13.3
Impact Factor
Theranostics 2016; 6(10):1672-1682. doi:10.7150/thno.16135 This issue Cite
Research Paper
RGD Peptide Cell-Surface Display Enhances the Targeting and Therapeutic Efficacy of Attenuated Salmonella-mediated Cancer Therapy
1. Department of Nuclear Medicine, Chonnam National University Medical School, Gwangju, Republic of Korea;
2. Department of Microbiology, Chonnam National University Medical School, Gwangju, Republic of Korea;
3. Department of Nuclear Medicine, Haikou Hospital Affiliated to Xiangya School of Medicine, Central South University, China;
4. Fraunhofer IZI, Leipzig, Germany;
5. Department of Biology, Kyung Hee University College of Science, Seoul, Republic of Korea;
6. Biological Resource Center, Korea Research Institute of Bioscience and Biotechnology (KRIBB), Jeongeup, Republic of Korea.
Received 2016-1-3; Accepted 2016-5-16; Published 2016-6-20
Abstract
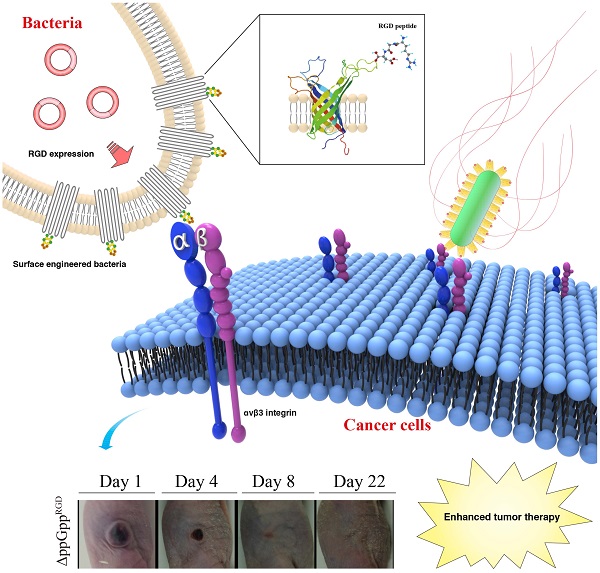
Bacteria-based anticancer therapies aim to overcome the limitations of current cancer therapy by actively targeting and efficiently removing cancer. To achieve this goal, new approaches that target and maintain bacterial drugs at sufficient concentrations during the therapeutic window are essential. Here, we examined the tumor tropism of attenuated Salmonella typhimurium displaying the RGD peptide sequence (ACDCRGDCFCG) on the external loop of outer membrane protein A (OmpA). RGD-displaying Salmonella strongly bound to cancer cells overexpressing αvβ3, but weakly bound to αvβ3-negative cancer cells, suggesting the feasibility of displaying a preferential homing peptide on the bacterial surface. In vivo studies revealed that RGD-displaying Salmonellae showed strong targeting efficiency, resulting in the regression in αvβ3-overexpressing cancer xenografts, and prolonged survival of mouse models of human breast cancer (MDA-MB-231) and human melanoma (MDA-MB-435). Thus, surface engineering of Salmonellae to display RGD peptides increases both their targeting efficiency and therapeutic effect.
Keywords: Salmonella typhimurium, RGD peptide, bacteria-mediated cancer therapy, surface display, bioluminescence imaging.
Introduction
Attenuated Salmonella typhimurium have been used for cancer therapy in animal models of breast cancer [1, 2], colon cancer [2, 3], hepatocellular carcinoma [4, 5], melanoma [2, 6], neuroblastoma [7], pancreatic cancer [8, 9], prostate cancer [10], and spinal cord glioma [11]. Some bacteria have the ability to target tumors, actively proliferate in tumors, and induce anticancer effects [12]. Attenuated S. typhimurium that are defective in the synthesis of ppGpp (strain ΔppGpp) suppress tumor growth by activating the immune system via the release of TNF-α and IL-1β[13, 14]. These tumor suppressing effects correlate with the persistence of Salmonella in tumor tissue[13]. Thus, the targeting efficiency and proliferation of bacteria in tumor tissue appear important. We found that ΔppGpp S. typhimurium showed variable targeting efficiency when tested in mouse xenograft models. Clinical trials in metastatic melanoma patients demonstrate that attenuated S. typhimurium (VNP20009) show weak targeting ability, with no induction of regression [15, 16]. Therefore, high tumor targeting efficiency is essential to increase the tumor suppressive effects of bacterial cancer therapy.
Microbial cell-surface display systems allow peptides and proteins to be displayed on the surface of microbial cells by fusing them with an anchoring motif; these motifs are usually cell-surface proteins or fragments thereof (carrier proteins). This system has both biotechnology and industry applications [17], including live vaccine development [18], screening-displayed peptide library construction [19], antibody production [20], bioadsorbent manufacture [21], and biosensor development [22]. Only a few studies have explored the influence of surface engineering, involving the attachment of tumor-specific ligands to outer membrane proteins, on tumor targeting efficiency by bacteria [23, 24]. Chang et al. showed that the display of anti-HER2/neu affibody on the surface of Escherichia coli resulted in the bacteria selectively targeting HER2-positive cancer cells in vitro [23]. In a separate study, Massa et al. displayed an anti-CD20 single domain antibody on attenuated S. typhimurium carrying a prodrug-converting enzyme and used it to treat mice bearing human lymphoma [24]. The previous studies were designed to study only the effects of bacteria on hematologic cancers [24] or were confined to an in vitro study of solid tumor cells [23]. Thus, there is a need to systematically evaluate the performance of engineered bacteria displaying tumor-specific ligands on their surfaces in diverse in vitro and in vivo models.
In this study, we used a novel, easy and straightforward approach to generate engineered Salmonella ΔppGpp strains displaying the arginine-glycine-aspartate (RGD) peptide on the outer membrane protein A (OmpA). The RGD peptide is a well-studied tumor homing peptide that specifically binds to alpha v beta 3 (αvβ3) integrin, which is widely overexpressed on cancer cells and blood vessels during cancer angiogenesis. Here, we demonstrate for the first time the successful engineering of attenuated Salmonella ΔppGpp strains that show enhanced tumor targeting and tumor regression in αvβ3-overexpressing tumor xenograft models.
