13.3
Impact Factor
Theranostics 2016; 6(10):1528-1541. doi:10.7150/thno.15246 This issue Cite
Research Paper
Pharmacokinetic Behaviors of Intravenously Administered siRNA in Glandular Tissues
1. Institute of Molecular Medicine, Peking University, Beijing 100871, China;
2. Suzhou Ribo Life Science Co. Ltd., Jiangsu 215300, China;
3. Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), Tianjin 300072, China;
4. National Chengdu Center for Safety Evaluation of Drugs, State Key Laboratory of Biotherapy, Collaborative Innovation Center for Biotherapy, West China Hospital, Sichuan University, Chengdu 610041, China.
Received 2016-2-12; Accepted 2016-5-18; Published 2016-6-18
Abstract
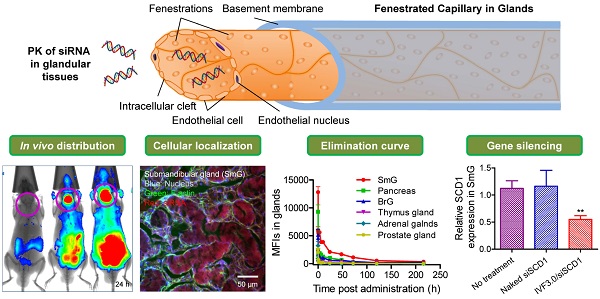
The pharmacokinetics of small interfering RNAs (siRNAs) is a pivotal issue for siRNA-based drug development. In this study, we comprehensively investigated the behavior of siRNAs in vivo in various tissues and demonstrated that intravenously-injected naked siRNA accumulated remarkably in the submandibular gland, bulbourethral gland, and pancreas, with a respective half-life of ~22.7, ~45.6, and ~30.3 h. This was further confirmed by gel separation of tissue homogenates and/or supernatants. In vivo imaging and cryosectioning suggested that delivery carriers significantly influence the distribution and elimination profiles of siRNA. Gene-silencing assays revealed that neither naked nor liposome-formulated siRNA resulted in gene knockdown in the submandibular and bulbourethral glands after systemic administration, suggesting that these glands function as drug reservoirs that enable slow siRNA release into the circulation. But robust gene-silencing was achieved by local injection of liposome-encapsulated siRNA into the submandibular gland. Our results enhance understanding of the pharmacokinetic properties of siRNAs and we believe that they will facilitate the development of siRNA therapy, especially for the submandibular gland.
Keywords: siRNA, pharmacokinetics, submandibular gland, biodistribution.
Introduction
RNA interference (RNAi) is a process by which RNA molecules inhibit gene expression, typically by cleaving messenger RNA (mRNA) or repressing mRNA translation. Through these processes, it compromises gene expression and regulates gene activity. It has not only become a powerful experimental tool for scientific research, but also provides a new approach to drug discovery and development [1, 2]. Almost 30 small interfering RNA (siRNA)-based therapeutics undergoing clinical studies, aiming to cure diseases such as transthyretin-mediated amyloidosis [3, 4], hypercholesterolemia [5], cancer [6, 7], Ebola [8], and hepatitis B [9].
Pharmacokinetics (PK) is a branch of pharmacology concerned with determining the fate (absorption, distribution, metabolism, and excretion) of substances administered to an organism. The pharmacokinetics of siRNA is one of the major issues that has to be elucidated during its therapeutic development. Although a dozen siRNAs are undergoing clinical trials, the exploration of their pharmacokinetic properties mainly focused on plasma and the targeted organs (e.g., liver, eye and tumor), few paid attention to the glandular tissues. It has been demonstrated that naked siRNAs are eliminated quickly from the circulatory system after intravenous injection into mice, rats, non-human primates, and humans, and the half-life (t1/2) can be as short as several minutes or ~ 0.5 h [6, 10-14]. T1/2 is the time required for the concentration of siRNA to reach half of its original value. The circulation time (or t1/2) of siRNA can be extended by forming complexes with delivery carriers [6, 11, 12, 15]. It has been shown that intravenously-administered naked siRNAs preferentially accumulate in the kidneys and are excreted in the urine. These siRNAs can selectively suppress gene function in renal proximal tubules [16]. Moreover, it has also been found that the amount of siRNA accumulating in the kidneys is much greater than in other tissues after intravenous injection into both normal rats and disease models [10]. However, the biodistribution profiles vary when different delivery carriers are used [12, 17-20].
The above studies mainly focused on the pharmacokinetics of siRNAs in the circulatory system and/or the main organs, such as the liver, heart, spleen, lung, kidneys, brain, or bladder, as well as tumors and targeted organs (such as the eyes); a few have examined the gastrointestinal tract, lymphoid tissue, adipose tissue, muscle, and skin. However, little work has focused on glandular tissue, such as the submandibular (salivary), thymus, adrenal, bulbourethral, and prostate glands, and the pancreas.
