13.3
Impact Factor
Theranostics 2016; 6(10):1603-1610. doi:10.7150/thno.15534 This issue Cite
Research Paper
Holographic Assessment of Lymphoma Tissue (HALT) for Global Oncology Field Applications
1. Center for Systems Biology, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02114;
2. Department of Radiology, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02114;
3. Department of Pathology, Massachusetts General Hospital, Boston, MA, 02114;
4. Center for Lymphoma, Massachusetts General Hospital, Boston, MA, 02114;
5. Massachusetts General Hospital Cancer Center, Boston, MA 02114;
6. Department of Medicine, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02114;
7. Global Oncology Initiative, Dana-Farber / Harvard Cancer Center, Boston, MA, 02115;
8. Department of Obstetrics, Gynecology and Reproductive Biology, Massachusetts General Hospital, Boston, MA 02114;
9. Department of Systems Biology, Harvard Medical School, Boston, MA 02115.
* These authors contributed equally to the paper.
Received 2016-3-14; Accepted 2016-5-9; Published 2016-6-18
Abstract
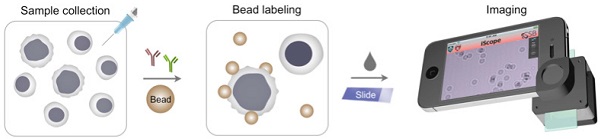
Low-cost, rapid and accurate detection technologies are key requisites to cope with the growing global cancer challenges. The need is particularly pronounced in resource-limited settings where treatment opportunities are often missed due to the absence of timely diagnoses. We herein describe a Holographic Assessment of Lymphoma Tissue (HALT) system that adopts a smartphone as the basis for molecular cancer diagnostics. The system detects malignant lymphoma cells labeled with marker-specific microbeads that produce unique holographic signatures. Importantly, we optimized HALT to detect lymphomas in fine-needle aspirates from superficial lymph nodes, procedures that align with the minimally invasive biopsy needs of resource-constrained regions. We equipped the platform to directly address the practical needs of employing novel technologies for “real world” use. The HALT assay generated readouts in <1.5 h and demonstrated good agreement with standard cytology and surgical pathology.
Keywords: Cancer diagnostics, Smartphone testing, Point-of-care diagnostics, global oncology.
Introduction
Cancer remains a global health challenge, claiming more than 7 million deaths per year [1]. The burden is disproportionally placed on low and middle income countries (LMICs). Missed diagnoses and delayed treatments result in high mortality even from preventable or treatable cancers. This occurs with virus-associated cancers, the so called “second wave of AIDS”, that have increasingly plagued sub-Saharan countries [2-4]. This epidemic has expanded as larger populations gain access to antiretroviral drugs. Patients are now surviving long enough to develop HIV associated cancers, including non-Hodgkin lymphoma, cervical cancer, Kaposi sarcoma and head and neck cancers. However, a substantial number of cases evade diagnosis and are inappropriately classified due to the lack of proper tissue specimens, diagnostic reagents and specialists, among other factors. Although a significant proportion are treatable cancers, windows of therapeutic opportunities are commonly missed [5,6].
Recent advances in nanomaterials and device miniaturization [7] promise effective cancer management in LMICs. Lens-free imaging systems for wide-field holographic microscopy have paved way for use of smartphones as mobile imaging alternatives to bulky microscopes [8, 9]. Smaller and portable sensors could reduce diagnostic costs, shorten turnaround times, circumvent logistic constraints, and improve diagnostic sensitivity in scant samples. Smartphones are increasingly adopted as a sensor base given their capacity for wireless communication, high resolution imaging, and portable computing. Indeed, smartphone-based medical devices and software applications have been developed for various clinical purposes [10-12] but molecular diagnostics have been more difficult to implement. To address these challenges, immunolabeling or capture methods have been adopted in the smartphone-based in-line holographic microscopy. For example, Stybayeva et. al. used antibody microarrays to capture target cells for imaging [13]; Wel et. al. used immunolabeling with gold and silver nanoparticles to generate optical contrasts on targeted cells [14]. We have previously demonstrated quantitative cellular profiling by adopting immunobead-based labeling [15].