Materials and methods
Cells
The MDA-MB-231 (human breast cancer), MDA-MB-435 (human melanoma), U87MG (human glioblastoma), MCF7 (human breast cancer), ASPC-1 (human pancreatic cancer), CT-26 (mouse colon carcinoma), and 4T1 (mouse breast cancer) cell lines were obtained from the American Type Culture Collection (HTB-26, HTB-129, HTB-14, HTB-22, CRL-1682, CRL-2638, and CRL-2539, respectively). The MC38 mouse colonic adenocarcinoma cell line was obtained from Dr. Je-Jung Lee (Chonnam National University Hwasun Hospital, Jeonnam, Republic of Korea). M21 and M21L human melanoma cells were kindly provided by Dr. Hak Soo Choi (Beth Israel Deaconess Medical Center of Harvard Medical School, Boston, USA). Cells were grown in α-MEM (U87MG), RPMI 1640 (MDA-MB-231, MDA-MB-435, MCF7, and ASPC-1), or high-glucose DMEM (M21, M21L, MC38, CT-26, and 4T1) (HyClone Lab, Inc., Logan, UT) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin, and cultured at 37°C in a humidified atmosphere of 5% CO2. Cells were counted using a hemocytometer and seeded into 6-well plates (3 × 105 cells per well) or culture dishes (1 × 106 cells per dish). The culture medium was changed every 2 days until cells reached 80% confluence.
Salmonella typhimurium strains and plasmids.
| Strains/plasmids | Description | Reference |
|---|---|---|
| Salmonella typhimurium | Wild type | |
| ΔppGpp (SHJ2037) | ΔrelA, ΔspoT | Song M et al. [25] |
| ΔppGpp ΔompA | ΔrelA, ΔspoT, ΔompA::cat | This study |
| ΔppGppAAA | ΔrelA, ΔspoT, OmpAAAA | This study |
| ΔppGppRGD | ΔrelA, ΔspoT, OmpARGD | This study |
| pRLu8 | pBAD-RLuc8 | Loening AM et al.[29] |
| pOmpA | pBAD-OmpA | This study |
| pOmpAAAA | pBAD-OmpA containing the AAA peptide | This study |
| pOmpARGD | pBAD-OmpA containing the RGD peptide | This study |
Bacterial strains and plasmids for surface display
The bacterial strains and plasmids used in this study are listed in the Table 1. ΔppGpp S. typhimurium, SHJ2037 (ΔrelA::km, Δspot::cm), has been previously described [25]. To image bioluminescence, the whole luciferase operon of Photorhabdus luminescens from S. typhimurium-Xen26 (Caliper Life Science, USA) was transduced into strain SHJ2037 by P22HT int transduction [26]. ΔppGpp ΔompA was constructed from ΔppGpp S. typhimurium using the linear DNA transformation method as described previously [27]. PCR amplification of pKD13 was performed to replace the ompA open reading frame with the cat gene. The primers were as follows: forward, 5'-ATGAAAAAGACAGCTATCGCGATTGCAGTGGCACTGGCTGGTTTCGCTACCGTAGCGCAGGTGTAGGCTGGAGCTGCTTCG-3'; and reverse, 5'-TTAAGCCTGCGGCTGAGTTACCACGTCTTTAACGCCTTTAACTTCGATCTCTACGCGACGCATATGAATATCCTCCTTA-3'. The cat gene was then removed to generate ΔompA, as described by Datsenko and Wanner [27].
The expression vectors for pOmpAAAA or pOmpARGD were constructed as follows. The ompA gene was amplified from S. typhimurium genomic DNA[28] using forward primer, 5'-CCATGGCAATGAAAAAGACAGCTATCGCGATTGCAGTG-3' and reverse primer, 5'-GTTTAAACTTAAGCCTGCGGCTGAGTTACAAC-3'. The PCR product was cloned directly into the pGEM-T Easy Vector (Promega, Madison, WI) to generate pOmpA. Recombinant DNA was cut by NcoI and PmeI, and used to replace RLuc8 at the same site in the pRLuc8 [3, 29] expression vector. To construct the plasmid encoding the ompA gene containing RGD-4C (ACDCRGDCFCG), amplified DNA was inserted via the synthetic oligomer RGD-4C (5'-GCCTGTGATTGTCGCGGCGATTGTTTCTGTGGC-3'). The synthetic oligomer AAA-4C (5'-GCCTGTGATTGTGCGGCGGCGTGTTTCTGTGGC-3') was used as a control. The oligomers were inserted into the third transmembrane domain of ompA gene to generate a sandwich fusion (Fig. 1A, B).
Preparation of Salmonellae for the in vitro and in vivo experiments
All bacterial strains were streaked on LB plates containing appropriate antibiotics. A single colony was picked, dipped into LB/antibiotic medium containing 0.2% glucose, and grown overnight in a shaking incubator (37°C, 200 rpm). The next day, the overnight culture was diluted 50-fold into fresh medium containing L-arabinose (final concentration, 0.2%) to express recombinant proteins and grown to early stationary phase (A600 = 2-2.5). The cells were then harvested by centrifugation (4000 × g for 10 minutes), washed with PBS, quantified by spectrophotometry, and diluted again in PBS to obtain the desired concentration of bacteria in an appropriate volume for the in vitro and in vivo experiments. For the animal experiments, each mouse received 3 × 107 colony-forming units suspended in 100 μL of PBS. The bacterial count was calculated as follows: 1.0 A600 = 0.8 × 109 CFU.
Western blot analysis
A bacterial pellet (40 μg of total proteins) was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 12% linear gradient gels and then transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA). The membranes were first probed with an anti-MOMP monoclonal antibody (1:2000; Geneway Biotech, San Diego, CA), followed by a horseradish peroxidase-conjugated anti-mouse secondary antibody (1:2000; Amersham, UK). Immunoreactive proteins were detected using Luminol reagent (Santa Cruz Biotechnology, Santa Cruz, CA) and visualized by a Fuji Film image reader LAS-3000 machine.
Invasion assays for bacterial infection
Human cancer cells (MCF7, M21L, U87MG, M21, MDA-MB-231, and MDA-MB-435) were seeded into the individual wells of 24 well plates at 5 × 105 cells per well. Bacteria strains prepared as described above were added to the cancer cells at a ratio of 100:1, and the mixture was incubated at 37°C under 5% CO2 for 12 h. Infected cells were washed three times with PBS, cancer cell line- optimal medium containing gentamycin (5 mg/mL, Sigma) was added, and the mixture was incubated for an additional 60 min. Intracellular bacteria were harvested by extraction with lysis buffer (0.05% Triton X-100 in PBS), and replica plated for colony counting on LB agar plates containing appropriate antibiotics.