Previously, we reported that some of the systemically-administered naked or formulated siRNAs are processed in the liver, secreted into the intestine via the common bile duct, and finally eliminated in excrement [21]. Moreover, for the first time, we showed that both naked and formulated siRNAs are distributed into the submandibular gland after intravenous administration. In this study, by using two RNase-resistant siRNAs, we set out to explore the distribution and elimination profiles of systemically-administered siRNAs not only in the main organs (heart, liver, spleen, lung, and kidney), but also in glandular tissues (submandibular, thymus, adrenal, bulbourethral, and prostate glands, as well as the pancreas). Also, the gene-silencing efficiency of siRNAs administered into the submandibular gland via both tail vein injection and local injection were further investigated.
Materials and methods
Materials
We used two Cy5-labeled siRNAs, Cy5-siRNA-1 (Catalog: SR-NC001) and Cy5-siRNA-2 (Catalog: SR1602), synthesized by Suzhou Ribo Life Science Co., Ltd. (Kunshan, China). The 5' end of the sense strand was labeled with the Cy5 fluorophore. Several bases of the sense and antisense strands were modified with a methoxy group or fluorine at the 2' site hydroxyl group so as to enhance the stability in serum. Both siRNA-1 and siRNA-2 are irrelevant siRNAs without any target gene in normal mouse, rat, and human. They are strongly RNase-resistant as they were very stable in mouse serum (Supplementary Figure S1). The sequences of siRNA-1 and siRNA-2 used in the gene silencing assays are the same with the sequences of Cy5-siRNA-1 and Cy5-siRNA-2, respectively. siSCD1 targeting stearoyl-CoA desaturase-1 (SCD1) (Catalog: SR3504) was also from Suzhou Ribo Life Science Co., Ltd. It was chemically modified at certain bases to enhance its stability and avoid a potential immune response. The sequences of PCR primer sets of SCD1 were as follows: forward: 5'-TGGTGAACAGTGCCGCGCAT-3', reverse: 5'-ACTCAGAAGCCCAAAGCTCAGCTAC-3'. The sequences of primer sets of β-actin were as follows: forward: 5'-AGCTTCTTTGCAGCTCCTTCGTTG-3', reverse: 5'-TTCTGACCCATTCCCACCATCACA-3'. All these primers were from CapitalBio Corp. (Beijing, China).
RNAlater® was from Sigma-Aldrich (St Louis, MO). The reverse transcription kit was from Promega Corp. (Fitchburg, WI) and the real-time PCR kit was from Beijing ComWin Biotech Co., Ltd. (Beijing, China). Golden View and Sybr Gold for staining nucleic acids in gels were from Beijing BioDee BioTech Corp., Ltd. (Beijing, China) and Life Technologies (Molecular Probes, Eugene, OR), respectively. Lipofectamine 2000 and Invivofectamine® 3.0 were also from Life Technologies. Loading buffer was provided by Takara Bio, Inc. (Kyoto, Japan). Agarose was from Oxoid (Hampshire, UK). Optimal cutting temperature (OCT) compound was from Sakura Finetek USA, Inc. (Torrance, CA). DAPI (4',6-diamidino-2-phenylindole, for staining nuclei) was from Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing, China, and fluorescein isothiocyanate-labeled phalloidin (for staining F-actin) was from Sigma-Aldrich (MO, USA).
Evaluation of siRNA stability
Mouse serum was used to investigate the stability of siRNA-1 and siRNA-2. We used two serum/siRNA incubation solutions, with 90% and 50% (v/v) serum. The serum was prepared by mixing six serum specimens collected from six normal C57BL/6 mice (three males and three females). Ten microliters of siRNA (20 μM) and 90 μl of serum were mixed to prepare 100 μl of serum/siRNA incubation solution (with 90% serum). Meanwhile, 10 μl of siRNA (20 μM), 50 μl of serum and 40 μl of 1×PBS were mixed to obtain another 100 μl incubation solution (with 50% serum). Then these solutions were incubated at 37°C, followed by collection of 10 μl samples at each desired time point and immediate freezing at -20°C until gel electrophoresis, in which all samples were separated on 12% native polyacrylamide gels for 70-80 min at a constant voltage of 100V (current ~40 mA). Finally, the gels were stained with Sybr Gold for 20 min and exposed with ChampGel (Beijing Sage Creation Science Co, Ltd., Beijing, China) to show the locations of the siRNAs.
Animals
Animals were purchased from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) and maintained in the Peking University Laboratory Animal Center (an AAALAC-accredited and specific pathogen-free (SPF) experimental animal facility). All procedures involving experimental animals were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Peking University.