We now report work undertaken to expand and position the technology for smartphone diagnostics of lymphoma (Holographic Assessment of Lymphoma Tissue or HALT) using fine needle aspirates within resource-constrained areas globally. Harvested cells of interests are labeled with molecular-specific microbeads and their diffraction patterns are imaged by a smartphone. The captured images are then wirelessly sent to a remote server for reconstruction and analyses. We optimized the platform for lymphoma diagnostics in field settings: 1) the platform uses cellular samples obtained by fine needle aspiration (FNA), an invariably less-invasive and more feasible method than core biopsies or surgical resection; 2) all reagents (e.g., beads, antibodies, buffers) could be stored and transported; 3) an automated algorithm was developed to provide quantitative readout. We applied the HALT platform to detect B-cell non-Hodgkin lymphoma, the most prevalent lymphoma subtype in LMICs. Our approach was fast (<1.5 h), required a small amount of samples (2 FNA passes) and matched well with conventional pathology readouts. HALT could be leveraged as a practical tool for front line cancer screening and management in LMICs where significant pathology bottlenecks exist [1].
Results
HALT assay
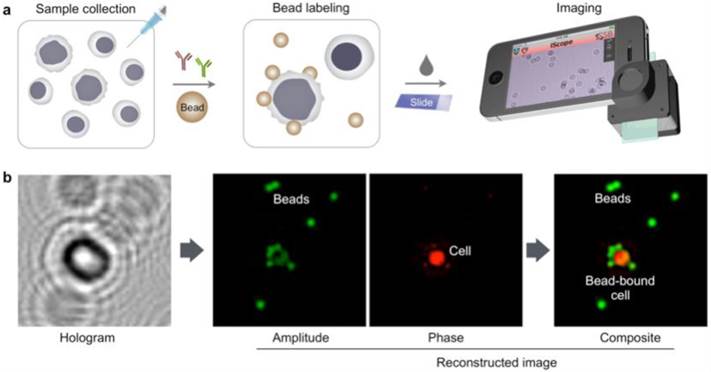
The HALT assay procedure is illustrated in Fig. 1a. Specimens are obtained through FNA and homogenized into a cell suspension. Target cells of interests are immuno-labeled with microbeads, and detected. The optical setup (Fig. S1), directly attached to a smartphone camera, comprises a light-emitting diode (LED) powered by a coin battery, a 100 µm pinhole, a mini-lens and a sample holder. With the attachment, the system generates a wide field-of-view (FOV) hologram over 10 mm2, simultaneously imaging >104 cells in a single shot11. The recorded holograms are then sent to a remote server via cloud storage for the reconstruction of amplitude and phase images (Fig. 1b), and a numerical algorithm distinguishes cells and beads according to cells' high phase contrast. The reconstructions and analyses are completed in <1 min through parallel computing by a remote server equipped with a graphic processing unit (GPU); results are immediately sent back to the smartphone. In addition to the reconstructed images, readouts contain the number of total cells, bead-bound cells and beads per cell. This server-client model is scaleable for large data storage and multiple-user connections. The measurement, data transfer and readout is controlled by a custom-built mobile App.
Assay optimization
We optimized the HALT-assay protocol for lymphoma detection, a major unmet need in sub-Saharan countries where palpable lymphadenopathy (HIV, TB, lymphoma) commonly occurs. In coupling microbeads to cells, we shifted away from our previously published protocol. Specifically, we opted for a two-step labeling method: cells were first targeted with a primary antibody, and then incubated with microbeads conjugated with secondary antibodies. This scheme simplified reagent preparation by obviating primary antibody modification (i.e., removes need for costlier customized antibodies) and using generic microbeads for all different markers. We first tested the procedure by targeting CD10 and CD20 in a lymphoma cell line (DB, human germinal center B-cell like diffuse large B-cell lymphoma, DLBCL). Cultured cells were blocked for non-specific binding (30 min), aliquoted, and labeled with each primary antibody (30 min). Cells were then incubated (30 min) with microbeads (4.5 µm) coated with secondary antibodies. The HALT assay automatically detected bead-bound cells (Fig. 2a), and provided analytical readouts (e.g., targeted cell numbers and bead counts per target cell). The average bead counts per cell were higher for CD20 than CD10 (Fig. 2b), which agreed with the expression profiles measured by flow cytometry (Fig. S2a). Bead counts could be further increased by applying a cocktail of CD10 and CD20 antibodies (Fig. 2b) to improve detection sensitivity.