Adhesion and competition assays
Human cancer cells were seeded into 6-well plates containing cover slips (to enable observation under a microscope). Bacteria strains were grown and prepared as described above. The harvested bacteria were washed twice with serum-free medium, diluted in serum-free medium, and added to each well to achieve the desired multiplicity of infection (MOI, 1:100). For the competition assay, cancer cells were incubated with the synthetic RGD peptide (final concentration, 1 μM; AnyGen Inc., Gwangju, Republic of Korea) for 2 h at 37°C before addition of RGD-displaying bacteria. The incubation was performed in a humidified atmosphere of 5% CO2 at 37°C for 2 h. The culture plates were then washed three times with PBS to remove unbound bacteria. The treated cells were examined by immunofluorescence staining (see below). Each condition was tested in triplicate, and at least three separate experiments were performed.
Immunofluorescence analysis
After bacterial infection, cells were washed with PBS and fixed in 3.7% paraformaldehyde for 10 minutes at room temperature. Cells were then washed three times with PBS and subsequently blocked with 5% BSA in PBS-T (containing 0.1% Triton X-100; Sigma-Aldrich, St. Louis, MO). For observation, cells were stained with 5 μg/mL phalloidin-555 (Molecular Probes, CA) and 1 μg/mL DAPI (Molecular Probes, CA). Alternatively, the BSA-blocked cells were stained with a mouse anti-αvβ3 primary antibody (1:100; ab7166, Abcam, Cambridge, UK) and a rabbit anti-Salmonella antibody (1:100; ab35156, Abcam, Cambridge, UK) overnight at 4°C. After washing three times with PBS-T, cells were incubated with anti-mouse IgG coupled to Alexa Fluor 555 (1:500; Molecular Probes) and anti-rabbit IgG coupled to Alexa 488 (1:500; Molecular Probes) for 2 h at room temperature. Stained cells were subsequently washed with PBS, mounted on glass slides, and analyzed under an FV1000D confocal laser scanning microscope (Olympus, Tokyo, Japan). For binding analysis, a composite image of 20 sections with a 0.5 μm shift in the z-axis was taken and combined using FV10-ASW 2.0 viewer. The number of adherent bacteria was determined by counting at least 100 high power fields (320 μm × 240 μm). Calculation of exact cell numbers was based on the measured areas and the overall size of the cover slip.
Animal models
Five- to six-week-old male nude (nu-/nu-) and C57BL/6 mice (20-25 g) were purchased from the Central Lab Animal Inc. (Republic of Korea). Animal care, experiments, and euthanasia were performed in accordance with protocols approved by the Chonnam National University Animal Research Committee (CNU IACUC-H-2014-1). Human cancer cell lines (MDA-MB-231, MDA-MB-435, M21, M21L, ASPC-1, and U87MG; 107 cells) and mouse cancer cell lines (MC38, CT-26, and 4T1; 106 cells) were harvested and suspended in 100 μL of PBS prior to subcutaneous injection into the right thigh of each mouse. When the tumor volume reached about 100 mm3, PBS, synthetic RGD peptide (5 μg), or bacteria strains (3 × 107 CFU) were injected intravenously. Mice were anesthetized with 2% isoflurane (for imaging) or a mixture of ketamine and xylazine (200 mg/kg) (for implantation). The subcutaneous volumes of the MDA-MB-231 and MDA-MB-435 xenografts were measured with calipers every 2-3 days from Days 0 to 40. Tumor volume (mm3) was estimated using the formula (L × H × W) / 2, where L is the length, W is the width, and H is the height of the tumor in millimeters. Mice with tumors ≥ 1000 mm3 were scheduled for euthanasia.
Optical bioluminescence imaging
To image bacterial bioluminescence, anesthetized animals were placed in a light-tight chamber within the IVIS imaging system (Caliper), which was equipped with a cooled charge-coupled device camera. Photons emitted from bioluminescent bacteria were collected and integrated over 1 minute periods [26]. Pseudocolor images indicating photon counts were overlaid on photographs of mice using Living Image software v. 2.50 (Caliper). A region of interest was selected manually. Based on the signal intensity, the average radiance within each region of interest was recorded.
Accumulation of engineered Salmonella
Animals were euthanized and sacrificed at the indicated dpi. Tissues were removed, placed individually in sterile tubes containing PBS at 4°C, and weighed. Samples were transferred to sterile homogenization tubes, homogenized, and then returned to their original tubes for serial dilution with PBS. Agar plates containing appropriate antibiotics were inoculated with diluted homogenate and incubated overnight at 37°C. Colonies were counted and the bacterial load was expressed as CFU g-1 tissue.
Clinical chemistry parameters
Blood samples were collected from all groups of animals at 4 days after the injection of bacteria. Samples were obtained by cardiac puncture using heparinized syringes. Blood samples were deposited in serum separator gel tubes (Microtainer, Becton Dickinson, Franklin, NJ) and centrifuged (9300 × g, 30 minutes at 4°C) to separate the serum. After centrifugation, serum samples were immediately subjected to biochemical analyses. Serum activity of aspartate aminotransferase and alanine transaminase, and the concentration of blood urea nitrogen, creatinine, CRP, and PCT, were measured in an automated analyzer (Hitachi instruments, Tokyo, Japan), according to the manufacturers' instructions. Standard controls were run before each measurement, and the values obtained were within the expect ranges.
Statistical analyses
Statistical analysis was performed using the SPSS 21.0 statistical package (SPSS, Chicago, IL). Two-tailed Student's t-tests and one-way ANOVA were used to determine the statistical significance of differences in primary tumor growth between control and treatment groups. The Kruskal-Wallis test was used to determine the statistical significance of differences in clinical chemistry parameters. A P-value < 0.05 was considered significant for all analyses. Survival analysis was performed using the Kaplan-Meier method and the log-rank test. All data are expressed as the mean ± standard deviation.