In vivo fluorescence imaging
Male C57BL/6 mice, 5-7 weeks old and weighing 18-22 g, were used to investigate the biodistribution profile of systemically-administered naked siRNA. The abdomen was depilated with depilatory cream one day before administration to facilitate observation. Formulations were administered to each mouse via tail vein injection at 2.5 mg/kg (for siRNA). The fluorescence signal of cyanine Cy5 from the whole body was recorded at given time points using an imaging system (Kodak In-Vivo Imaging System FX Pro, Carestream Health, Toronto, Canada). In this experiment, 630 nm excitation and 700 nm emission filters were selected. According to the manufacturer's specifications, the bandpasses of the excitation and emission filters were 20 nm and 60 nm (wide-angle), i.e., the excitation and emission spectra were 620-640 nm and 670-730 nm. The other conditions were as follows: exposure time, 60.0 sec; X-binning, 2× binning; Y-binning, 2× binning; f-stop, 2.50; field of view (FOV), 180 mm; focal plane, 13.0 mm. Mice were anesthetized during imaging with a mixture of oxygen and isoflurane using a vaporizer (Matrix VIP3000 Isoflurane Vaporizer, Midmark Corporation, Ohio, USA). At the end-point, mice were sacrificed by cervical dislocation, and the major organs were isolated and examined. If perfusion of the blood is needed, the abdomen of the animal was quickly opened and the postcava was cut after cervical dislocation. Then 10 mL normal saline was injected into the heart of the animal under a constant speed. Then the tissues were isolated. The above experiments were independently replicated three times. Semi-quantitative analyses were performed using a molecular imaging software package (Carestream Health, Toronto, Canada).
Tissue cryosectioning
Tissues were placed in Omnisette tissue cassettes, embedded in OCT, and frozen on a foam floater on liquid nitrogen in a pre-chilled Dewar flask for ~1 min until the OCT turned white and opaque. Then, the specimens were cut into 6-10 μm sections on a cryostat. Each section was picked up on a glass slide, stained with DAPI to visualize the nucleus and with phalloidin to visualize F-actin in order to display the rough outline of the cell. Finally, cryosections were examined under a confocal microscope (LSM 510 or 700, Carl Zeiss, Oberkochen, Germany).
Gel separation of in vivo specimens
In order to confirm the finding that siRNAs accumulate in glands after systemic administration, tissue homogenates and supernatants were further separated on gels. Male C57BL/6 mice weighing ~25 g were used in this assay. Cy5-labeled siRNA-2 was dissolved to 1 mg/ml with DEPC (diethylpyrocarbonate)-treated water. Normal saline was used to adjust the siRNA solution to an injectable volume. Then the solution was intravenously injected at 5 mg/kg (in 200 μl). We are intended to determine the presence or absence, rather than the amount of siRNA in this assay. In consideration of the experience that siRNA usually coprecipitates with unknown proteins when centrifuging because of its negative charge (data not shown), and the phenomenon that tissue specimens of siRNA are hard to run into the gel during electrophoresis, we used a relative higher dose of siRNA in this test (5 mg/kg) than the dose used in the biodistribution assays (2.5 mg/kg).
After sacrifice by cervical dislocation at 0.5 h, blood, urine, submandibular gland, bulbourethral gland, pancreas, and kidneys were collected. Gland tissues (~100 μg) were cut up with a knife-blade and immediately transferred into 0.5 ml DEPC water followed by homogenization (Superfine Homogenizers, Fluko®, Essen, Germany), ultrasonication with a cell disruptor (Uibra-cellTM, Sonics, CT, USA) and freeze-thawing three times in liquid nitrogen/water at 37°C. Samples were slowly thawed at 4°C for the last thawing cycle. Meanwhile, 3 μg of pure Cy5-labeled siRNA-2 was dissolved to 20 ng/μl with DEPC water, then treated with the same procedures as for the tissue samples. Then one half of the tissue homogenates and the pure siRNA solutions were kept at -20°C until gel separation. Subsequently, the other half of the tissue homogenates were centrifuged for 3 min at 12000 rpm and 4°C (Eppendorf® Centrifuge 5417R, Eppendorf, Germany), and the first supernatants (S1s) were collected (not pellucid). These supernatants were further centrifuged for 12 min at 12000 rpm and 4°C, then the second supernatants (S2s) were harvested (typically pellucid) and stored at -20°C until gel separation. Furthermore, sera were obtained by centrifuging blood samples for 15 min at 3000 rpm at room temperature (Eppendorf® Centrifuge 5415D, Eppendorf, Germany), and also kept at -20°C.
Finally, each sample of serum, urine, tissue homogenate, and second supernatants (S2s) was mixed with loading buffer, loaded into 5% native PAGE (polyacrylamide gel electrophoresis) gel and separated for ~1 h at a constant current of 20 mA. The electrophoresis chamber was placed in a container surrounded by ice and covered by black film in order to protect the siRNA from RNase attack and the Cy5 fluorophore from light quenching. Pure siRNA-2 with and without treatments were included as controls. The loading volume of serum, tissue homogenate, and supernatant was 20-30 μl, and that of urine was 1-5 μl (depending on the siRNA concentration). Finally, the siRNA bands were analyzed with a fluorescence imaging system (Kodak In-Vivo imaging system FX Pro, Cearthstream Health, Toronto, Canada).
