving 281,533 SNPs [22, 23] was performed and fifteen SNPs targeted to the intronic regions of the ATP6V1H gene were selected. These variants were compared among the three groups. Multiple testing identified 4 of them (26.7%) having a nominal significant association with spine BMD, P<0.05 (Table 2).
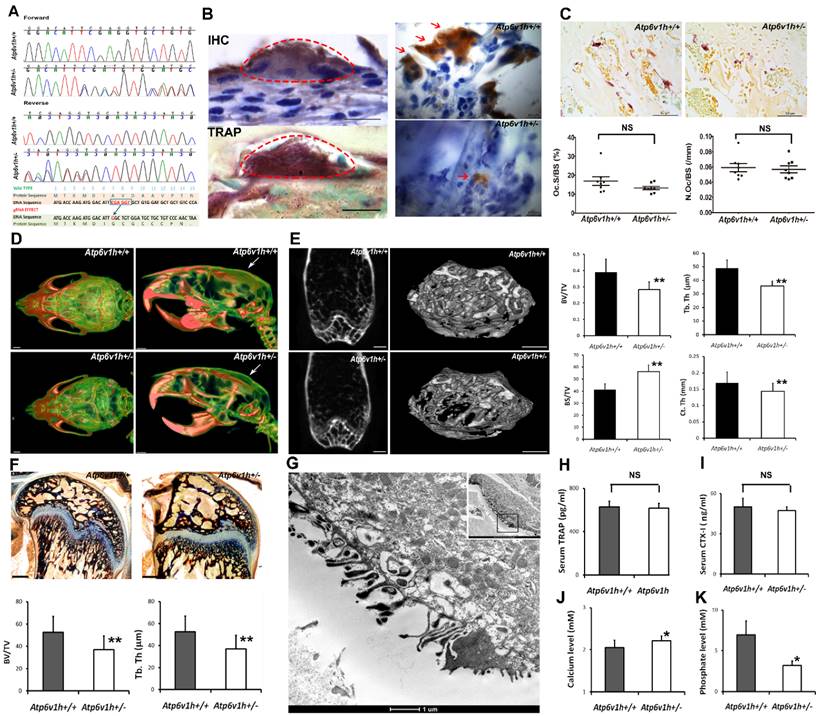
Bone loss and decreased bone formation in Atp6v1h+/- mice. We used the CRISPR/Cas9 technique, designed guide RNA (gRNA) to target exon 2 of the Atp6v1h gene, and created a 5bp (CGAGG) deletion and one base replacement (T>G). The mutations caused an early stop codon (Fig.1A). The gRNA has less chance of off-target events according to the results from MIT CRISPR design and analysis website (http://crispr.mit.edu/). We tested 7 genes (Aldh16a1, Tbrg1, Wdr4, Atp2b3, Gcsh, Zscan, and Kcnt11) with 4%~5% predicted occurrence ratio of off-target events, but none of them showed off-target effects. All homozygous Atp6v1h-/- embryos died before embryonic day 10.
Atp6v1h+/- heterozygous mice were born at the expected Mendelian ratio and exhibited a normal growth rate. There were no obvious differences in weight or size between wild-type and heterozygous mice. IHC staining and tartrate-resistant acidic phosphatase (TRAP) staining indicated that ATP6V1H was localized in osteoclasts. The positive staining of ATP6V1H in osteoclasts became weak in heterozygous mice (Fig. 1B). TRAP staining for osteoclasts showed an equal number of osteoclasts and same size of osteoclasts in the distal femurs of Atp6v1h+/- mice compared with control littermates (Fig.1 C). High-resolution micro-CT analysis in calvaria (Fig. 1D), femurs (Fig.1E and Table 3) and lumbar vertebrae (Table 3), however, demonstrated obvious decreased bone mass in Atp6v1h+/- mice. Ponceau red staining of calcified bone tissues further showed decreased bone volume and trabecular thickness (Fig.1 F). Transmission electron microscope (TEM) analysis found that Atp6v1h+/- osteoclasts had normal ruffled borders (Fig. 1G). The serum CTX-I and TRAP, which reflected osteoclastic resorptive activity, were not altered in Atp6v1h+/- mice (Fig. 1H, I). Serum calcium concentrations were slightly higher (Fig. 1J), and serum phosphate levels were lower in Atp6v1h+/- mice compared to the wild-type (Fig. 1K).
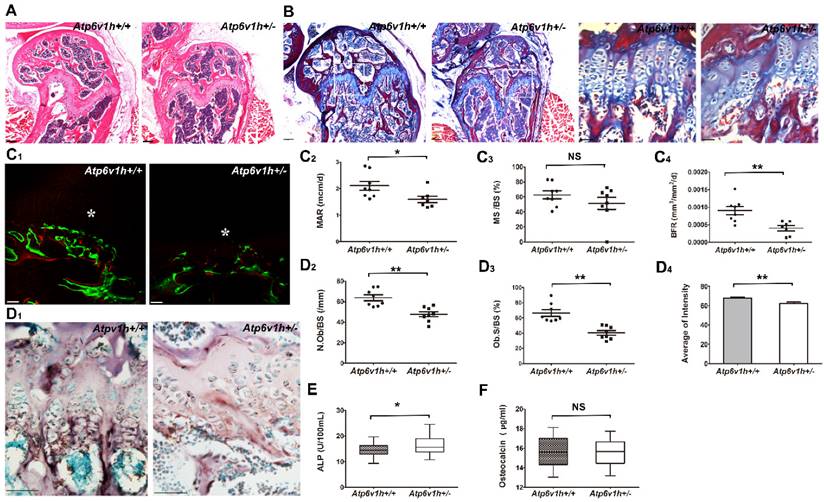
The Atp6v1h+/- mice exhibited less cartilage and bone formation (Fig. 2A, B). Dynamic histomorphometric analysis detected the reduced bone formation rate (BFR) and mineral apposition rate (MAR) in Atp6v1h+/- mice (Fig. 2C1~C4). The numbers of osteoblasts and area of osteoblast surfaces were reduced in the distal femurs of Atp6v1h+/- mice. ALP staining was weak and uneven (Fig. 2D1~D4), whereas the serum level of ALP was increased in Atp6v1h+/- mice (Fig. 2E). Serum osteocalcin was not altered in Atp6v1h+/- mice (Fig. 2F).
The changes of bone loss and decreased bone formation co-existed in Atp6v1h+/- mice. This suggests that the osteoporotic changes of Atp6v1h+/- mice might result from an imbalance of osteoclasts and osteoblasts activities.