Results
Engineering Salmonellae to express the RGD sequence on their surfaces
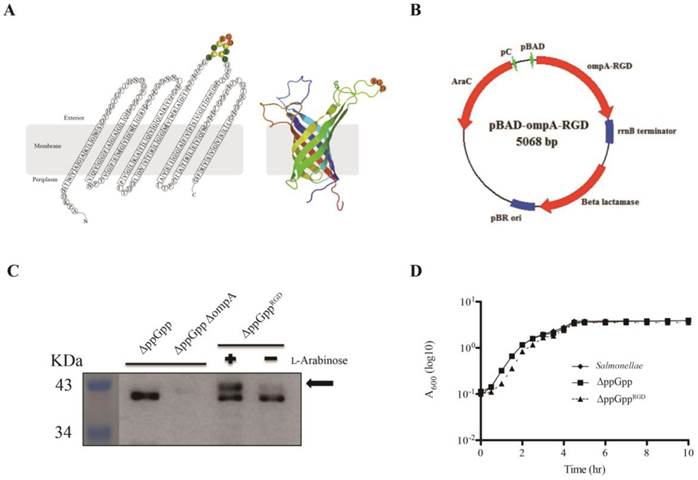
Although the tumor tropism of ΔppGpp Salmonellae is well documented [3, 5], we observed variable levels of accumulation of bacteria depending on tumor type; for example, accumulation was higher in solid tumor xenografts (ASPC-1, MC38, CT26, and 4T1) than in MDA-MB-231, MDA-MB-435, M21, and U87MG tumor xenografts (P < 0.01, Fig. S1A, B). Therefore, to increase tumor specificity we engineered ΔppGpp Salmonellae to express the RGD peptide, a specific ligand for αvβ3 integrin expressed by tumor cells and tumor endothelial cells. The RGD sequence (ACDCRGDCFCG) was expressed within the third loop of OmpA (Fig. 1A). The fusion gene, OmpARGD, was then substituted for the RLuc8 gene in the pRLuc8 plasmid [3] to generate the pOmpARGD plasmid (Fig. 1B). ΔppGpp Salmonellae were electro-transformed with the pOmpARGD vector and then examined by Western blotting with an anti-OmpA antibody. The results confirmed successful expression of the fused OmpARGD protein (which was larger than the endogenous OmpA protein (37 kDa)) only after L-arabinose induction (Fig. 1C). Comparison with the parental strain confirmed that growth of transformed Salmonellae was not affected by OmpARGD induction (Fig. 1D).
L-Arabinose-induced OmpARGD expression by surface-engineered S. typhimurium (ΔppGppRGD). (A) A topological model of S. typhimurium OmpA after insertion of the RGD sequence (ACDCRGDCFCG), and its secondary structure. The image is based on the expected RGD sequence/OmpA structure (Protein Data Bank (PDB) ID code 2GE4). (B) A map of the bacterial expression plasmid (pOmpARGD). (C) The ppGpp-defective S. typhimurium strain (ΔppGpp) was transformed with pOmpARGD (ΔppGppRGD). Protein production by ΔppGpp, ΔppGpp ΔompA, and ΔppGppRGD strains was induced by L-arabinose. The expression of OmpA (37 kDa) was analyzed by Western blotting with an anti-major outer membrane protein (MOMP ) antibody. The position of OmpARGD is indicated by an arrow. (D) The growth of S. typhimurium was assessed using the wild type or in untransformed ΔppGpp or transformed ΔppGppRGD after induction of OmpARGD expression. To induce the pBAD system in cultured bacteria, 0.2% L-arabinose was added to cultures at mid-log phase (1 h after starting the culture). The A600 was determined every 1 or 1.5 h for a period of 10 h. Results are representative of at least three independent experiments.
RGD-displaying Salmonellae show enhanced binding to cancer cells overexpressing αvβ3 integrin
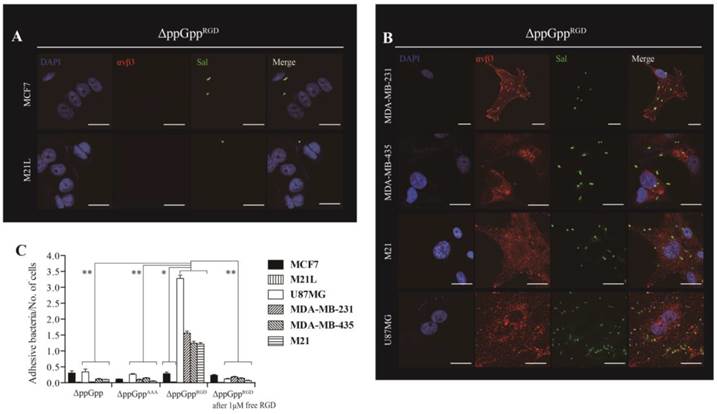
Next, we asked whether expressing RGD on the surface of Salmonellae would direct the bacteria to cells expressing αvβ3 integrin on their membranes. Thus, αvβ3-positive (MDA-MB-231, MDA-MB-435, M21, and U87MG) and -negative (MCF7 and M21L) cells were incubated for 2 h with ΔppGpp Salmonellae displaying RGD (ΔppGppRGD strain). ΔppGpp Salmonellae (the parental ΔppGpp strain) and ΔppGpp Salmonellae expressing a triple alanine peptide (AAA; ΔppGppAAA strain) in OmpA were used as controls. The ΔppGppRGD strain bound MDA-MB-231, MDA-MB-435, M21, and U87MG cancer cells to a greater extent than MCF7 and M21L cells (P = 0.008, Fig. 2). Binding was significantly inhibited by the addition of 1 µM free synthetic RGD peptide (P = 0.001), suggesting that bacterial binding to cells was dependent upon surface expression of RGD. Control strains showed negligible binding to αvβ3-positive and -negative cell lines (Fig. 2C, S2). Binding was further confirmed by Z section analysis using a confocal microscope. The merged image clearly indicated co-localization of the ΔppGppRGD strain with αvβ3 integrins expressed by MDA-MB-231 cells (Fig. S3). Despite the high binding affinity, the ΔppGppRGD strain showed extremely low invasion of cancer cells expressing αvβ3; less than 0.1% of the cells were invaded compared with WT Salmonellae (Fig. S4). Taken together, these results indicate that displaying the RGD peptide on the bacterial surface results in strong binding to αvβ3 integrin expressed by cancer cells.