Molecular detection with holography. (a) Schematic of HALT assay for lymphoma cells collected by fine needle aspiration. (b) A hologram image is reconstructed for amplitude and phase contrast images. While both cells and beads are visible in the amplitude image, the phase-contrast image identifies cells only. Using both image acquisitions, the algorithm detects bead-bound target cells and the number of beads on the cells. The amplitude (green) and phase contrast (red) images are pseudocolored for better visualization of cells and beads.
Optimization of HALT platform for high-throughput analyses. (a) Composite images of cells immunolabeled with microbeads (4.5 μm) and imaged using HALT. The amplitude (green) and the phase (red) were pseudocolored for clarification. (b) Using an antibody cocktail improves HALT sensitivity. The average number of beads per cell is higher in the presence of cocktail of CD10 and CD20 antibodies as compared to CD10 or CD20 used individually in DB cells. (c) The robust specificity of the system can help in decreasing processing times by omitting the washing steps. No decrease in the signal was observed for CD20 antibody without washing the Daudi cells' samples. (d) In order to increase the portability and shelf-life of our system we tested the system with lyophilized antibodies. Lyophilized antibodies (CD3 and CD20) were tested and compared with parent solution antibodies after 2 weeks of storage at 4℃ by HALT platform and exhibited similar activity as the original antibodies (Exp1: experiment 1, Exp:2 experiment 2 show data from antibodies lyophilized at two different occasions).
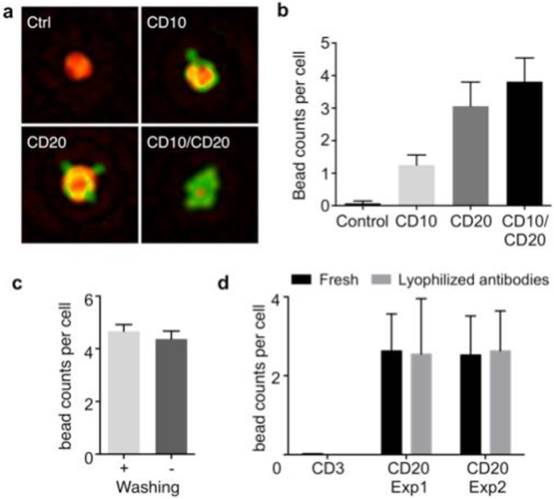
We further optimized HALT for operations in resource-limited settings. By adjusting the ratio between primary antibody and microbead concentrations, we improved the labeling protocol to remove all washing steps. The optimal ratio was found to be 0.25 μg antibody : 1x106 beads; the number of beads per cell was statistically identical among samples prepared with or without washing steps (Fig. 2c, P = 0.46; unpaired t test). We next tested antibody storage via lyophilization to confer durability. Two antibodies (CD3 and CD20) were lyophilized and stored. After two weeks of storage, we resuscitated antibodies and used them for cell profiling. Both flow cytometry (Fig. S2b) and HALT (Fig. 2d) confirmed that antibodies maintained their reactivity with negligible degradation.