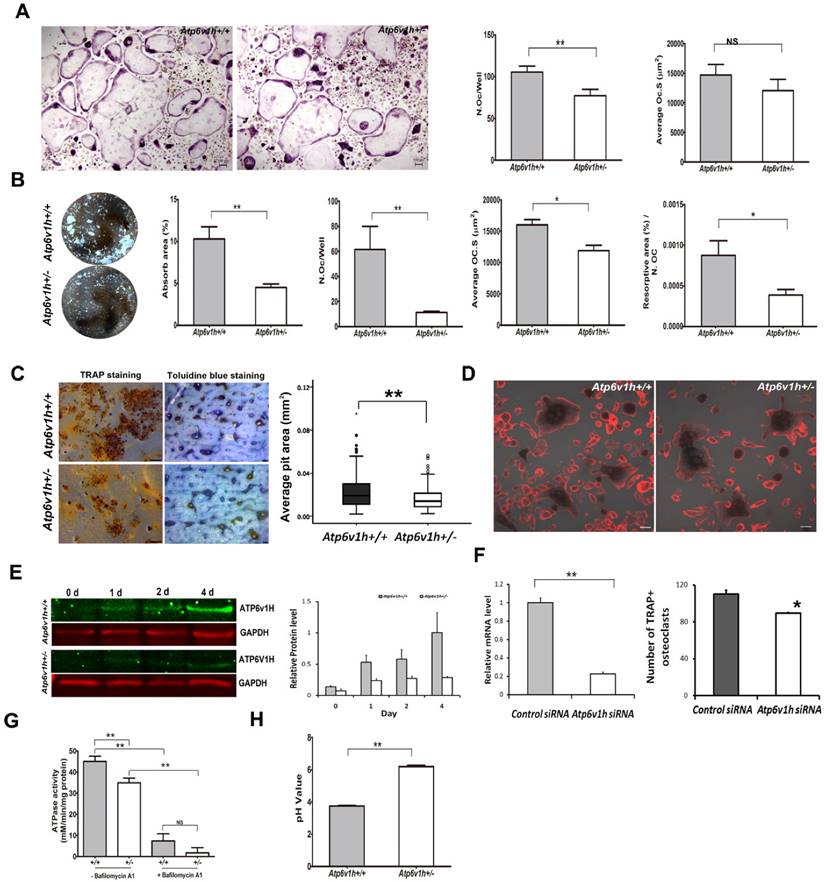
Impaired osteoclast formation and function in Atp6v1h+/- mice. To characterize the osteoclasts of Atp6v1h+/- mice in vitro, we stimulated primary bone marrow cells (BMs) with receptor activator of nuclear factor kappa-B (NF-κ B) ligand (RANKL) and macrophage colony-stimulating factor (M-CSF). The number of TRAP-positive Atp6v1h+/-osteoclasts was reduced and the size of Atp6v1h+/- osteoclasts was not significantly changed when the cells were cultured in a general 96-well plate (Fig.3A). When BMs were cultured on an osteoassay microplate with the same RANKL/M-CSF stimulating conditions, we observed fewer and smaller osteoclasts in the Atp6v1h+/- group, and the total resorptive area, as well as the osteoclast number normalized to the resorptive area, were also decreased (Fig. 3B). Consistent with the above results, the number of pits per bone slice (4mm×4mm) was decreased as was the average pit area of Atp6v1h+/- osteoclasts compared with those of osteoclasts from wild type mice (Fig. 3C); this indicates that the Atp6v1h defect influenced bone resorptive activity. Comparing the formation of actin ring between wild-type and Atp6v1h+/- osteoclasts detected no difference (Fig. 3D).
Clinical Characteristics of 1625 Han Chinese Subjects.
| Sex No (%) | Yr of age Median (range) | Weight (kg) Average(SD) | Height (cm) Average(SD) | Lumbar Spine BMD Average (SD) | Spine Z-score Average (SD) | ||
|---|---|---|---|---|---|---|---|
| Male | Female | ||||||
| Group 1 | 683 (49.4) | 699 (50.6) | 28 (18~82) | 60.4 (10.6) | 164.7 (8.1) | 0.98 (0.11) | 0.27 (0.82) |
| Group 2 | 108 (51.7) | 101 (48.3) | 34 (20~72) | 58.6 (10.0) | 162.2 (7.8) | 0.78 (0.04) | -1.36 (0.26) |
| Group 3 | 10 (30.3) | 23 (69.7) | 62 (25~75) | 58.2 (13.9) | 157.8 (8.3) | 0.64 (0.07) | -2.41 (0.43) |
Group1: Spine Z score >-1; Group 2: ≤-1, >-2; Group3: Spine Z score ≤-2.
Association of 15 Tag SNPs in ATP6V1H with Spine Z-score in 1625 Han Chinese.
| SNP No. | Position (hg18) | A1 / A2 | Total | Female | Male | |||
|---|---|---|---|---|---|---|---|---|
| X-squared | P | X-squared | P | X-squared | P | |||
| rs3735833 | 54791289 | C/A | 1.198 | 0.879 | 3.534 | 0.473 | 8.364 | 0.079 |
| rs16919505 | 54796072 | G/A | 5.470 | 0.065 | 5.315 | 0.070 | 1.069 | 0.586 |
| rs16919507 | 54796534 | G/C | 7.438 | 0.115 | 7.308 | 0.121 | 2.843 | 0.585 |
| rs16919508 | 54802986 | G/A | 7.746 | 0.101 | 7.489 | 0.112 | 2.285 | 0.684 |
| rs4737554 | 54809631 | C/T | 7.188 | 0.126 | 5.511 | 0.239 | 4.861 | 0.302 |
| rs4737558 | 54810978 | G/A | 8.723 | 0.068 | 8.426 | 0.077 | 4.849 | 0.303 |
| rs2376011* | 54813858 | T/C | 10.573 | 0.032 | 7.607 | 0.107 | 7.712 | 0.103 |
| rs4738884* | 54820708 | T/C | 10.830 | 0.029 | 7.176 | 0.127 | 9.199 | 0.056 |
| rs10435587* | 54825832 | C/T | 10.052 | 0.040 | 4.312 | 0.366 | 10.674 | 0.030 |
| rs41321146* | 54826259 | A/C | 9.655 | 0.047 | 7.134 | 0.129 | 7.191 | 0.126 |
| rs12056719 | 54826538 | A/G | 9.038 | 0.060 | 7.176 | 0.127 | 6.195 | 0.185 |
| rs16919544 | 54834944 | A/G | 0.351 | 0.839 | 0.178 | 0.915 | 0.173 | 0.917 |
| rs1552148 | 54837276 | G/G | 9.305 | 0.054 | 6.797 | 0.147 | 7.157 | 0.128 |
| rs6991513 | 54908091 | G/C | 1.591 | 0.811 | 1.922 | 0.750 | 5.413 | 0.248 |
| rs6994851 | 54910994 | G/A | 5.918 | 0.205 | 4.093 | 0.394 | 3.304 | 0.508 |
| rs16919588 | 54914244 | T/C | 1.714 | 0.788 | 2.116 | 0.714 | 0.649 | 0.958 |
A1: Minor allele; A2: Major allele. A linear regression implemented in PLINK was fitted to test for association assuming an additive inheritance model.
Structural Parameters Measured by Microcomputed Tomography.
| Site | Mice | No. | BV/TV (%) | Tb.N | Tb.Th (µm) | Tb.S (µm) | BS/TV (mm2/mm3) | Ct.Th (mm) |
|---|---|---|---|---|---|---|---|---|
| Femur | Wild type | 8 | 0.389± 0.081 | 7.892± 1.136 | 49.075± 5.918 | 80.489± 24.056 | 41.290± 4.780 | 0.169± 0.034 |
| Heterozygous | 8 | 0.264± 0.049 ** | 7.311± 0.937 | 35.903± 3.594 ** | 103.050± 19.637 | 56.211± 5.559** | 0.144± 0.025** | |
| Vertebrae (L3) | Wild type | 8 | 0.249± 0.033 | 7.379± 0.752 | 33.687± 2.832 | 103.147± 15.077 | 59.734± 4.968 | NA |
| Heterozygous | 7 | 0.174± 0.038 * | 5.934± 0.817 | 30.506± 2.629 | 142.559± 27.656* | 65.916± 5.517 | NA |
BV/TV: Bone volume; Tb.N: Trabecular number; Tb. Th: Trabecular thickness; Tb.S.: Trabecular separation; BS/TV: Bone surface; Ct.Th: Cortical thickness; **P < 0.05; ** P < 0.01 (Only the columns marked with * show statistical differences); NA: not available.