In vivo gene silencing
Real-time PCR was used to determine whether the naked siRNAs that accumulated in glands triggered gene silencing. Anti-SCD1 siRNA (siSCD1) was used in this assay. Normal male C57BL/6 mice, weighing ~20 g, were randomly divided into five groups (6 mice per group), followed by intravenous injection of following formulations: (1) normal saline; (2) naked siRNA-1 (without Cy5-labeling); (3) naked siSCD1; (4) IVF3.0/siRNA-1; and (5) IVF3.0/siSCD1. IVF3.0/siRNA-1 and IVF3.0/siSCD1 were prepared with siRNA and Invivofectamine® 3.0 (IVF3.0, Life Technologies, Carlsbad, CA) according to the manufacturer's protocol. All of the other carriers used in the biodistribution assays can efficiently bind siRNA and form regular nanoparticles. They displayed high potency in transfecting siRNA into cells in vitro, however, they showed unsatisfactory delivery efficiency in vivo (some of them showed ideal tumor-targeted delivery efficiency in vivo, such as the amphiphilic dendrimer). In contrast, IVF3.0 is a novel lipid-based in vivo transfection reagent that has exhibited robust siRNA delivery efficacy in the liver [22], demonstrating it can smoothly reach hepatocytes, induce endocytosis and efficiently escape form the endosome/lysosome. Hence, it was included as a control in this assay. The formulations were administered as 10 mL/kg body weight at 2.5 mg/kg (for siRNA). Tissue samples were subjected to RNAlater® (Sigma-Aldrich) at 72 h after administration. Subsequently, tissues were homogenized, followed by adding RNAVzol (Vigorous Biotechnology Beijing Co., Ltd., Beijing, China) and extracting total RNA according to the manufacturer's protocol. cDNA was prepared by incubating the reaction mixture [(4 μl MgCl2 (500 mM), 2 μl reverse transcription buffer (10×), 2 μl dNTP mixture (10 mM), 0.5 μl recombinant RNasin, 0.5 μl AMV, 1 μl Oligo d(T), and 10 μl total RNA)] at 42°C for 15 min, 95°C for 5 min, and 4°C for 5 min. Then the real-time PCR reaction system [(10 μl reaction mix, 1 μl forward primer (5 μM), 1 μl reverse primer (5 μM), 1 μl cDNA template, and 7 μl ddH2O)] was prepared and first hot-started for 10 min at 95°C before 40 cycles of 30 sec at 95°C, 30 sec at 60°C, and 30 sec at 72°C. After the melting procedure was completed, samples were stored at 4°C. The expression level of SCD1 was analyzed using the Ct (cycle threshold) values with the standard protocol. β-actin was selected as the reference gene, and its PCR reaction conditions were the same as those for SCD1.
Local injection of siRNA into the submandibular gland
To evaluate whether naked and carrier-formulated siRNA administered by local injection could mediate gene silencing in vivo, we further investigated the submandibular gland. Male C57BL/6 mice weighing 18-22 g were divided into three groups and the neck was depilated. Twenty-four hours later, normal saline, naked siSCD1, or IVF3.0/siSCD1 was locally injected into the submandibular gland. The siRNA dose of naked siSCD1 was 2.0 mg/kg and of IVF3.0/siSCD1 was 0.3 mg/kg. Submandibular glands were collected 72 h later. Extraction of total RNA, cDNA preparation, and real-time PCR were performed according to the above protocols to analyze the gene knockdown efficiency.
Statistical analysis
Data are expressed as mean ± SEM. Statistical variance was calculated by t-test and P <0.05 was considered statistically significant.
Results and discussion
Dynamic distribution of systemically-administered naked siRNA
Many studies have explored the pharmacokinetic (PK) properties of siRNA in the circulation, particularly the concentration-time curve in plasma [6, 10-14]. A few studies have also focused on the concentration-efficacy relationship in targeted organs, such as the liver (via intravenous or subcutaneous injection) [23, 24] and the eyes (via intravitreal injection) [14]. However, the distribution and kinetic properties in other organs or tissues remain to be further characterized. In this study, we investigated the biodistribution profiles of intravenously-administered naked siRNAs. Here, siRNAs were labeled with the Cy5 fluorophore, injected at 2.5 mg/kg, and visualized with an in vivo fluorescence imaging system to reveal their distribution in both living animals and isolated organs/tissues. Two siRNAs (siRNA-1 and siRNA-2) were used in parallel to exclude the possibility of a sequence-specific distribution pattern. At each time point, three animals with each treatment were sacrificed by cervical dislocation to collect fluorescence intensity data.
Dynamic distribution of naked siRNA injected via tail vein. (a) Whole-body imaging of intravenously administered siRNA. Pink circles, submandibular glands. (b) Fluorescence images of isolated tissues harvested at each time point. The time points are shown in the lower right corners of the images. (c) Semi-quantitative analyses of (b) using the software package included with the in-vivo imaging system (mean ± SEM, n = 3). B, brain; SmG, submandibular gland; Pa, pancreas; Th, thymus gland; A, adrenal gland; BrG, bulbourethral gland; Pr, prostate gland; H, heart; Lu, lung; Li, liver; S, spleen; K, kidney; Lip, lipid; SM, skeletal muscle; Te, testes.