Enhanced targeting efficiency of RGD-displaying Salmonellae in xenograft mouse models
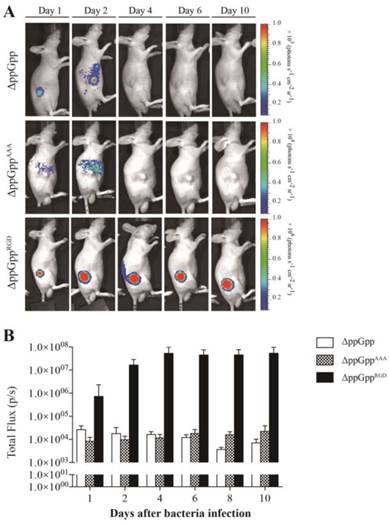
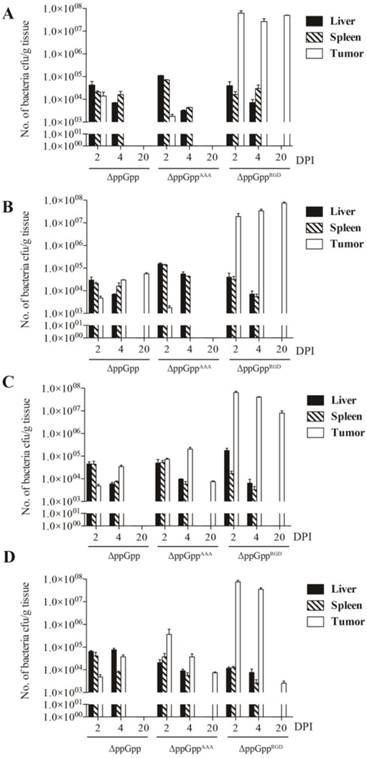
We next investigated whether the ΔppGppRGD strain accumulated in αvβ3-positive cancer (U87MG and M21)-and αvβ3-negative cancer (M21L) bearing nude mice. To monitor its distribution in vivo, the p22 bacteriophage [26] was used to engineer bacteria expressing bacterial luciferase (Lux). In vivo bioluminescence imaging revealed significant initial accumulation of ΔppGppRGD in U87MG and M21 xenografts. Continued monitoring revealed that the emission levels were stable, and were approximately 1000~5000-fold higher in animals that received ΔppGppRGD strain than in those that received control bacteria (ΔppGpp strain or ΔppGppAAA strains) (Fig. 3A, B, Fig. S5A-C). Direct comparison of targeting efficiency between αvβ3-positive (M21) and -negative melanoma (M21L) demonstrated significantly higher accumulation of the ΔppGppRGD strain in M21 melanoma than in M21L melanoma during the whole observation period (Fig. S5A-C). Counting the actual number of bacteria showed that, consistent with the results of the imaging studies, the number of ΔppGppRGD cells in the tumor was significantly higher (> 1000-fold) than those of control strains after initial injection; these numbers remained constant for the duration of the experiment (until 10 ~ 20 dpi) (Fig. 4, Fig. S5C).
αvβ3 integrin expression and tumor cell targeting efficiency of RGD-displaying Salmonella. ΔppGppRGD was cultivated and OmpARGD expression was induced by L-arabinose. Tumor cells were infected with ΔppGppRGD for 2 h at an MOI of 1:100. For visualization by confocal microscopy, cell nuclei (blue), αvβ3 (red), and Salmonella (green) were stained with DAPI and antibodies specific for αvβ3 and Salmonella, respectively. Scale bar, 20µm. (A) Low level and (B) high level αvβ3 integrin expression by tumor cells incubated with ΔppGppRGD. (C) Quantitative analysis of the tumor targeting ability of RGD-displaying Salmonella and competition assay results. ΔppGpp and ΔppGppAAA were induced to express OmpAAAA by addition of L-arabinose (triple alanine instead of RGD) and used as controls. Infection of tumor cells with either ΔppGpp or ΔppGppAAA for 2 h at a MOI of 1:100. For the competition assay, cells were pre-incubated for 2 h with 1 μM synthetic RGD peptide (ACDCRGDCFCG), washed with PBS, and then infected with ΔppGppRGD (MOI, 1:100). *P = 0.008 (low level vs. high level of αvβ3 integrin expression of tumor cells in ΔppGppRGD); **P = 0.001 (ΔppGppRGD vs. ΔppGpp, ΔppGppAAA or ΔppGppRGD after addition of 1 μM free RGD in U87MG, MDA-MB-231, MDA-MB-435, and M21 tumor cells). The results are representative of at least three independent experiments.
In vivo imaging of RGD-displaying S. typhimurium in the U87MG xenograft models. BALB/c athymic nu-/nu- mice (n = 7 per group) were injected subcutaneously with U87MG (1 × 107) cells. When the tumors reached approximately 100 mm3, mice were intravenously injected with bioluminescent bacteria (ΔppGpp, ΔppGppAAA, or ΔppGppRGD). (A) Non-invasive in vivo imaging of bacterial bioluminescence in representative mice. (B) Signal intensity in tumor regions of interest was assessed by measuring the toral flux. Regions of interest were selected manually within each tumor and results are shown as a bar graph after bacterial injection.
Tumor suppressive effects of RGD-displaying Salmonellae in tumor xenograft-bearing mice
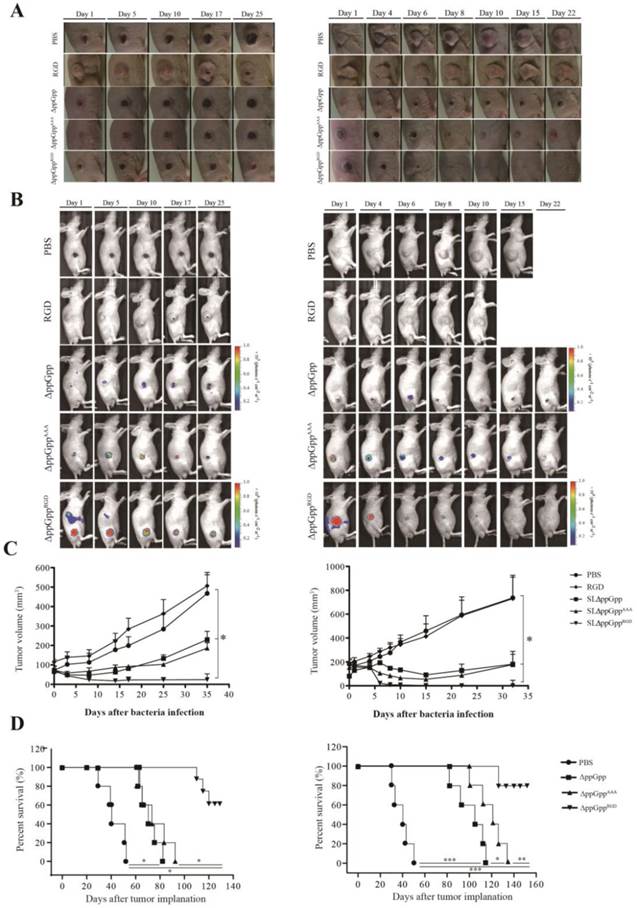
αvβ3-expressing cancer cells (MDA-MB-231 or MDA-MB-435) were injected subcutaneously into the right thigh of 5-week-old male nu/nu mice (107 cells/100 μL per mouse). To explore bacterial anti-tumor activity, tumor-bearing mice were treated with strains ΔppGppRGD, ΔppGppAAA, and ΔppGpp, or with PBS or synthetic RGD peptide (vehicle control). Strain ΔppGppRGD showed the strongest anti-tumor effect, followed by ΔppGppAAA and ΔppGpp. There was no statistically significant difference in the anti-tumor efficacy of strains ΔppGppAAA and ΔppGpp, although both were significantly more effective than the PBS control. At the end of the experiment (33-35 days), the mean volume of MDA-MB-231 and MDA-MB-435 tumors was 24 mm3 and 10 mm3, respectively, in the ΔppGppRGD group, and 186 mm3 and 177 mm3, respectively, in the ΔppGppAAA group (P < 0.05, Fig. 5A-C). In addition, animals receiving ΔppGppRGD survived for significantly longer than animals in the other groups (P < 0.01, Fig. 5D). Taken together, these results suggest that RGD-displaying Salmonella significantly suppress tumor growth in mouse xenograft models.