Increased bone resorption in Atp6v1h+/- mice. A. Atp6v1h+/- mouse with a 5bp (CGAGG) deletion and one base replacement (T>G) which caused an early stop codon. B. Right: sABC immunohistochemical (IHC) staining for ATP6V1H in wild-type osteoclasts, parallel to TRAP-positive staining. Bars=20μm. Left: Strong IHC staining for ATP6V1H in wild-type osteoclasts (red arrow). Bars=10μm. C. TRAP staining of the distal femurs from 3-month-old Atp6v1h+/- mice and control littermates. Bars = 50μm. The number of TRAP-positive osteoclasts (N. Oc/BS,) and osteoclast surface area (OC.S/BS) were not altered in Atp6v1h+/- mice. BS. Bone surface. n=8. D. Three-dimensional micro-CT images of calvarial bones. Right: Whole appearance. Left: Middle-sagittal view of a 3D image. Atp6v1h+/- mouse showed decreased bone density (white arrow). Bars=1mm. E. Micro CT images of the distal region of femurs from3-month-old Atp6v1h+/- mice showed decreased bone mass. Bars=0.5mm. BV/TV, Tb.Th, and Ct.Th were decreased and BS/TV was increased in Atp6v1h+/- mice compared with wild type. n=8. F. Modified Ponceau red staining of non-mineralized sections from the 4-week old wild-type and Atp6v1h+/- mice. The Atp6v1h+/- mouse showed less bone mass and decreased BV/TV and Tb.Th. Bars = 100μm. n=8. G. TEM images of Atp6v1h+/- osteoclasts showed a normal ruffled border. Bars=1μm (large image), 5 μm (small image). H and I. ELISA results of TRAP and CTX-I in sera. There was no significant difference between wild-type and Atp6v1h+/- mice. n=9. J and K. The serum level of calcium was higher and that of phosphate was lower in Atp6v1h+/- mice than in wild-type. n=8. All data represent mean ±s.d. ** P<0.01, *P<0.05. NS: not significant difference.
The protein level of ATP6V1H in wild-type osteoclasts gradually increased during days 0~4 of RANKL stimulation, whereas the level of ATP6V1H in Atp6v1h+/- osteoclasts was lower than that of wild-type osteoclasts throughout (Fig. 3E). Knockdown of Atp6v1h by siRNA reduced the formation of TRAP+ osteoclasts obtained from BMs stimulated with M-CSF and RANKL (Fig. 3F).
Atp6v1h deficiency also affected ATPase activities in Atp6v1h+/- osteoclasts (Fig. 3G), and the intracellular environment of Atp6v1h+/- osteoclasts became more alkaline compared to the wild-type cells (Fig. 3H).
These findings suggest that Atp6v1h deficiency affects the formation of osteoclasts and the resorptive ability of osteoclasts by impairing the V-ATPase activity and increasing the intracellular pH.
Decreased interaction between osteoblast and osteoclast in Atp6v1h+/- mice. The lower rate of bone formation prompted us to study whether the Atp6v1h deficiency also affected the osteoblasts directly. The immunohistochemical staining did not detect ATP6V1H protein in osteoblasts from wild-type and Atp6v1h+/- mice, although comparable mRNA levels of Atp6v1h could be detected in the cranial bone of new born mice and primary cultured osteoblasts of wild-type and Atp6v1h+/- mice (Fig. 4A). The screening of the mRNA levels of osteogenic markers in cranial bones and osteoblasts detected lower Alp, Col1a1 and Bsp expression in osteogenic-induced osteoblasts (Fig. 4A). When the mRNA level of Atp6v1h was downregulated in primary cultured osteoblasts or the osteoblast MC3T3-E1 cell line by Atp6v1h siRNA, only the levels of Ocn and Opn were altered (Fig. 4B), whereas the Atp6v1h +/- osteoblasts had the normal ability of calcified node formation (Fig. 4C). Thus we believe that a cell-autonomous effect of ATP6V1H haploinsufficiency on the osteoblast function is unlikely, and the osteoblastic effect is probably secondary to the reduced osteoclast formation and function.
Since TGF-β1 activation is pH sensitive [24, 25] and TGF-β1 could regulate the functions of osteoclasts and osteoblasts [26, 27], we hypothesized that the changes in intracellular pH in Atp6v1h +/- osteoclasts alter the level and activity of TGF-β1. Indeed, the protein level of TGF-β1 in Atp6v1h +/- osteoclasts was reduced more than half, whereas in Atp6v1h +/- osteoblasts, it was comparable to that of wild-type cells (Fig. 4D). The active TGF-β1 level in sera was also reduced in Atp6v1h +/- mice (Fig. 4E). Using ELISA to evaluate the secretion of active TGF-β1 from osteoclasts and osteoblasts, we observed that Atp6v1h +/- osteoclasts cultured on bone slices secreted less TGF-β1 (Fig. 4F), and that the osteogenic-induced Atp6v1h +/- osteoblasts produced more TGF-β1 (Fig. 4G). The lower levels of TGF-β1 have been reported to downregulate the differentiation of mesenchymal stem cells into osteoblasts. Thus the reduced number and size of osteoblasts in Atp6v1h +/- mice might result from a lower rate of osteoblasts formation.
Decreased bone formation in Atp6v1h+/- mice. A.H&E staining of femurs from 3-month-old Atp6v1h+/- mice and control littermates. Bars = 200μm. B. Modified Ponceau red staining of femurs from 3-month-old male Atp6v1h+/- mice and control littermates. The two images on the right are magnified presentations of the white outline in the left two images. Bars= 200μm (left two images), 50μm (right two images). C. Dynamic analysis of femurs from 4-week-old Atp6v1h+/- mice and control littermates. C1: Images of laser scanning confocal microscope. Stars represent growth plate. Bars=50 μm. C2~C4: Analysis results. The rate of mineral apposition (MAR, mcm/d) and bone formation rate (BFR, mm3/mm2/d) were decreased in Atp6v1h+/- mice, while the mineralizing surface (MS, %) was not altered. n=8. D. ALP staining of femurs from 3-month-old Atp6v1h+/- mice and control littermates. D1: ALP staining was reduced and unevenly distributed in Atp6v1h+/- mice. Bars = 50μm. D2, D3: The number of ALP+ osteoblasts (N. Ob/BS, /mm) and the osteoblast surface (Ob. S/BS, %) were significantly reduced in Atp6v1h+/- mice. n=9. D4: The average intensity of ALP-positive staining was reduced in Atp6v1h+/- mice by Image J software analysis. n=9. E. The serum ALP level of Atp6v1h+/- mice was higher than for controls. n=9. F. There was no significant difference in serum osteocalcin levels between wild type and Atp6v1h+/- mice. n=9. All data represent mean ±s.d. ** P<0.01, *P<0.05. NS: not significant difference.
Reduced formations and function of Atp6v1h+/- osteoclasts. A. Fewer TRAP+ Atp6v1h+/- osteoclasts with normal size were induced in a general 96 well plate. Bars=100μm. n≥100. B. When osteoclasts were induced in osteoassay plate, the number of osteoclasts per well, average size of osteoclasts, resorptive area (%), and resorptive area (%)/N.Oc (number of osteoclasts) were all decreased in Atp6v1h+/- group. n=3. C. TRAP staining (left) and toluidine blue staining (right) of bone slices. The decreased number of pits matched the reduced number of TRAP+ osteoclasts. The diameter of Atp6v1h+/- pits was smaller than for wild-type. n ≥100. D. Merged images for TRAP and F-actin staining. Both Atp6v1h+/- and wild-type cells could form actin rings. Bars=50μm. F. Western blot analysis showed the decreased ATP6V1H protein in Atp6v1h+/- cells induced by RANKL and M-CSF 1~ 4 day. F. Atp6v1h siRNA reduced the mRNA level of Atp6v1h in wild type osteoclasts and the formation of TRAP+ osteoclasts. n=3. G. Atp6v1h+/- osteoclasts showed a decreased V-ATPase activity which was inhibited by 10nM Bafilomycin A1. n=3. H. The intracellular pH of Atp6v1h+/- osteoclasts was increased by staining with lysosensor DND-160. n≥50. All data represent mean ±s.d. ** P<0.01, *P<0.05. NS: not significant difference.