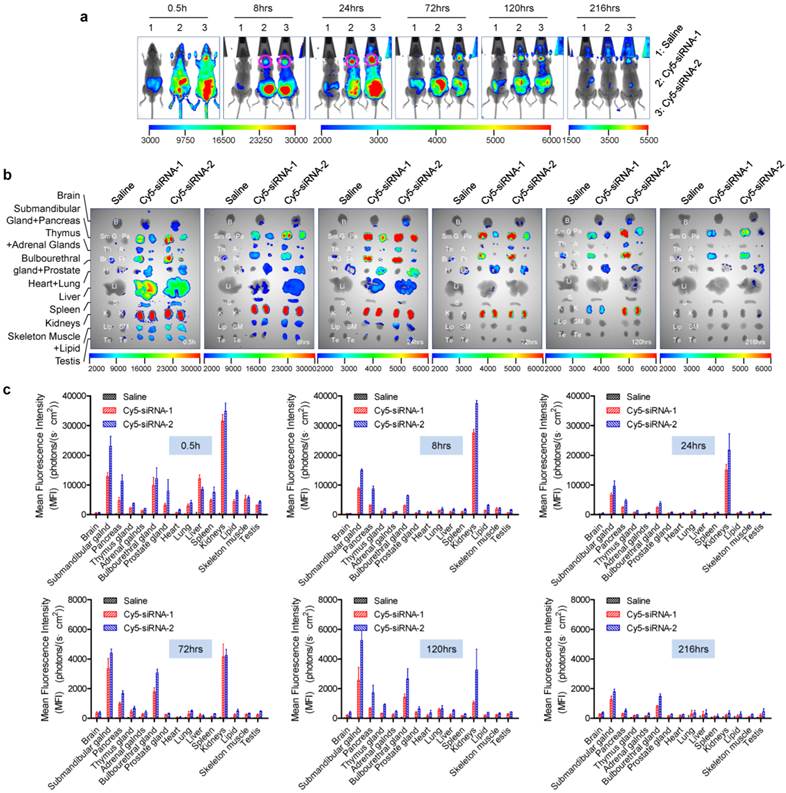
In accord with our previous findings [21, 25, 26] and other reports [16], a large proportion of intravenously-injected naked siRNAs rapidly accumulate in the kidneys and bladder. Thirty minutes after administration, naked siRNAs were widely distributed throughout the body via the circulation (Figure 1a, b). Some were also entrapped by the liver during the first few hours [27, 28] (Figure 1). Particularly, a strong siRNA fluorescence signal was detected in the submandibular gland, and this was sustained for >216 h (9 days) (Figure 1a, as the pink circles indicated). In addition, we found that naked siRNA also significantly accumulated in the bulbourethral gland and pancreas for a long period (>216 h for the bulbourethral gland). Since the bulbourethral gland is anatomically close to the bladder, it was hard to distinguish the two signals in whole-body imaging. However, the distribution pattern was much clearer in the images of isolated organs (Figure 1b).
Based on the different structural properties of capillaries, endothelial systems can be divided into three classes: (i) continuous and non-fenestrated capillaries (e.g., heart, lung, muscle, and brain), (ii) continuous and fenestrated capillaries (e.g., gastrointestinal tract, exocrine/endocrine glands, and renal glomeruli), and (iii) discontinuous and fenestrated endothelium (e.g., liver sinusoidal capillaries) [29]. There is no opening in the walls of continuous capillaries and they are lined continuously by endothelial cells. Fenestrated capillaries have small openings, called fenestrae, between the endothelial cells (80-100 nm in diameter). These cells are spanned by a small, non-membranous, permeable diaphragm of radially-oriented fibrils and allow the diffusion of small molecules and limited amounts of protein. The basement membrane of endothelial cells is continuous over the fenestrae. Discontinuous capillaries, also called sinusoids, are a special type of open-pore capillary that has larger openings (30-40 µm in diameter) in the endothelium. They have a large lumen, many fenestrations with no diaphragm and a discontinuous or absent basal lamina [30]. The issue of siRNA accumulation in the liver and kidneys has been much discussed. On one hand, both the liver and kidneys are processing organs for siRNA excretion from the body [21]. On the other hand, siRNA accumulation in these tissues may be aided by their unique capillary properties. Actually, in the fenestrated and sinusoidal endothelial systems, naked or formulated siRNAs can penetrate blood vessel and reach the corresponding tissues. This is probably the primary reason for the accumulation of siRNA in glands.
Furthermore, siRNAs in other organs, including the kidneys, were barely detectable at 216 h after injection. We found that brain, heart, fatty tissue, testes, and skeletal muscle had no fluorescence signal or a much weaker signal at all time-points compared with the organs described above. According to the semi-quantitative data shown in figure 1c, siRNA concentration-time curves for two siRNAs were further plotted (supplementary Figure S2). Then areas under the curves (AUCs) (supplementary Table S1) were calculated by DAS2.0 software, which also suggested that a lot of siRNAs accumulated in kidneys, submandibular gland, bulbourethral gland and pancreas. Additionally, no significant difference was found between siRNA-1 and siRNA-2, suggesting that this distribution pattern is a general phenomenon that is independent of the siRNA sequences.