The biodistribution studies after infection of mice with RGD-displaying Salmonella. Quantification of engineered S. typhimurium in different organs from mice bearing high αvβ3 integrin-expressing tumors (n = 5 per group). (A) U87MG, (B) M21, (C) MDA-MB-231, and (D) MDA-MB-435.
Systemic toxicity of RGD-displaying Salmonella
As with all treatments, ΔppGppRGD may cause systemic toxicity. Therefore, we measured plasma C-reactive protein (CRP) and procalcitonin (PCT) levels (suggestive of toxicity/damage to normal organs such as the liver and kidney) at 5 dpi following intravenous (i.v.) injection of bacteria into nu/nu mice bearing MDA-MB-231. Serum aspartate aminotransferase and alanine aminotransferase levels were within the normal ranges (Fig. S7). Markers of inflammation or infection (CRP and PCT) were also within normal ranges in all experimental groups (Fig. S7). These results confirm those published in a previous report [5], i.e., that surface-engineered bacteria do not result in serious inflammatory reactions.
In vivo imaging and tumor suppressive effects of RGD-displaying Salmonella. BALB/c athymic nu-/nu- mice received a subcutaneous injection of MDA-MB-231 (1 × 107) or MDA-MB-435 (1 × 107) cells. When the tumors reached approximately 100 mm3, mice were intravenously injected with PBS, synthetic RGD peptide (5 µg), or bioluminescent bacteria (ΔppGpp, ΔppGppAAA, or ΔppGppRGD). Mice bearing MDA-MB-231 or MDA-MB-435 tumors were treated with PBS (n = 8) or ΔppGpp (n = 10). A separate group of tumor-bearing mice received a daily intraperitoneal injection of 60 mg L-arabinose, starting on day 4 post-injection with RGD (n = 8), ΔppGppAAA (n = 15 for MDA-MB-231; n = 15 for MDA-MB-435) or ΔppGppRGD (n = 15 for MDA-MB-231; n = 15 for MDA-MB-435) (A) Photographs of subcutaneous tumors in representative mice. (B) Non-invasive in vivo imaging of bacterial bioluminescence. (C) Changes in tumor volume. Left panel: effect on MDA-MB-231 tumor growth. Right panel: effect on MDA-MB-435 tumor growth. *P = 0.0001; results from one-way ANOVA (PBS or RGD vs ΔppGpp or ΔppGppAAA vs ΔppGppRGD in MDA-MB-231 and MDA-MB-435) (D) Kaplan-Meier survival curves for mice bearing MDA-MB-231 (*P < 0.001) or MDA-MB-435 (*P = 0.058, **P = 0.009, and ***P < 0.001) tumors.
Discussion
Some strains of bacteria naturally target tumors, including those of the breast, colon, pancreas, and prostate, as well as hepatocellular carcinoma, melanoma, neuroblastoma, and spinal cord glioma [1, 12, 30-32]. However, we found that the targeting efficiency of attenuated S. typhimurium varied depending on the tumor. To compensate for this limitation, bacteria were engineered to display a specific binding ligand (e.g., an affibody or single domain antibody) on the membrane protein with a view to enhancing targeting and therapeutic efficacy [23, 24].
The RGD peptide, either radiolabeled or conjugated to various drugs, enables imaging and treatment of αvβ3 integrin-positive cells [33]. Therefore, we engineered bacteria to display the RGD-4C peptide (ACDCRGDCFCG; also known as cyclic RGD), which harbors disulfide bridges between the first (Cys1) and fourth (Cys4), and the second (Cys2) and third (Cys3), cysteine residues [34], on the surface of an attenuated Salmonella strain (ΔppGpp). To display the RGD-4C peptide on the third transmembrane domain of OmpA, we developed an attenuated S. typhimurium strain carrying a sandwich fused-gene expression cassette encoding OmpA bearing the RGD-4C peptide [28, 35] (Fig. 1). The RGD-displaying bacteria showed strong initial targeting and subsequent proliferation in αvβ3-overexpressing xenograft tumors, and consequently suppressed tumor growth (Fig. 5). Thus, the results indicate that the bacteria acquired strong targeting efficiency and therapeutic efficacy against human tumor xenografts through the surface display of a tumor targeting ligand.
Adhesion and competition analyses indicated that the RGD surface-display bacteria accumulated in the cancer cells in a αvβ3-specific manner. Binding capacity was significantly reduced by free synthetic RGD-4C peptide (1 µM), suggesting that the RGD-4C peptide is a crucial factor in the interaction between the bacteria and cancer cells. We examined the ability of surface-engineered bacteria to bind three different cancer cell lines with very high (U87MG, M21), medium (MDA-MB-231, MDA-MB-435) and no (MCF7, M21L) expression of ανβ3. As expected, the surface-engineered bacteria bound to the cells in the order U87MG > M21 > MDA-MB-231 or MDA-MB-435, showing a strong correlation with ανβ3 expression levels. Despite the high binding affinities, the ΔppGppRGD strain showed extremely low invasiveness of cancer cells expressing αvβ3. The invasion and intracellular growth of Salmonella is mediated by a bacterial type III secretion system (TTSS) encoded by genes on Salmonella pathogenicity island I and II (SPI1 and SPI2), respectively. Because SPI1 and SPI2 genes are expressed in a ppGpp-dependent manner [25, 36], ΔppGpp Salmonellae are defective in intracellular growth as well as invasiveness even when they are engineered for RGD surface display.