Decreased interactions between osteoblasts and osteoclasts in Atp6v1h+/- mice. A. Comparison of osteogenic markers in newborn calvarial bone and its derived osteoblasts with Q-PCR analysis. Primary cultured Atp6v1h +/- osteoblasts with osteogenic induction had decreased Col1a1, Alp, and Bsp. n≥3. B. Atp6v1h siRNA decreased the mRNA level of Atp6v1h more than 50 percent in primary cultured osteoblasts (OB) and Mc3T3-E1 cells (M), and the mRNA level of Ocn and Opn were changed. n≥3. C. Alizarin red S and ALP staining showed that calvarial osteoblasts from wild-type and Atp6v1h+/- mice formed mineralized nodules but no difference existed between them. n≥3. D. Western results showed the decreased TGF-β1 in Atp6v1h+/- osteoclasts and unchanged TGF-β1 in Atp6v1h+/- osteoblasts, respectively. n=3. E. ELISA detection revealed the decreased active TGF-β1 in sera of Atp6v1h+/- mice. n≥8. F. The active TGF-β1 level in the supernatant of Atp6v1h+/- osteoclasts cultured on the bone slices was also decreased. Control: control medium. n≥3. G. Atp6v1h+/- osteoblasts with osteogenic induction secreted more active TGF-β1 by ELISA detection. No difference existed between wild-type and Atp6v1h+/- osteoblasts under a non-osteogenic induction condition. n≥3. H. Western blot results showed the decreased RANKL and unchanged OPG in Atp6v1h+/- osteoblasts. n≥3. I. ELISA results revealed the less soluble RANKL (sRANKL) in sera and the supernatant of Atp6v1h+/- osteoblasts with osteogenic induction. n≥3. J. OPG level was increased in sera of Atp6v1h+/- mice and the supernatant of Atp6v1h+/- osteoblasts with osteogenic induction. n≥3. K. Comparison of the number of osteoclasts in the co-culture system of osteoblasts and osteoclasts. When co-cultured with wild-type osteoblasts, wild-type and Atp6v1h+/- monocytes formed the same amount TRAP+ osteoclasts. When co-cultured with Atp6v1h+/- osteoblasts, fewer TRAP+ osteoclasts were formed in Atp6v1h+/- group. n≥3. All data represent mean ±s.d. ** P<0.01, *P<0.05. NS: not significant difference.
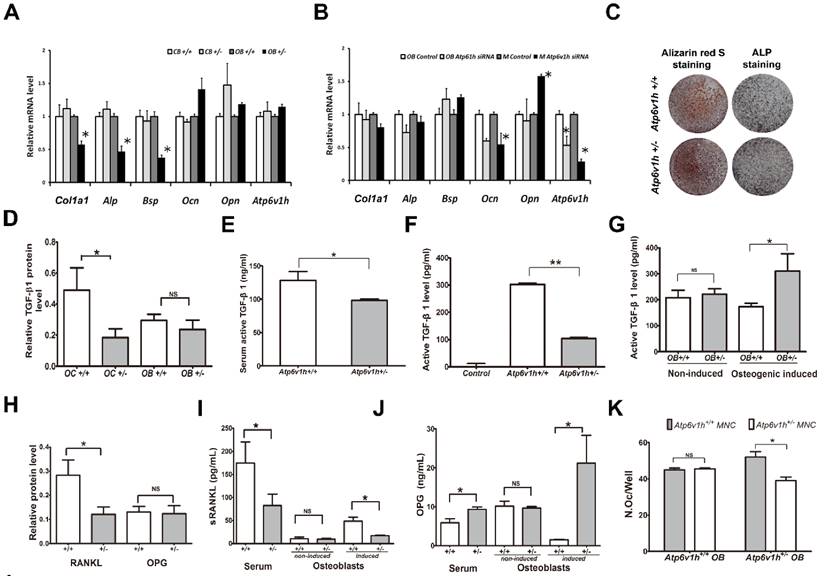
RANKL/OPG pathway was also analyzed to determine whether a lower bone formation rate downregulated the osteoclasts function. The protein level of RANKL was decreased in Atp6v1h +/- osteoblasts (Fig. 4H). The serum levels of RANKL and OPG were respectively decreased and increased in Atp6v1h +/- mice (Fig. 4I, J). The secretion of RANKL and OPG from Atp6v1h +/- osteoblasts following osteogenic induction was also respectively decreased and increased (Fig. 4I, J). The levels of RANKL and OPG from primary cultured osteoblasts were unchanged under non-osteogenic inducing conditions. In a co-culture system using different combinations of monocytes and osteoblasts from wild-type and heterozygous mice, co-culture of wild-type osteoblasts with wild-type and Atp6v1h+/- monocytes induced the same amount of TRAP-positive osteoclasts, whereas Atp6v1h+/- osteoblasts induced fewer osteoclasts from Atp6v1h+/- monocytes (Fig.4 K).
Discussion
Several genes are known to influence bone mass, including the vitamin D receptor, estrogen receptor, and collagen type I alpha I gene [19, 20]. In the present study, we report that defects of ATP6V1H are associated with bone matrix loss in mice and humans.
V-ATPase plays important roles in various cellular functions, such as acidification, endocytosis, and zymogen activation. The subunits of V-ATPase previously known to be critical for osteoclast function include those encoded by TCIRG1, ATP6V0D2, ATP6V1C1, and ATP6V1B2. Dysfunctions of these subunits manifest in osteopetrosis or osteopetrosis-related changes. The mechanisms involve changes in extracellular acidification, a fusion of preosteoclasts, and cytoskeletal F-actin assembly of osteoclasts [3, 8, 11, 17, 28, 29]. In this context, we expected that defects in ATP6V1H would impair osteoclast function; indeed, we discovered that ATP6V1H was highly expressed in osteoclasts and the expression level of ATP6V1H was decreased in Atp6v1h+/- osteoclasts. The reduced level of ATP6V1H in osteoclasts did not affect the formation of F-actin rings or ruffled borders of osteoclasts, but inhibited the number and size of TRAP+ osteoclasts, impaired V-ATPase activity, increased intracellular pH, and impaired osteoclastic bone resorption. Thus it appears that the key role of ATP6V1H is to promote osteoclast formation and maintain local acidification for bone resorption, rather than to affect osteoclast structure or movement.
We also anticipated that mutations of ATP6V1H homologs would impair osteoclast function and result in osteopetrosis, but instead detected that the in vivo effect of Atp6v1h+/- mice was bone loss. Atp6v1h+/- mice showed decreased bone density and serum phosphate level and increased serum calcium level. Bone formation and bone calcification were decreased with fewer and smaller osteoblasts and reduced ALP activity. One possibility to explain the bone loss in Atp6v1h+/- mice involves an imbalance of bone resorption and bone formation, altering bone homeostasis through coordinated cellular and molecular events. When the decrease in bone formation exceeds the decrease in bone resorption, the net effect is osteoporosis or osteopenia. Indeed, the impaired bone remodeling and a negative bone balance, causing profound osteoporosis, have been reported in other gene knockout mice [30-32], emphasizing the complex interactions and the delicate balance required for bone remodeling.
Although Atp6v1h mRNA could be detected in cranial bones, in primarily cultured osteoblasts, and in the osteoblastic cell line MC3T3-E1, we did not detect differences in the expression of osteogenic genes between wild-type and heterozygous mice, or when Atp6v1h was knocked down by siRNA. Thus, the decreased bone formation is probably secondary to osteoclasts instead of a cell-autonomous effect of ATP6V1H haploinsufficiency in osteoblasts. Research on ATP6V0D2 also revealed the importance of interactions between osteoclasts and osteoblasts [8, 9]. Thus, ATP6V1H is another subunit of V-ATPase that affects the coordination of osteoclast and osteoblastic functions as well as bone formation.