It is noteworthy that the MFIs defined here actually represented the amount of siRNA accumulated in both the tissue and the blood present in the tissue, because we did not perfuse the circulatory system before isolating the organs in this assay. However, the siRNA signal contributed by the blood accounted for a very small proportion because the first observation time point in this assay was 0.5 h and siRNAs were quickly excreted from the circulation (t1/2 <0.5 h), also because the glands were not rich in blood.
siRNA concentration-time curves for tissues
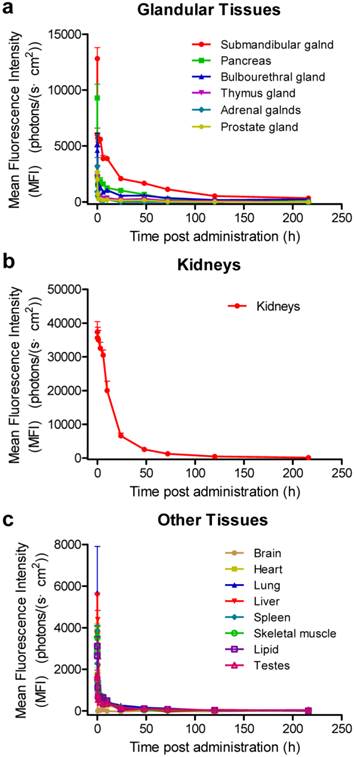
In order to explore the detailed distribution and clearance properties of siRNA in various tissues, we further recorded the fluorescence intensities of the Cy5-labeled siRNA-2 at 12 time points (1 min, 5 min, 0.5 h, 1 h, 3 h, 6 h, 10 h, 24 h, 48 h, 72 h, 120 h and 216 h) post administration. In consideration of that siRNAs entrapped by the tissues may affect the fluorescence imaging in those tissues rich in blood (such as the heart), especially at the initial two time points, the blood was washed out of the body and perfused with 10 mL saline for each mouse before tissue-isolating and imaging. Data revealed similar biodistribution profiles to the patterns showed in figure 1 (supplementary Figure S3, a and b). siRNA concentration-time curves were further plotted with the semi-quantitative data shown in figure S3b, and the pharmacokinetic parameters were analyzed by DAS2.0 software.
The results suggested that the distribution and elimination patterns varied among these tissues (Figure 2 and Table 1). Intravenously-administered naked siRNAs are quickly eliminated from the circulation and accumulate in the liver, kidneys, bladder, submandibular gland, bulbourethral gland, and pancreas, suggesting a process of waxing and waning in different metabolic compartments. The distribution half-lives (t1/2αs) of all tissues except kidneys are less than 1 h. More importantly, siRNA in the submandibular, bulbourethral, thymus and prostate gland, as well as pancreas, showed long retention times, with elimination half-lives (t1/2βs) of ~22.7, ~45.6, ~38.9, ~37.1 and ~30.3 h, respectively. The AUC(0-t)s for the submandibular gland, pancreas and bulbourethral gland were 189650.0, 80447.6 and 72754.9, respectively, only smaller than that for the kidneys (685839.9). The AUC is of particular use in estimating the bioavailability and the total clearance of drugs. These data further demonstrated a large proportion of siRNAs accumulated in the glands and remained for long time, suggesting the glandular tissues, especially for the submandibular gland, pancreas and bulbourethral gland, played pivotal roles in siRNA pharmacokinetic behaviors in vivo.
In addition, the MFI of the kidneys stayed at the highest level over 72 h than that of other tissues, and the MFIs of the submandibular and bulbourethral glands have become to be higher than that of the kidneys at 120 h and 216 h (Figure 1c, S3 and S4). Given that the amount of siRNA in circulation at those time points was negligible, the strong signal in kidneys might be due to the slow and continuous release of siRNA from the glandular tissues, especially from the submandibular gland.
Half-lives and AUCs of intravenously-administered naked siRNA in tissues with perfusion before imaging
| Tissues | t1/2α (h) | t1/2β (h) | AUC(0-t) {[photons/(s·cm2)]·h} | AUC(0-∞) {[photons/(s·cm2)]·h} |
|---|---|---|---|---|
| Brain | 0.030 | 17.567 | 1006.9 | 1099.0 |
| Submandibular gland | 0.018 | 22.656 | 189650.0 | 190622.0 |
| Pancreas | 0.017 | 30.309 | 80447.6 | 81278.3 |
| Thymus gland | 0.110 | 38.895 | 25877.7 | 26532.5 |
| Adrenal glands | 0.022 | 2.750 | 2854.9 | 2898.5 |
| Bulbourethral gland | 0.236 | 45.583 | 72754.9 | 75365.2 |
| Prostate gland | 0.195 | 37.088 | 13508.5 | 13733.6 |
| Heart | 0.043 | 9.379 | 9510.1 | 9567.9 |
| Lung | 0.065 | 8.371 | 12492.1 | 12581.4 |
| Liver | 0.862 | 13.008 | 17716.1 | 17801.0 |
| Spleen | 0.049 | 3.783 | 8344.3 | 8404.1 |
| Kidneys | 11.926 | 11.927 | 685839.9 | 686447.9 |
| Skeletal muscle | 0.298 | 12.035 | 16638.5 | 16691.5 |
| Lipid | 0.258 | 16.445 | 17659.7 | 17712.4 |
| Testes | 0.548 | 69.315 | 8321.8 | 14886.8 |
Notes: The the pharmacokinetic parameters, such as half-life (t1/2) and AUC, of siRNA were calculated by DAS2.0 software. t1/2α, distribution half-life; t1/2β, elimination half-life; AUC(0-t), area under the siRNA concentration-time curve from time zero to the last quantifiable value; AUC(0-∞), area under the siRNA concentration-time curve from time zero to infinity, which represents the total drug exposure over time. h, hour(s).