Assay validation
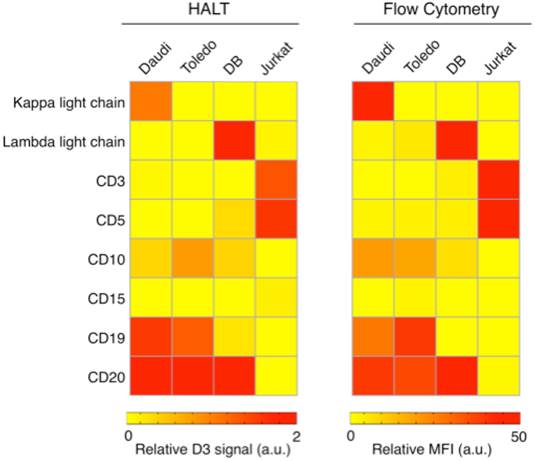
We next examined marker sets for B-cell lymphoma detection. We used the following representative B-cell lymphoma cell lines: Daudi (Burkitt lymphoma); Toledo and DB (germinal center B-cell like diffuse large B-cell lymphoma). As a control, we used a T-cell leukemia cell line (Jurkat). Modern lymphoma diagnostics commonly use ~5-25 antibody markers (in addition to morphology) to provide WHO classification type diagnoses [16, 17]. Given the cost and complexity of this approach, it is deemed impractical for many LMICs. We thus chose a panel of markers (kappa light chain, lambda light chain, CD3, CD5, CD10, CD19 and CD20) to diagnose B-cell non-Hodgkin lymphomas, since these comprise the majority of non-Hodgkin lymphomas. CD3, CD5, CD10, CD19 and CD20 markers were chosen to differentiate B cells from T cells. CD20 and CD19 served as B-cell specific markers; CD3 and CD5 (to a lesser extent) reflected T cell signatures. To determine if the tested B-cell populations represented lymphoma, we evaluated the clonality of these cells using kappa and lambda light chains. Lymphomas are monoclonal in nature and will generally over-express one of these two clonal markers. CD10 is a marker for DLBCL of germinal center origin, but is also expressed in other germinal center-derived B-cell malignancies including Burkitt lymphoma and follicular lymphoma.
To further examine HALT, we measured the expression levels of the above mentioned markers in four lymphoma cell lines using 4.5 µm beads (Fig. 3). As expected the average bead-count per cell was highest for B-cell markers (CD19 and CD20) in the lymphoma cell lines. Moreover, these cell lines were negative for CD3 expression (T-cell marker), which exhibited increased signal in Jurkat cells (T-cell leukemia); these results were consistent with the results obtained from the gold standard (flow cytometry).
HALT provides a simple, reliable and robust platform for detection of lymphoma biomarkers Lymphoma cells (Daudi, Burkitt lymphoma; Toledo and DB, germinal center B-cell like diffuse large B-cell lymphoma; and Jurkat, T-cell leukemia) were immunobead-labeled for a panel of biomarkers (kappa light chain, lambda light chain, CD3, CD5, CD10, CD15, CD19 and CD20) and tested in HALT system. The results obtained with our HALT platform were in concordance with flow cytometry data.