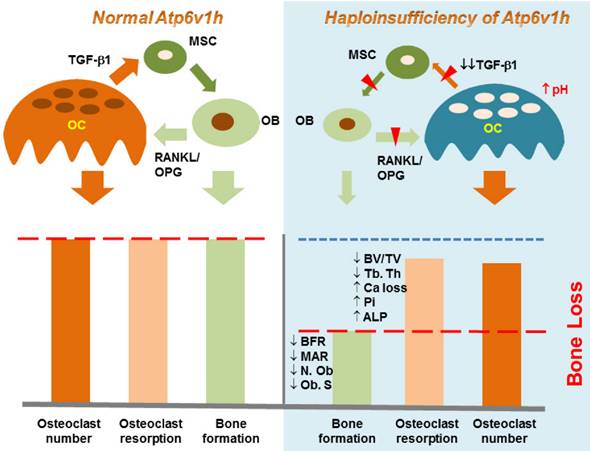
There are several pathways by which osteoclasts interact with osteoblasts, which arise from mesenchymal stem cells. TGF-β1 may stimulate the proliferation of mesenchymal stem cells and induce their differentiation into osteoblasts [33, 34]; the low concentration of TGF-β1 promotes the maturation of osteoclasts [35]. These process couple bone resorption with bone formation [26, 36, 37]. Our data showed decreased levels of active TGF-β1 in the sera of Atp6v1h+/- mice and the supernatant of Atp6v1h+/- osteoclasts cultured with bone slices. The reduced level of TGF-β1 may explain the differences between impaired Atp6v1h+/- osteoclasts in vitro and unexpected bone loss in Atp6v1h+/- mice in vivo. Reduced levels of TGF-β1 negatively regulate osteoblast differentiation, resulting in the fewer and smaller osteoblasts and finally causing the decreased bone formation in Atp6v1h+/- mice. Since the decrease in bone formation was greater than the decrease in osteoclast function, the final in vivo net effect was the bone loss (Fig. 5). pH is one of the important factors that can activate TGF-β [24, 25]. The increased intracellular pH resulted in decreased secretion of TGF-β1 from Atp6v1h+/- osteoclasts. We propose that TGF-β1 is the key factor that regulates the ATP6V1H-dependent osteoporosis changes.
The RANKL/OPG signaling pathway is important for the osteoclast differentiation. TGF-β1 level may trigger the expression of RANKL in osteoblasts. High concentrations of TGF-β1 upregulate the expression of OPG, which may inhibit RANKL-induced osteoclast differentiation [37]. In Atp6v1h+/- mice, the amount of RANKL and OPG secreted by osteoblasts was not changed when the cells were cultured in a non-osteogenic medium, but Atp6v1h+/- osteoblasts generated reduced RANKL and enhanced OPG signal under osteogenic conditions. This suggested that Atp6v1h+/- osteoblasts sent decreased signals to induce osteoclast formation only under calcifying conditions. Thus the bone remodeling of heterozygous mice was impaired.
Our population data identified alleles of ATP6V1H gene as novel risk factors for osteopenia/ osteoporosis implying ATP6V1H alleles might also influence the risk for osteopenia. The relationship between ATP6V1H and bone density suggests that Atp6v1h serves as a regulator of osteoclast formation and function. In summary, the ATP6V1H gene could be an important target for therapeutic interventions aimed at preventing bone resorption and treating osteoporosis [38].
Schematic map of haploinsufficiency of Atp6v1h and bone loss. TGF-β1 stimulates the proliferation of mesenchymal stem cells (MSC) and induces MSC to differentiate into osteoblasts (OB); TGF-β1 also promotes the maturation of osteoclasts (OC). The increased intracellular pH resulted in less secretion of TGF-β1 from Atp6v1h+/- osteoclasts. Decreased TGF-β1 negatively regulates osteoblasts differentiation, results in the fewer and smaller osteoblasts and finally causes the decreased bone formation in Atp6v1h+/- mice. The low level of TGF-β1 is associated with reduced RANKL and increased OPG signal, sending decreased signals to induce the osteoclasts formation. The decreased bone formation is more than decreased osteoclast function. Thus, the final in vivo net effect is bone loss.
Methods
Population study. A total of 1625 Chinese volunteers were recruited from Midwestern Chinese Han adults living in Xi'an and Changsha cities. The study was approved by Xi'an Jiaotong University. Signed informed consent documents were obtained from all study participants before entering the study. The following subjects were excluded from the study: Subjects with chronic diseases and conditions that might potentially affect bone mass, structure, or metabolism; subjects taking anti-bone resorptive or bone anabolic agents/drugs, such as bisphosphonates. BMD values at spine were measured using Hologic 4500W machines (Hologic Inc., Bedford, MA, USA). The coefficient of variation (CV) values of the dual-energy X-ray absorptiometry (DXA) measurements for spine was approximately 1.01% [22, 23]. Three groups were generated based on Z-score. Group 1 represented those with a Z-score above -1, Group 2 between -1 and -2 and Group 3 below -2.
Genomic DNA was extracted from peripheral blood leukocytes and SNP genotyping was performed using Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA) according to the manufacture's protocol. Data management and analyses were conducted using the Genotyping Command Console. The minimum call rate was above 95%. A total of 15 tag SNPs within the intronic region of ATP6V1H gene were included in subsequent association analyses. PLINK was used to test for association and meta-analysis and calculations were done using the METAL software package. A raw P value of < 0.05 in our study was considered nominally significant.
Atp6v1h knockout mouse generation using CRISPR/Cas9 mediated genome editing. The mouse Atp6v1h gene (Transcript: ENSMUSG00000033793) is located on mouse chromosome 1 and has 14 exons with an ATG start codon in exon 2 and a TGA stop codon in exon 14. Atp6v1h knockout mice were generated using the CRISPR/Cas9 system. Exon 2 of the Atp6v1h gene was chosen to introduce frame shift changes and create an early stop codon. The sequence of gRNA (5'-GAT GGA CAT TCG AGG TGC TGT GG-3') was selected based on the ZiFiT tool. The gRNA expression vectors were constructed using a gRNA Cloning Vector from the Church Lab [39] by Gibson Assembly and a diagram of the resulting gRNA expression vector was produced. Atp6v1h-specific gRNA and Cas9 mRNA were generated according to mMESSAGE mMACHINE T7 Kit (Life Technologies), and then co-injected into fertilized eggs of C57BL/6 mice to create knockout mice (Cyagen, China). PCR genotyping and sequencing confirmation were performed to verify the frame shift changes for Atp6v1h knockout mouse generation. PCR primers for mouse tail genotyping were: gRNA1-Forward: 5'-AGTCAATAGAAAGCCTTTTATGGGG-3', gRNA1-Reverse: 5'-CTGAGGGCACTCGCATTCAGGT-3'. The F0 founders had a 5bp (CGAGG) deletion and one base replacement (T>G) by the gRNA1/Cas9 procedure. All animals were treated in accordance with the ethical guidelines of the School of Stomatology, the Fourth Military Medical University (Xi'an, China).
Micro CT scanning and analysis. A micro CT scanner and Inveon Research Workplace (Siemens Inveon, Germany) were used for live animal scanning, tissue scanning and three-dimensional reconstruction. The femurs and vertebrae of mice were scanned with the parameters 80kV and 500μA, a 500 ms exposure time, and a scan angle of 360°. Three-dimensional images were reconstructed using standard methods. The bone density and other bone parameters were calculated and compared in the metaphyseal regions of distal femurs. The relative bone density, bone volume (BV/TV), bone surface (BS/TV), trabecular thickness (Tb.Th), trabecular number (Tb.N), trabecular separation (Tb.Sp), and cortical thickness (Ct. Th) of the femoral mid-diaphysis were calculated. The above trabecular morphometry measurements were also calculated in the third lumbar vertebrae of mice. Each group contained 8 mice.