Although the mechanism underlying Salmonellae-mediated suppression of tumor growth is likely multifaceted, there is evidence to suggest that the immune-activating characteristics of Salmonellae are sufficient to induce tumor regression[13]. Release of lipopolysaccharide (LPS) within the tumor upon Salmonella infection may induce production of tumor necrosis factor α (TNF-α) by immune cells [37]. Indeed, TNF-α released by macrophages, along with IL-1β released by dendritic cells and macrophages, appears to be responsible for the anticancer effects of S. typhimurium [13]. The main pathway responsible for IL-1β processing is the inflammasome, which in turn activates caspase-1, followed by the cleavage and secretion of active IL-1β and IL-18[13]. According to the results of the present study, the reduction in tumor size was highly correlated with the persistence of Salmonella in the tumor. When the number of tumor-colonizing bacteria decreased, the tumor re-grew. These results suggest the importance of bacterial targeting efficiency and their rate of proliferation within the tumor tissue. RGD-displaying Salmonellae accumulated in tumors at significantly higher (> 1000-fold) levels than control strains after the initial bacterial injection, and the bacterial titers in the tumors were maintained until the end of the experiment (20 dpi). The avidity of the surface-engineered bacteria for the tumor cells explains the excellent anti-tumor effect observed in MDA-MB-231 and MDA-MB-435 xenograft mouse models.
Conclusions
We demonstrated the in vitro and in vivo targeting and therapeutic effects of surface-engineered bacteria carrying the RGD-4C peptide. The display of RGD-4C on OmpA proteins expressed on the surface of bacteria increased tumor targeting efficiency, as demonstrated by the results of adhesion and competition assays. In vivo studies showed that surface-engineered bacteria strongly suppressed the growth of MDA-MB-231 and MDA-MB-435 tumors in nude mice, and that this targeting efficiency (1000-fold higher than that of control bacteria) was maintained for up to 20 days. Taken together, these results suggest that surface-engineered bacteria have a potential for the delivery of high amounts of protein drugs to specific tumors in vivo.
Supplementary Material
Supplementary figures S1-S6.
Abbreviations
RGD: arginine- glycine-aspartic acid; OmpA: outer membrane protein A; TNF-α: tumor necrosis factor alpha; IL-1β: interlukin 1 beta; MOMP: major outer membrane protein; MOI: multiplicity of infection; BSA: bovine serum albumin; DAPI: 4',6-diamidino-2-phenylindole; CFU: colony forming unit; ALT: alanine transaminase; AST: aspartate aminotransferase; BUN: blood urea nitrogen; CREA: creatinine; CRP: c-reactive protein; PCT: procalcitonin; LPS: lipopolysaccharide.
Acknowledgements
This research was supported by the Pioneer Research Center Program (2015M3C1A3056410) and the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2014M3A9B5073747). M.S. was supported by the Leading Foreign Research Institute Recruitment Program (2011-0030034) of the NRF. S.-H.P. was supported by the Basic Science Research Program through the NRF funded by the Ministry of Education (NRF-2014R1A6A3A04057492). H.E.C. was supported by a National Research Foundation of Korea(NRF) grannt funded by the Korea government (NRF-2014R1A2A1A10051664). D.-Y.K. was supported in part by the National Research Foundation of Korea (NRF-2015M2B2A9031798).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Zhao M, Yang M, Ma H, Li X, Tan X, Li S. et al. Targeted therapy with a Salmonella typhimurium leucine-arginine auxotroph cures orthotopic human breast tumors in nude mice. Cancer Res. 2006;66:7647-52
2. Friedlos F, Lehouritis P, Ogilvie L, Hedley D, Davies L, Bermudes D. et al. Attenuated Salmonella targets prodrug activating enzyme carboxypeptidase G2 to mouse melanoma and human breast and colon carcinomas for effective suicide gene therapy. Clin Cancer Res. 2008;14:4259-66
3. Nguyen VH, Kim HS, Ha JM, Hong Y, Choy HE, Min JJ. Genetically engineered Salmonella typhimurium as an imageable therapeutic probe for cancer. Cancer Res. 2010;70:18-23
4. Lee CH, Wu CL, Shiau AL. Salmonella choleraesuis as an anticancer agent in a syngeneic model of orthotopic hepatocellular carcinoma. Int J Cancer. 2008;122:930-5
5. Jiang SN, Park SH, Lee HJ, Zheng JH, Kim HS, Bom HS. et al. Engineering of bacteria for the visualization of targeted delivery of a cytolytic anticancer agent. Mol Ther. 2013;21:1985-95
6. Jazowiecka-Rakus J, Szala S. Antitumour activity of Salmonella typhimurium VNP20047 in B16(F10) murine melanoma model. Acta Biochim Pol. 2004;51:851-6
7. Barnett SJ, Soto LJ 3rd, Sorenson BS, Nelson BW, Leonard AS, Saltzman DA. Attenuated Salmonella typhimurium invades and decreases tumor burden in neuroblastoma. J Pediatr Surg. 2005;40:993-7 discussion 7-8
8. Nagakura C, Hayashi K, Zhao M, Yamauchi K, Yamamoto N, Tsuchiya H. et al. Efficacy of a genetically-modified Salmonella typhimurium in an orthotopic human pancreatic cancer in nude mice. Anticancer Res. 2009;29:1873-8
9. Maletzki C, Linnebacher M, Kreikemeyer B, Emmrich J. Pancreatic cancer regression by intratumoural injection of live Streptococcus pyogenes in a syngeneic mouse model. Gut. 2008;57:483-91
10. Zhang L, Gao L, Zhao L, Guo B, Ji K, Tian Y. et al. Intratumoral delivery and suppression of prostate tumor growth by attenuated Salmonella enterica serovar typhimurium carrying plasmid-based small interfering RNAs. Cancer Res. 2007;67:5859-64