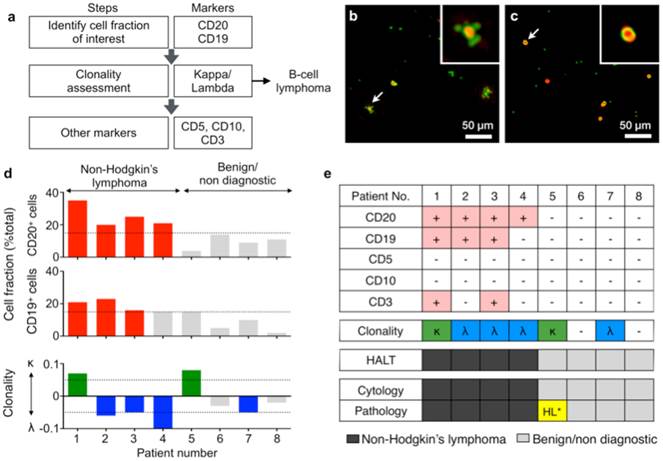
Diagnosis with HALT platform in patients suspected with lymphoma. (a) Choice of markers and algorithm used for diagnosing non-Hodgkin lymphoma. (b, c) Representative composite images of cells from fine needle aspirates (FNAs) of patients with suspected lymphoma. Cells were immunolabeled with microbeads (4.5 μm) and imaged using HALT. Amplitude and phase contrast are pseudocolored to green and red, respectively. Insets are respective zoomed-in images of the cells highlighted with white arrows. Cells from NHL (non-Hodgkin lymphoma) exhibit increased bead binding (b) for the lymphoma marker, which is absent or comparatively less in sample obtained from benign adenopathy (c). (d) Sample consisting of more than 15% of a particular cluster designation (CD) marker was considered positive. The clonality of the population was determined by using the formula: (κ-λ)/(κ+λ) where κ and λ are average bead counts per cell for kappa and lambda light chains, respectively. Clonality was defined in the range of -1 to +1 where value >0 means kappa is more dominant while the value <0 means lambda is more prominent in the population. (e) FNAs from 8 patients were tested individually for a panel of relevant lymphoma biomarkers (CD20, CD19, CD5, CD10, CD3, kappa light chain, lambda light chain) and compared to pathology results. The results obtained from HALT platform were in concordance with flow cytometry and surgical pathology results. (HL: Hodgkin lymphoma).
Analysis of clinical samples
Finally, we tested the HALT assay with clinical samples. Cells were obtained from patients with abnormal lymphadenopathy using lymph node FNAs (see Materials and Methods for details). Fine needle aspirated cells were suspended in PBS, and profiled for a panel of biomarkers. To detect non-Hodgkin lymphoma (NHL), we set up the following detection flow (Fig. 4a): CD19 and CD20 to identify B cells; kappa and lambda screening to determine B cell clonality; CD10 as additional marker for germinal center-derived B-cell lymphomas; CD3 and CD5 to detect T cells. Figs. 4b and c show representative images of samples from NHL and benign adenopathy, respectively. The relative fraction of CD19 or CD20-positive cells was higher (>15%) in NHL than in benign adenopathy patients (Fig. 4d). After identifying the cell fraction of interest (CD19+/CD20+/CD3-/CD5-) we further measured the clonality of the population using a metric: (κ-λ)/(κ+λ) where κ and λ are average bead counts per cell for kappa and lambda light chains, respectively. The metric has a range of -1 to +1, where non-zero values indicate a dominance of either clonality (Fig. 4d). We observed that NHL patients were generally associated with a preferential expansion of either clone (Fig. 4d, bottom). These results were in concordance with official flow cytometry and surgical pathology reports. Of note, both HALT and the cytology missed a case of a Hodgkin lymphoma that was evident in a surgical pathology specimen. This can be attributed to the paucity of neoplastic cells in Hodgkin lymphoma, which typically requires excisional biopsy for definitive diagnosis.
Discussion
In sub-Saharan Africa access to diagnostic pathology services is limited, yet smartphones and cellular services are abundant. Leveraging smartphones as a mobile diagnostic terminal could empower resource-poor communities with complex laboratory tests. This work addresses the practical diagnostic needs of LMICs and reflects the type of technologies that may gain sustainable traction in such settings. Major advantages of HALT include the simplicity of the method, accuracy, and sustainability. The assay system can be operated by non-specialists with brief training. Test results are generated from quantitative molecular analyses thus minimizing potential human errors. Furthermore, the system can readily link to central pathology laboratories for expert second opinions - HALT can thus serve as a gateway to expand pathology support in LMICs.