Histology and histomorphometry. For dynamic histomorphometric analysis, 3-week-old littermate male wild-type and Atp6v1h +/- mice were used. Calcein at 10 mg/kg body weight and xylenol orange at 90 mg/kg were injected on days 1 and 8, respectively. The mice were sacrificed 2 days after the second injection. The femurs were removed and fixed in 70% ethanol at 4°C, then subjected to standard tissue calcification procedures. Plastic sections were cut at a thickness of 50 μm with a Leica SP2600 (Leica Microsystems, Germany). Images were captured by FluoView™ FV1000 Confocal Microscope (Olympus Corporation, Japan), and the mineral apposition rate (MAR, mcm/d), the mineralizing surface (MS/BS, %) and bone formation rate (BFR/BS, mcm3/mcm2/d) were measured. Each group contained 6 to 8 mice.
Plastic sections were stained using the modified ponceau red method. Trabecular morphometry parameters, such as bone volume (BV/TV) and Tb.Th, were calculated and compared in the distal region of the femur. In each group, five 4-week-old littermate male wild-type and Atp6v1h +/- mice were used. Each group contained 6 to 8 mice.
Femur samples from 3-months -old male wild-type and Atp6v1h +/- mice were fixed with 4% paraformaldehyde and demineralized with 10% EDTA in PBS (pH 7.2~7.4) for 20~30 days at 37℃, followed by gradient dehydration with ethanol, embedded in paraffin and sectioned at a thickness of 5~7 μm. The slides went through with hematoxylin & eosin staining and modified Ponceau red staining and were stained for ALP using a commercial kit (Jiancheng Bioengineering Institute, Nanjing, China). The distribution and intensity of ALP staining in the distal region of the femur were measured and compared. Each group contained 8 mice. ALP+ osteoblasts adjacent to bone surface (N.Ob/mm2) and the osteoblast surface (Ob.S/BS, %) were calculated following the rules recommended by the Nomenclature Committee of the American Society of Bone and Mineral Research [40].
Commercial TRAP kits (Sigma, US) were used for staining osteoclasts. TRAP+ osteoclasts adjacent to bone surface (N.Oc/T.A, /mm2) and the osteoclast surface (Oc.S/BS, %) were calculated following the recommended rules (2). Each group contained 8 mice.
Femur samples from 4-week-old male Atp6v1h +/- mice were fixed with 2.5% glutaraldehyde at 4℃ for 3 hours and 1% osmium tetroxide at 4℃ about 1 hours, respectively, then demineralized with 10% EDTA in PBS (pH 7.2~7.4) for 20~30 days at 37℃, followed by gradient dehydration and routine TEM method. The images were acquired by transmission electron microscope (FEI G2 Sprit, USA).
Immunohistochemistry staining. The sABC immunohistochemistry staining kit (Boster, Wuhan, China) was used for immunohistochemical staining of ATP6V1H following the manufacturer's protocols. Rabbit polyclonal IgG anti-ATP6V1H (Origene, China was used in this experiment at a dilution of 1:400).
In vitro assay of osteoclasts. Bone marrow (BM) cells were separated from the femur and tibia of 6-weeks-old male wild-type and Atp6v1h +/- mice and were quickly treated with Red Blood Cell Lysis Buffer (150 mM NH4Cl, 10mM KNCO3, 0.1 mM EDTA, pH 7.4) for 5min. The cells were cultured in the presence of M-CSF (50ng/ml) (R&D, USA) for 24 hours. After floating cells had been removed, the attached cells were used as osteoclast precursors and treated with M-CSF (50ng/ml) and RANKL (100ng/ml) (R&D, USA) for 7 days for the TRAP staining. The TRAP-positive multinucleated cells (≥5 nuclei per cells; >100μm in diameter) were counted. The cellular numbers (N.OC/well) and average areas (OC.S) were compared in the heterozygous and control groups.
Cells were fixed with 4% paraformaldehyde in PBS buffer and stained with rhodamine-phalloidin (Molecular Probes) for F-actin rings. In some experiments, TRAP staining was performed before F-actin ring staining.
BM cells were seeded on cattle bone slices (4mm×4mm×100μm) for 10-14 days with the RANKL and M-CSF treatment. The bone slices were fixed with 2.5% glutaraldehyde, stained with 0.1% toluidine blue and observed under a Leica DFC 550 Stereomicroscope (Leica Microsystems, Germany). The bone slices were also observed by scanning electron microscopy (Hitachi S-4800 high-resolution SEM, Japan). The number of resorptive lacunae of each bone slice was counted and the areas of the lacunae were measured using ImageJ software (NIH, USA) for at least 50 lacunae in each group.
The BM cells were also seeded on osteoassay surface polystyrene microplates (Corning, US) for 10 days with the RANKL and M-CSF treatment. The images of the wells were captured using a Leica DMI6000B microscope (Leica Microsystems, Germany). The number and size of osteoclasts areas, as well as the resorptive area (%) per well and osteoclast number normalized resorptive area (%) were measured using NIH Image J software (NIH, USA).
siRNA transfection. Atp6v1h siRNA against the nucleotide 324 position of mouse Atp6v1h (NM_133826.4) was designed and synthesized (GenePharma, China). The sequence of Atp6v1h siRNA is: sense (5' to 3'): AAG CAA AUG CUU CAA ATT; antisense (5' to 3'): UUU GAA GCA UCU CUU GCU UTT. The negative control sequence is: sense (5' to 3'): UUC UUC GAA CGU GUC ACG UTT; antisense (5' to 3'): ACG UGA CAC GUU CGG AGA ATT. The BM cells from 6-week -old male wild-type and Atp6v1h +/- mice were grown to 50% to 60% confluence and then transfected with siRNA using lipofectamine 2000 according to the manufacturer's protocols. The number of TRAP+ osteoclasts was determined and Q-PCR was performed.
Osteoblast functional assay. Calvarial osteoblasts (1 × 104 cells/well) were cultured in high glucose DMEM medium in the presence of 10% FBS (Gibco, US). For the osteogenic induction experiments, calvarial osteoblasts were grown in DMEM containing 10% FBS, 10mM β-glycerophosphate, 50 μg/ml ascorbic acid, and 10-7mM dexamethasone for 7 to 21 days. Osteogenic activity and mineralization nodule formation were assessed by staining with an ALP kit (Jiancheng Bioengineering Institute, Nanjing, China) and an alizarin red S kit (10 mg/ml, Sigma, US), respectively. Cell culture plates were photographed under a phase-contrast microscope using a Canon EF 100mm f/2.8 USM camera lens.
The third passage of wild-type osteoblasts as well as MC3T3-E1 cells were treated with Atp6v1h siRNA (same sequence as above), respectively. After 72 hours, the mRNA was collected and Q-PCR was performed as above. The mRNA of calvarial bone tissues from the newborn wild-type and heterozygous mice were also collected and Q-PCR was performed as above.
Co-culture of osteoblast and osteoclasts. Calvarial osteoblasts (1 × 103 cells/well) from wild-type and Atp6v1h+/- mice were cultured in 96 well plate high glucose DMEM medium in the presence of 10% FBS (Gibco, US). The monocytes from wild-type and Atp6v1h+/- mice were seeded on the osteoblasts with a ratio of 1:200 in the presence of 10-8 M 1α,25(OH)2D3 and 10-6 M PGE2 (Sigma, US). After 9 days, TRAP staining was performed and TRAP+ osteoclasts were calculated.