As mentioned above, the blood of the animals has been perfused with saline before organ-isolating and imaging when collecting the data of figure 2, S3 and table 1. Meanwhile, we have also obtained a data set in parallel from the animals without perfusion, as shown in supplementary figure S4, S5 and table S2. The distribution and elimination profiles identified from these data are very similar to those concluded from the perfused organs. The glandular tissues also displayed strong MFIs, long elimination half-lives (t1/2β) and large AUCs.
Kidney-bladder-urine is a well-known excretion pathway for systemically-administered siRNA. In addition, liver-gallbladder-intestine-excrement is another recently-identified elimination route [21, 27]. Therefore, if the circulation is defined as the first drug “warehouse”, several glands, including the submandibular gland, bulbourethral gland and pancreas, may be considered as the second natural drug reservoir. These results are important for siRNA-based drug development because they offer a better understanding of siRNA pharmacokinetics and could greatly facilitate the optimization of dosage regimens.
Concentration-time curves of siRNAs in various organs. The mice were sacrificed and the fluorescence intensities were recorded at 1 min, 5 min, 0.5 h, 1 h, 3 h, 6 h, 10 h, 24 h, 48 h, 72 h, 120 h and 216 h after intravenous injection of Cy5-labeled siRNA-2. Based on the semi-quantitative data shown in supplementary figure S3b, siRNA concentration-time curves were plotted. The blood was perfused with 10 mL saline before each time of fluorescence imaging for each mouse. Data are mean ± SEM (n = 3).
siRNA stability
The above assays were performed using two Cy5-labeled siRNAs. It was extremely important that tracking of the Cy5 signal represented the behavior of the siRNAs. Hence, we first evaluated the stability of the siRNAs in mouse serum (rich in RNase), and second, we separated tissue specimens in gels to confirm the location of the siRNAs. The data showed that siRNA-2 was stable in both 90% and 50% serum, as full-length siRNA bands were clearly evident even after 144 h (6 days) of incubation at 37°C (Supplementary Figure S1), revealing its strong resistance to RNase attack. siRNA-1 was stable in serum for at least 24 h in 90% serum. Only one or two bases of siRNA-1 (probably the overhang dTdT) were cleaved in 50% serum at 144 h (Supplementary Figure S1). Given that the half-life of intravenously-administered naked siRNA is <0.5 h (5-30 min) in the circulation [6, 11, 14], siRNA-1 and siRNA-2 (particularly siRNA-2) were sufficiently stable to permit the data collection and the corresponding interpretations in the above and following experiments.
Validation of siRNA accumulation in glands
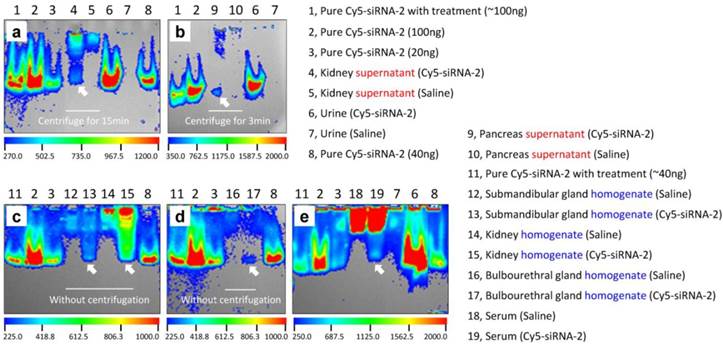
In addition, in vivo specimens (urine, blood, and tissues) were collected or harvested at 0.5 h after intravenous administration of Cy5-labeled siRNA-2. Then tissue homogenates and/or supernatants, as well as sera, were prepared before separation on 5% native PAGE. Tissue homogenates were obtained by successive homogenization, ultrasonication, and repeated freezing-thawing. An additional centrifugation step was required for preparing tissue supernatants. We found that pure naked siRNA, regardless of treatment, showed clear bands in the gels (Figure 3, lanes 1, 2, 3, 8, and 11 in a-e). In line with expectations, urine collected from siRNA-treated animals displayed a pattern similar to naked siRNA at the same location in the gels (Figure 3, lane 6 in a, b, and e). In contrast, urine collected from saline-treated animals showed no bands (Figure 3, lane 7 in a, b, and e). Also as expected, kidney specimens from siRNA-treated animals, both supernatants and homogenates, revealed distinct siRNA bands in the gels (Figure 3, lanes 4 and 15 in a and c). The same band was observed for serum samples (Figure 3, lane 19 in e). More importantly, visible fluorescence bands located at the same site as naked siRNA were found in homogenates or supernatants of the submandibular gland, bulbourethral gland, and pancreas (Figure 3, lanes 13, 17, and 9 in b-d). These results convincingly demonstrated that intravenously-administered siRNA accumulates in these glands. Moreover, all samples with complicated compositions, such as serum and all tissue homogenates and supernatants, showed some autofluorescence in the gel (Figure 3, lanes 4, 5, 9, 10, and 12-19 in a-e), which might be due to unknown proteins or molecules that were excited under these experimental conditions. However, these signals were located in the upper part of the gels, revealing molecular weights much larger than siRNA. In addition, the fluorescence intensities in these gels were not comparable because we did not unify the loading amount of siRNA while performing gel separation, as we intended to determine the presence or absence, rather than the amount of siRNA (see Figure 1).