In the current study, we have adopted HALT for lymphoma detection. Lymphoma is a major health challenge in sub-Saharan LMICs, further driven by the high prevalence of AIDS. Incidence of aggressive lymphoma in the region continues to rise, but its diagnosis and treatment are hampered by limited resources, poor sample quality, and lack of specialists (<1 pathologist per 500,000 people). We focused on optimizing HALT to overcome such challenges: 1) the assay process is simple (“mix-and-shake”) with no washing steps; 2) the reagents are in a dry powder form (lyophilized) to facilitate transport and storage; 3) the assay requires small amounts of cells obtained via FNA, a procedure more amenable in LMICs than core biopsies; 4) disease subtypes can be determined through multiple-marker screening. Coupled with low cost requirements (~$5 USD for reagents and antibodies per test before wholesale discounts; Table S1), these features render HALT an affordable, practical tool to augment pathology testing. The approach performed well with clinically procured FNAs of patients with suspicious lymphadenopathy; we observed a good concordance between HALT and cytology. Note that the marker sets employed in HALT align with current clinical testing. As such, the “bar” is lower compared to validation studies of novel biomarker panels that may or may not be grounded in clinical reality. While additional field study of a larger numbers of cases is needed for further validation, the HALT showed a promise as a new approach that could effectively triage cases and prompt expedited confirmation by overwhelmed pathologists in resource-constrained areas.
Further work is in progress to advance HALT technology. We are developing a next generation version that will feature 1) improved cartridges to simplify sample handling and user operation, and 2) a new App built on an open-source framework (e.g., ResearchKit) to enable more efficient data collection and sharing. We are also expanding the assay portfolio to diagnose other cancer types (e.g., breast, cervical) that are prevalent in LMICs. Finally, the data presented here has led us to design a field study of HALT in lymphoma diagnosis in regions of Botswana. This study will critically test the feasibility of mobile technology for point-of-care diagnostics. With such efforts, we envision further global penetration of medical diagnostics as seen with the information revolution.
Materials and Methods
Cell lines and growth conditions
The germinal center B-cell like diffuse large B-cell lymphoma (DLBCL) cell lines DB and Toledo were kind gifts from Dr. Anthony Letai (Dana Farber Cancer Institute, Boston, MA). Burkitt lymphoma cell line (Daudi) and T-cell leukemia cell line (Jurkat) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Cell lines were maintained in RPMI-1640 media (Cellgro, Mediatech, Mannassas, VA) supplemented with 10% heat-inactivated fetal bovine serum, 100 IU penicillin and 100 µg/mL streptomycin at 37 °C in a humidified atmosphere of 5% CO2. All cell lines used for experiments were tested regularly for mycoplasma contamination using (MycoAlert™ Mycoplasma Detection Kit, Lonza, Switzerland).
Flow cytometry for molecular profiling of cell lines
Cells (0.5x106/ antibody marker) were fixed with 1x BD Phosflow Lyse/Fix buffer (BD Biosciences, San Jose, CA) for 10 min at at 37°C and then washed thrice with PBS (0.5% BSA). Fixed cells were then blocked for 30 min at room temperature and then incubated with primary antibodies (10 µg/mL; Table S1) for 30 min at room temperature. Following this, cells were washed, incubated with fluorophore conjugated secondary antibody (2 µg/mL, Table S1) for 30 min at room temperature, then washed again. The fluorescence signals from the labeled cells were measured using BD LSRII Flow Cytometer (BD Biosciences). The analyses were performed using FlowJo software. Mean fluorescent intensities (MFIs) recorded were normalized using the formula (Signal- IgG isotype control)/ secondary. A heat map was generated using MATLAB (MathWorks, Natick, MA) and scaled appropriately. Every washing step comprised three 5 min washes at 300g with PBS (0.5% BSA).
Clinical sample processing
The study was approved by the Institutional Review Board at the Dana-Farber/Harvard Cancer Center, and informed consent was obtained from all subjects (n = 8). Patients with clinical suspicion for new or recurrent lymphoma were referred to the MGH Radiology Abdominal Intervention suites for FNA and core biopsy. Correct needle location within a suspicious lymph node was confirmed by ultrasound or computed tomography imaging. Prior to the core biopsy, two extra fine needle aspirate (FNA) passes, in addition to the routine FNA pass for cytology and flow cytometry, were obtained for HALT purposes. FNA samples were centrifuged at 300g for 10min to remove excess saline. Collagenase (0.2 mg/mL, Sigma-Aldrich) was used to breakdown any clumps present in the sample prior to fixation. Cells were fixed using 1x BD Phosflow Lyse/Fix buffer (BD Biosciences) for 10 min at 37°C and then washed twice (300g for 5min) with PBS (2% BSA). Samples were then stored at -80°C in PBS (2% BSA). A total of 8 samples were prepared and analyzed independently via the HALT method.