Measurement of V-ATPase activity. V-ATPase activity was determined according to the previously reported methods [41, 42]. Briefly, membrane proteins were extracted from osteoclasts after M-CSF/RANKL 4-day stimulation and quantified with BCA methods. 4 μg soluble membrane proteins reacted in 160 µATP reaction buffer (50 mM Tris-MOPS, 3 mM 2-mercaptoethanol, 20 mM KCl, 0.003% C12E10, 1 mM MgCl2, 20 mM NaCl, 3 mM Tris-HCl, 0.1 mM EDTA, 0.1 mM sodium orthovanadate, 0.5 mM sodium azide, 0.3% BSA, 1mM ATP, pH8.0). 10nM Bafilomycin A1 (Sigma) were used to inhibit V-ATPase specific activities. 2mM malachite solution was used to determine the final liberated phosphate in the reaction mixture by absorbance at 625 nm in a Biotek Synergy HTX (US). The final V- ATPase activity was expressed as mM/min/mg protein.
Analysis of intracellular pH. BM cells from wild-type and Atp6v1h+/- mice were treated with M-CSF and RANKL for 7 days, then stained with lysoSensor™ DND-160 (Invitrogen, US), and a pH calibration curve was performed at the end of each experiment to calibrate fluorescence intensity to actual pH values according to modified methods [43]. In each group, at least 50 cells were calculated and the experiment was repeated at least three times.
Biochemical and ELISA assay. Sera were collected from 3-month-old or 4-week-old male wild-type and Atp6v1h +/- mice after overnight fasting. The following items were measured as the methods described by the manufacturers: the concentrations of calcium (Ca2+) and phosphate ions (Pi) in sera (Jiancheng Bioengineering Institute, Nanjing, China), alkaline phosphatase (Jiancheng Bioengineering Institute, Nanjing, China), osteocalcin, CTX-I and TRAP (AMEKO, Shanghai, China), active TGF-β1, soluble RANKL (sRANKL), and OPG (R&D, US). Each group contained 8 to 10 mice.
The supernatant of osteoclasts cultured on the bone slices in 96-well plates was collected and the amount of active TGF-β1 was assayed by ELISA methods. The supernatant of cultured osteoblasts in a regular medium or an osteogenic induction medium were collected for the assays of active TGF-β1, soluble RANKL (sRANKL), and OPG as above.
Quantitative real-time polymerase chain reaction (Q-PCR). Total RNA from different cells or tissues were extracted using an RNA extraction kit (Omega, China). PrimeScriptTM RT Reagent Kit (Perfect RealTime) (TaKaRa, Japan) and SYBR® Premix Ex Tag TM Ⅱ (TaKaRa, Japan) were used to synthesize complementary DNA (cDNA) and quantitative PCR was performed with the ABI 7500 RT-PCR system (ABI, USA). All the primers involved in this study are listed in Table 4.
Western blot. Western blots were performed as described [44]. Antibodies included rabbit polyclonal IgG anti-ATP6V1H (1:1000, Origene, China), mouse anti-RANKL (1:500, Abcam, US) , rabbit anti-OPG (1:400, Abcam, US), rabbit anti TGF-β(1:500, Cell signaling, US), mouse anti-GAPDH monoclonal antibody (at a dilution of 1:1000, Comwin, Beijing, China), mouse anti-TUBLIN monoclonal antibody (at a dilution of 1:1000, Comwin, Beijing, China), and fluorophore-labelled goat anti-mouse or goat anti-rabbit secondary antibody (at a dilution of 1:10000, LI-COR, Lincoln, NE, USA). Bands were detected and quantified using an Odyssey image system (LI-COR, Lincoln, NE, USA).
PCR Primers.
| Gene Name | Forward primer (5'-3') | Reverse primer (5'-3') | Accession number | PCR product size (bp) |
|---|---|---|---|---|
| Gapdh | CATGTTCCAGTATGACTCCACTC | GGCCTCACCCCATTTGATGT | NM_001289726 | 136 |
| Atp6v1h | GGATGCTGCTGTCCCAACTAA | TCTCTTGCTTGTCCTCGGAAC | NM_133826 | 164 |
| Alp | CCAACTCTTTTGTGCCAGAGA | GGCTACATTGGTGTTGAGCTTTT | NM_001287172 | 110 |
| Col1a1 | TGACTGGAAGAGCGGAGAGT | GTTCGGGCTGATGTACCAGT | NM_007742 | 151 |
| Opn | TCTGATGAGACCGTCACTGC | AGGTCCTCATCTGTGGCATC | NM_001204201 | 170 |
| Bsp | CAGGGAGGCAGTGACTCTTC | AGTGTGGAAAGTGTGGCGTT | NM_008318 | 158 |
| Ocn | GGTAGTGAACAGACTCCGGC | ATAGCTCGTCACAAGCAGGG | NM_031368 | 107 |
Acknowledgements
This work was supported by National Natural Science Foundation of China (81470728) (X. D.). We thank Dr. Cornelius F. Boerkoel at the University of British Columbia and Dr. William A. Gahl at the National Human Genome Research Institute for their helpful comments.
Competing Interests
The authors declare no conflict of interest.
References
1. Cipriano DJ, Wang Y, Bond S, Hinton A, Jefferies KC, Qi J. et al. Structure and regulation of the vacuolar ATPases. Biochim Biophys Acta. 2008;1777:599-604
2. Yao G, Feng H, Cai Y, Qi W, Kong K. Characterization of vacuolar-ATPase and selective inhibition of vacuolar-H(+)-ATPase in osteoclasts. Biochem Biophys Res Commun. 2007;357:821-7
3. Frattini A, Orchard PJ, Sobacchi C, Giliani S, Abinun M, Mattsson JP. et al. Defects in TCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis. Nat Genet. 2000;25:343-6
4. Susani L, Pangrazio A, Sobacchi C, Taranta A, Mortier G, Savarirayan R. et al. TCIRG1-dependent recessive osteopetrosis: mutation analysis, functional identification of the splicing defects, and in vitro rescue by U1 snRNA. Hum Mutat. 2004;24:225-35
5. Scimeca JC, Quincey D, Parrinello H, Romatet D, Grosgeorge J, Gaudray P. et al. Novel mutations in the TCIRG1 gene encoding the a3 subunit of the vacuolar proton pump in patients affected by infantile malignant osteopetrosis. Hum Mutat. 2003;21:151-7
6. Sobacchi C, Frattini A, Orchard P, Porras O, Tezcan I, Andolina M. et al. The mutational spectrum of human malignant autosomal recessive osteopetrosis. Hum Mol Genet. 2001;10:1767-73
7. Li YP, Chen W, Liang Y, Li E, Stashenko P. Atp6i-deficient mice exhibit severe osteopetrosis due to loss of osteoclast-mediated extracellular acidification. Nat Genet. 1999;23:447-51
8. Lee SH, Rho J, Jeong D, Sul JY, Kim T, Kim N. et al. v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat Med. 2006;12:1403-9
9. Kim T, Ha H, Kim N, Park ES, Rho J, Kim EC. et al. ATP6v0d2 deficiency increases bone mass, but does not influence ovariectomy-induced bone loss. Biochem Biophys Res Commun. 2010;403:73-8
10. Wu H, Xu G, Li YP. Atp6v0d2 is an essential component of the osteoclast-specific proton pump that mediates extracellular acidification in bone resorption. J Bone Miner Res. 2009;24:871-85
11. Feng S, Deng L, Chen W, Shao J, Xu G, Li YP. Atp6v1c1 is an essential component of the osteoclast proton pump and in F-actin ring formation in osteoclasts. Biochem J. 2009;417:195-203
12. Lee BS, Holliday LS, Krits I, Gluck SL. Vacuolar H+-ATPase activity and expression in mouse bone marrow cultures. J Bone Miner Res. 1999;14:2127-36
13. Vitavska O, Wieczorek H, Merzendorfer H. A novel role for subunit C in mediating binding of the H+-V-ATPase to the actin cytoskeleton. J Biol Chem. 2003;278:18499-505