11. Kimura H, Zhang L, Zhao M, Hayashi K, Tsuchiya H, Tomita K. et al. Targeted therapy of spinal cord glioma with a genetically modified Salmonella typhimurium. Cell Prolif. 2010;43:41-8
12. Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer. 2010;10:785-94
13. Kim JE, Phan TX, Nguyen VH, Dinh-Vu HV, Zheng JH, Yun M. et al. Salmonella typhimurium Suppresses Tumor Growth via the Pro-Inflammatory Cytokine Interleukin-1beta. Theranostics. 2015;5:1328-42
14. Phan TX, Nguyen VH, Duong MT, Hong Y, Choy HE, Min JJ. Activation of inflammasome by attenuated Salmonella typhimurium in bacteria-mediated cancer therapy. Microbiol Immunol. 2015;59:664-75
15. Toso JF, Gill VJ, Hwu P, Marincola FM, Restifo NP, Schwartzentruber DJ. et al. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J Clin Oncol. 2002;20:142-52
16. Heimann DM, Rosenberg SA. Continuous intravenous administration of live genetically modified salmonella typhimurium in patients with metastatic melanoma. J Immunother. 2003;26:179-80
17. Lee SY, Choi JH, Xu Z. Microbial cell-surface display. Trends Biotechnol. 2003;21:45-52
18. Lee JS, Shin KS, Pan JG, Kim CJ. Surface-displayed viral antigens on Salmonella carrier vaccine. Nat Biotechnol. 2000;18:645-8
19. Westerlund-Wikström B, Tanskanen J, Virkola R, Hacker J, Lindberg M, Skurnik M. et al. Functional expression of adhesive peptides as fusions to Escherichia coli flagellin. Protein engineering. 1997;10:1319-26
20. Martineau P, Charbit A, Leclerc C, Werts C, O'Callaghan D, Hofnung M. A genetic system to elicit and monitor anti-peptide antibodies without peptide synthesis. Nat Biotechnol. 1991;9:170-2
21. Xu Z, Lee SY. Display of Polyhistidine Peptides on theEscherichia coli Cell Surface by Using Outer Membrane Protein C as an Anchoring Motif. Appl Environ Microb. 1999;65:5142-7
22. Dhillon JK, Drew PD, Porter AJ. Bacterial surface display of an anti-pollutant antibody fragment. Lett Appl Microbiol. 1999;28:350-4
23. Chang CH, Cheng WJ, Chen SY, Kao MC, Chiang CJ, Chao YP. Engineering of Escherichia coli for targeted delivery of transgenes to HER2/neu-positive tumor cells. Biotechnol Bioeng. 2011;108:1662-72
24. Massa PE, Paniccia A, Monegal A, de Marco A, Rescigno M. Salmonella engineered to express CD20-targeting antibodies and a drug-converting enzyme can eradicate human lymphomas. Blood. 2013;122:705-14
25. Song M, Kim HJ, Kim EY, Shin M, Lee HC, Hong Y. et al. ppGpp-dependent stationary phase induction of genes on Salmonella pathogenicity island 1. J Biol Chem. 2004;279:34183-90
26. Min JJ, Nguyen VH, Kim HJ, Hong Y, Choy HE. Quantitative bioluminescence imaging of tumor-targeting bacteria in living animals. Nat Protoc. 2008;3:629-36
27. Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640-5
28. Smith SG, Mahon V, Lambert MA, Fagan RP. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol Lett. 2007;273:1-11
29. Loening AM, Fenn TD, Wu AM, Gambhir SS. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng Des Sel. 2006;19:391-400
30. Zhao M, Geller J, Ma H, Yang M, Penman S, Hoffman RM. Monotherapy with a tumor-targeting mutant of Salmonella typhimurium cures orthotopic metastatic mouse models of human prostate cancer. Proc Natl Acad Sci U S A. 2007;104:10170-4
31. Agrawal N, Bettegowda C, Cheong I, Geschwind JF, Drake CG, Hipkiss EL. et al. Bacteriolytic therapy can generate a potent immune response against experimental tumors. Proc Natl Acad Sci U S A. 2004;101:15172-7
32. Akin D, Sturgis J, Ragheb K, Sherman D, Burkholder K, Robinson JP. et al. Bacteria-mediated delivery of nanoparticles and cargo into cells. Nat Nanotechnol. 2007;2:441-9
33. Wang H, Chen K, Cai W, Li Z, He L, Kashefi A. et al. Integrin-targeted imaging and therapy with RGD4C-TNF fusion protein. Molecular cancer therapeutics. 2008;7:1044-53
34. Assa-Munt N, Jia X, Laakkonen P, Ruoslahti E. Solution structures and integrin binding activities of an RGD peptide with two isomers. Biochemistry. 2001;40:2373-8
35. Zitzmann S, Ehemann V, Schwab M. Arginine-glycine-aspartic acid (RGD)-peptide binds to both tumor and tumor-endothelial cells in vivo. Cancer Res. 2002;62:5139-43
36. Song M, Kim HJ, Ryu S, Yoon H, Yun J, Choy HE. ppGpp-mediated stationary phase induction of the genes encoded by horizontally acquired pathogenicity islands and cob/pdu locus in Salmonella enterica serovar Typhimurium. J Microbiol. 2010;48:89-95
37. Pawelek JM, Low KB, Bermudes D. Tumor-targeted Salmonella as a novel anticancer vector. Cancer Res. 1997;57:4537-44
Author contact
![]() Corresponding authors: Jung-Joon Min, M.D., Ph.D. Department of Nuclear Medicine, Chonnam National University Hwasun Hospital, 322 Seoyang-ro, Hwasun, Jeonnam 58128, Republic of Korea. Tel: +82-61-379-7271 Fax: +82-61-379-7280 e-mail: jjminac.kr Or Hyon E. Choy, PhD. Department of Microbiology, Chonnam National University Medical School, 264 Seoyang-ro, Hwasun, Jeonnam 58128, Republic of Korea. Tel +82-61-379-2742 Fax +82-61-379-2753 E-mail: hyonchoyac.kr.
Corresponding authors: Jung-Joon Min, M.D., Ph.D. Department of Nuclear Medicine, Chonnam National University Hwasun Hospital, 322 Seoyang-ro, Hwasun, Jeonnam 58128, Republic of Korea. Tel: +82-61-379-7271 Fax: +82-61-379-7280 e-mail: jjminac.kr Or Hyon E. Choy, PhD. Department of Microbiology, Chonnam National University Medical School, 264 Seoyang-ro, Hwasun, Jeonnam 58128, Republic of Korea. Tel +82-61-379-2742 Fax +82-61-379-2753 E-mail: hyonchoyac.kr.
 Global reach, higher impact
Global reach, higher impact