Sample preparation for HALT
Cell lines: Cells (50,000 cells/ sample) were blocked with PBS (0.5% BSA) for 30 min. Cells were then labeled with primary antibodies (10 µg/mL) for 30 min, washed and incubated with goat anti-mouse IgG secondary antibody coated Dynabeads (Invitrogen, Grand Island, NY) for 30 min. All washing steps comprised three 5 min washes at 300g with PBS (0.5% BSA).
Clinical samples: Samples were washed once with PBS (0.5% BSA) and then blocked for 30 min in FcR blocking solution (4 parts PBS with 0.5% BSA and 1 part FcR blocking reagent, human, Miltenyi Biotech, San Diego, CA) at room temperature. Following this, samples were labeled with primary antibodies (10 µg/mL) by incubating at room temperature for 30 min. After being washed with FcR blocking solution, samples were incubated with Dynabeads coated with goat anti-mouse IgG secondary antibody (Invitrogen) for 30 min at room temperature. All the centrifugation and washing steps were performed at 300g for 5 min.
HALT sensor
The imaging module is equipped with a light-emitting diode at 405 nm, a 100-µm pinhole, a mini lens with a focal length of 4 mm, a switch and a coin battery11. The case is made of black acrylic plastic. A 10 µL of bead-labeled cell sample was placed between two glass cover slips (Fisher Scientific Inc.) and imaged by a camera. A reference image was taken without any sample for image normalization. Uncompressed images were sent to a remote server via a designated cloud storage. Following image reconstruction and bead detection, a log file containing the total cell counts, bead-labeled cell counts and number of beads on each cell was sent back to a user.
Custom HALT App and server program
The remote server is based on HP xw4600 workstation (Hewlett Packard) equipped with a Tesla C-2070 graphic processing unit (GPU, Nvidia). The GPU has 448 CUDA (compute unified device architecture) cores with 6GB memory for rapid parallel image processing. The image reconstruction and detection algorithm was written in C++ language using modules provided by Nvidia Inc (CUDA extensions, CUDA driver 5.0, CUFFT library). The server periodically checked new images in a designated cloud folder (Dropbox) and executed image reconstruction and post-imaging analyses. A custom-designed iPhone App was used to control the camera module (ISO, exposure time), export image files in gray-scale RAW images and communicate with the remote server. Xcode version 6 (Apple Inc) was used for the App design.
Antibody lyophilization
Anti-human primary antibodies for CD3 and CD20 (Table S2) were lyophilized overnight using VirTis freeze dryer/ lyophilizer (SP Scientific, Warminster, PA). Lyophilized antibodies were stored at 4°C for indicated time periods. Following incubation, the lyophilized antibodies were resuspended in nuclease-free water (Life Technologies, Grand Island, NY) and used for flow cytometry and HALT experiments as mentioned above.
Supplementary Material
Supplementary Tables S1-S2 and Figures S1-S2.