14. Perez-Sayans M, Suarez-Penaranda JM, Barros-Angueira F, Diz PG, Gandara-Rey JM, Garcia-Garcia A. An update in the structure, function, and regulation of V-ATPases: the role of the C subunit. Braz J Biol. 2012;72:189-98
15. Holliday LS, Lu M, Lee BS, Nelson RD, Solivan S, Zhang L. et al. The amino-terminal domain of the B subunit of vacuolar H+-ATPase contains a filamentous actin binding site. J Biol Chem. 2000;275:32331-7
16. Chen SH, Bubb MR, Yarmola EG, Zuo J, Jiang J, Lee BS. et al. Vacuolar H+-ATPase binding to microfilaments: regulation in response to phosphatidylinositol 3-kinase activity and detailed characterization of the actin-binding site in subunit B. J Biol Chem. 2004;279:7988-98
17. Yuan Y, Zhang J, Chang Q, Zeng J, Xin F, Wang J. et al. De novo mutation in ATP6V1B2 impairs lysosome acidification and causes dominant deafness-onychodystrophy syndrome. Cell Res. 2014;24:1370-3
18. Kortum F, Caputo V, Bauer CK, Stella L, Ciolfi A, Alawi M. et al. Mutations in KCNH1 and ATP6V1B2 cause Zimmermann-Laband syndrome. Nat Genet. 2015;47:661-7
19. Raisz LG. Pathogenesis of osteoporosis: concepts, conflicts, and prospects. J Clin Invest. 2005;115:3318-25
20. Peacock M, Turner CH, Econs MJ, Foroud T. Genetics of Osteoporosis. Endocrine Reviews. 2002;23:303-26
21. Tan LJ, Wang ZE, Wu KH, Chen XD, Zhu H, Lu S. et al. Bivariate Genome-Wide Association Study Implicates ATP6V1G1 as a Novel Pleiotropic Locus Underlying Osteoporosis and Age at Menarche. The Journal of Clinical Endocrinology & Metabolism. 2015;100:E1457-E66
22. Yang TL, Guo Y, Liu YJ, Shen H, Liu YZ, Lei SF. et al. Genetic variants in the SOX6 gene are associated with bone mineral density in both Caucasian and Chinese populations. Osteoporos Int. 2012;23:781-7
23. Yang TL, Guo Y, Li J, Zhang L, Shen H, Li SM. et al. Gene-gene interaction between RBMS3 and ZNF516 influences bone mineral density. J Bone Miner Res. 2013;28:828-37
24. Pircher R, Jullien P, Lawrence DA. Beta-transforming growth factor is stored in human blood platelets as a latent high molecular weight complex. Biochem Biophys Res Commun. 1986;136:30-7
25. Miyazono K, Hellman U, Wernstedt C, Heldin CH. Latent high molecular weight complex of transforming growth factor beta 1. Purification from human platelets and structural characterization. J Biol Chem. 1988;263:6407-15
26. Tang Y, Wu X, Lei W, Pang L, Wan C, Shi Z. et al. TGF-[beta]1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat Med. 2009;15:757-65
27. Zhen G, Wen C, Jia X, Li Y, Crane JL, Mears SC. et al. Inhibition of TGF-[beta] signaling in mesenchymal stem cells of subchondral bone attenuates osteoarthritis. Nat Med. 2013;19:704-12
28. Qin A, Cheng TS, Pavlos NJ, Lin Z, Dai KR, Zheng MH. V-ATPases in osteoclasts: structure, function and potential inhibitors of bone resorption. Int J Biochem Cell Biol. 2012;44:1422-35
29. Vitavska O, Merzendorfer H, Wieczorek H. The V-ATPase subunit C binds to polymeric F-actin as well as to monomeric G-actin and induces cross-linking of actin filaments. J Biol Chem. 2005;280:1070-6
30. Delany AM, Amling M, Priemel M, Howe C, Baron R, Canalis E. Osteopenia and decreased bone formation in osteonectin-deficient mice. J Clin Invest. 2000;105:915-23
31. Zhao H, Ito Y, Chappel J, Andrews NW, Teitelbaum SL, Ross FP. Synaptotagmin VII regulates bone remodeling by modulating osteoclast and osteoblast secretion. Dev Cell. 2008;14:914-25
32. Qiu N, Xiao Z, Cao L, Buechel MM, David V, Roan E. et al. Disruption of Kif3a in osteoblasts results in defective bone formation and osteopenia. J Cell Sci. 2012;125:1945-57
33. Wang X, Zhen L, Miao H, Sun Q, Yang Y, Que B. et al. Concomitant Retrograde Coronary Venous Infusion of Basic Fibroblast Growth Factor Enhances Engraftment and Differentiation of Bone Marrow Mesenchymal Stem Cells for Cardiac Repair after Myocardial Infarction. Theranostics. 2015;5:995-1006
34. Rahmi G, Pidial L, Silva AK, Blondiaux E, Meresse B, Gazeau F. et al. Designing 3D Mesenchymal Stem Cell Sheets Merging Magnetic and Fluorescent Features: When Cell Sheet Technology Meets Image-Guided Cell Therapy. Theranostics. 2016;6:739-51
35. Karsdal MA, Hjorth P, Henriksen K, Kirkegaard T, Nielsen KL, Lou H. et al. Transforming growth factor-beta controls human osteoclastogenesis through the p38 MAPK and regulation of RANK expression. J Biol Chem. 2003;278:44975-87
36. Nishimura R. A novel role for TGF-[beta]1 in bone remodeling. IBMS BoneKEy. 2009;6:434-8
37. Kasagi S, Chen W. TGF-beta1 on osteoimmunology and the bone component cells. Cell Biosci. 2013;3:4
38. Millard M, Odde S, Neamati N. Integrin targeted therapeutics. Theranostics. 2011;1:154-88
39. Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE. et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823-6
40. Dempster DW, Compston JE, Drezner MK, Glorieux FH, Kanis JA, Malluche H. et al. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 2013;28:2-17
41. Huss M, Ingenhorst G, Konig S, Gassel M, Drose S, Zeeck A. et al. Concanamycin A, the specific inhibitor of V-ATPases, binds to the V(o) subunit c. J Biol Chem. 2002;277:40544-8
42. Wieczorek H, Cioffi M, Klein U, Harvey WR, Schweikl H, Wolfersberger MG. Isolation of goblet cell apical membrane from tobacco hornworm midgut and purification of its vacuolar-type ATPase. Methods in Enzymology Biomembranes Part W: Cellular and Subcellular Transport: Epithelial Cells. 192 ed: Academic Press. 1990:608-16
43. McCurley N, Mellman I. Monocyte-Derived Dendritic Cells Exhibit Increased Levels of Lysosomal Proteolysis as Compared to Other Human Dendritic Cell Populations. PLoS One. 2010;5:e11949
44. Yang T, Zhang Y, Li Y, Hao Y, Zhou M, Dong N. et al. High amounts of fluoride induce apoptosis/cell death in matured ameloblast-like LS8 cells by downregulating Bcl-2. Arch Oral Biol. 2013;58:1165-73
Author contact
![]() Corresponding author: Xiaohong Duan, State Key Laboratory of Military Stomatology, National Clinical Research Center for Oral Diseases, Shaanxi Key Laboratory of Oral Diseases, Department of Oral Biology, Clinic of Oral Rare and Genetic Diseases, School of Stomatology, the Fourth Military Medical University, 145 West Changle Road, Xi'an 710032, P. R. China. Tel: 86-29-84776169; Fax: 86-29-84776169; E-mail: xhduanedu.cn.
Corresponding author: Xiaohong Duan, State Key Laboratory of Military Stomatology, National Clinical Research Center for Oral Diseases, Shaanxi Key Laboratory of Oral Diseases, Department of Oral Biology, Clinic of Oral Rare and Genetic Diseases, School of Stomatology, the Fourth Military Medical University, 145 West Changle Road, Xi'an 710032, P. R. China. Tel: 86-29-84776169; Fax: 86-29-84776169; E-mail: xhduanedu.cn.
 Global reach, higher impact
Global reach, higher impact