Acknowledgements
We especially thank the members of the MGH Interventional Radiology Division who assisted with sample collection as well as the clinical providers within the MGH Center for Lymphoma. This work was supported in part by US NIH Grants R01-HL113156 (to H.L.), R01-EB010011 (to R.W.), R01-EB00462605A1 (to R.W.), T32CA79443 (to R.W.), K12CA087723-11A1 (to C.M.C.), K99CA201248-01 (to. H.I.), MGH Physician Scientist Development Award (to C.M.C.) and Department of Defense Ovarian Cancer Research Program Award W81XWH-14-1-0279 (to H.L.). AK was supported by the Mac Erlaine Research Scholarship from the Academic Radiology Research Trust, St. Vincent's Radiology Group, Dublin, Ireland.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sayed S, Lukande R, Fleming KA. Providing pathology support in low-income countries. J Global Oncol. 2015;1:3-6
2. Varmus H, Kumar HS. Addressing the growing international challenge of cancer: a multinational perspective. Sci Transl Med. 2013;5:175cm2
3. Livingston J. Cancer in the shadow of the AIDS epidemic in southern Africa. Oncologist. 2013;18:783-786
4. Chabner BA, Efstathiou J, Dryden-Peterson S. Cancer in Botswana: the second wave of AIDS in sub-saharan Africa. The oncologist. 2013;18:777-778
5. Naresh KN, Raphael M, Ayers L. et al. Lymphomas in sub-Saharan Africa-what can we learn and how can we help in improving diagnosis, managing patients and fostering translational research. Br J Haematol. 2011;154:696-703
6. Mwamba PM, Mwanda WO, Busakhala N. et al. AIDS-Related Non-Hodgkin's Lymphoma in Sub-Saharan Africa: Current Status and Realities of Therapeutic Approach. Lymphoma. 2012;2012:1-9 (Article ID 904367)
7. Langer R, Weissleder R. Nanotechnology. JAMA. 2015;313:135-136
8. Tseng D, Mudanyali O, Oztoprak C. et al. Lensfree microscopy on a cellphone. Lab Chip. 2010;10:1787-1792
9. Mudanyali O, Tseng D, Oh C. et al. Compact, light-weight and cost-effective microscope based on lensless incoherent holography for telemedicine applications. Lab Chip. 2010;10:1417-1428
10. Greenbaum A, Zhang Y, Feizi A. et al. Wide-field computational imaging of pathology slides using lens-free on-chip microscopy. Sci Transl Med. 2014;6:267ra175
11. D'Ambrosio MV, Bakalar M, Bennuru S. et al. Point-of-care quantification of blood-borne filarial parasites with a mobile phone microscope. Sci Transl Med. 2015;7:286re4
12. Laksanasopin T, Guo TW, Nayak S. et al. A smartphone dongle for diagnosis of infectious diseases at the point of care. Sci Transl Med. 2015;7:273re1
13. Stybayeva G, Mudanyali O, Seo S. et al. Lensfree holographic imaging of antibody microarrays for high-throughput detection of leukocyte numbers and function. Anal Chem. 2010;82:3736-3744
14. Wei Q, McLeod E, Qi H, Wan Z, Sun R, Ozcan A. On-chip cytometry using plasmonic nanoparticle enhanced lensfree holography. Scientific reports. 2013;3:1699
15. Im H, Castro CM, Shao H. et al. Digital diffraction analysis enables low-cost molecular diagnostics on a smartphone. Proc Natl Acad Sci U S A. 2015;112:5613-5618
16. Jaffe ES. The 2008 WHO classification of lymphomas: implications for clinical practice and translational research. Hematology Am Soc Hematol Educ Program. 2009;1:523-531
17. LaCasce AS, Kho ME, Friedberg JW. et al. Comparison of referring and final pathology for patients with non-Hodgkin's lymphoma in the National Comprehensive Cancer Network. J Clin Oncol. 2008;26:5107-5112
Author contact
![]() Corresponding authors: C. M. Castro, MD; H. Lee, PhD; R. Weissleder, MD, PhD. Center for Systems Biology, Massachusetts General Hospital, 185 Cambridge St, CPZN 5206, Boston, MA 02114, 617-726-8226 (Telephone) / 617-643-6133 (Fax) castro.cesarharvard.edu or hleeharvard.edu or rweisslederharvard.edu.
Corresponding authors: C. M. Castro, MD; H. Lee, PhD; R. Weissleder, MD, PhD. Center for Systems Biology, Massachusetts General Hospital, 185 Cambridge St, CPZN 5206, Boston, MA 02114, 617-726-8226 (Telephone) / 617-643-6133 (Fax) castro.cesarharvard.edu or hleeharvard.edu or rweisslederharvard.edu.
 Global reach, higher impact
Global reach, higher impact