Gel separation of tissue homogenates and supernatants. (a-e) Fluorescence images of samples of kidney, urine, pancreas, submandibular gland, bulbourethral gland, and serum in five gels. Clear siRNA bands (as the arrows indicated) occurred in the gels, which robustly demonstrated that siRNA indeed accumulated in these tissues after intravenous administration. 'Pure Cy5-siRNA-2 with treatment' means Cy5-siRNA-2 was homogenized, ultrasonicated, and repeatedly frozen and thawed under the same conditions as the tissue samples. Bands marked 'homogenate' indicate that these samples were homogenized, ultrasonicated, and repeatedly frozen and thawed, but not centrifuged.
Influence of delivery carrier on siRNA biodistribution
Having demonstrated that naked siRNA accumulates in the submandibular gland, bulbourethral gland, and pancreas, we then used various siRNA delivery carriers to investigate whether they affected the siRNA distribution profile.
Lipofectamine 2000 is widely used for nucleic acid transfection. In vivo fluorescent imaging data (supplementary Figure S6) suggested that siRNAs complexed with Lipofectamine 2000 accumulated less in the submandibular gland than naked siRNA, and they were excreted much more rapidly than naked siRNA. Along with this, more siRNA was transported into the liver and spleen. Another in vivo delivery system, Invivofectamine® 3.0 (IVF3.0), also delivered more siRNA into the liver and less siRNA into the glands (data not shown). IVF3.0 is a lipid-based liver-targeting delivery system, which showed a high silencing efficiency in hepatocytes after combination with siRNA [22] (Figure 6a).
RGD10-10R, with the amino-acid sequence DGARYCRGDCFDGRRRRRRRRRR (from the N terminal to the C terminal) and a disulfide linkage between the two cysteines, has been tested for integrin-targeted siRNA delivery in vitro and in vivo [31]. Taking advantage of their electrostatic interaction with polyarginine (10R), negatively-charged siRNAs are readily complexed by RGD10-10R peptides at different molar ratios, forming spherical RGD10-10R/siRNA nanoparticles with a narrow size distribution [31]. Nanoparticle-treated animals showed less siRNA accumulation in the submandibular gland, bulbourethral gland, and pancreas than naked siRNA-treated mice (Supplementary Figure S7a-b). Tissue cryosections revealed that RGD10-10R/siRNA nanoparticles were located in the acinar compartment as well as in duct cells, primarily in the form of peptide/siRNA granules (Supplementary Figure S7c). Another bifunctional peptide, NGR-10R, also delivers a considerable mass of siRNA into the submandibular gland, pancreas, and bulbourethral gland after systemic administration into mice [32].
In addition, our previous studies based on cationic polymers [25, 26, 33-35] and mesoporous silica [36] also show that delivery carriers play a pivotal role in both the distribution profile and elimination pattern. siRNA complexed with PEA [PEGylated poly(2-aminoethyl methacrylate)] and PEAG [guanidinylated PEGylated poly(2-aminoethyl methacrylate)] at various N/P ratios accumulate less in the submandibular gland and pancreas than naked siRNA [33]. The N/P ratio of polymer to siRNA is calculated as the molar ratio of the amino groups (N) on the polymer to the phosphate groups (P) on the siRNA. Additional imaging and cryosection data also showed that the fluorescence intensities of the PEA/siRNA and PEAG/siRNA complexes were lower than that of naked siRNA in submandibular glands (Supplementary Figure S8). Complexes of siRNA and PDMAEMA [poly(2-dimethylaminoethyl methacrylate)]-based polymers displayed various distribution profiles [25, 26, 34, 35] (Supplementary Figure S9). It is difficult for naked siRNAs to penetrate cell membranes. In contrast, polymers can deliver them into the cytoplasm, as revealed by pancreas cryosections (Supplementary Figure S10). Here, siRNAs were found surrounding the nuclei and dispersed in the cytoplasm. siRNAs loaded in PDMAEMA-functionalized mesoporous silica nanoparticles (ssCP-MSNs) accumulate more in the pancreas, thymus, and adrenal gland in mice than naked siRNA [36]. ssCP-MSNs deliver many siRNAs into the adrenal gland [36], but this does not occur with the RGD10-10R/siRNA complex [31].
Further investigations using cationic and amphiphilic dendrimers also suggested irregular gland distribution patterns. Naked siRNA and a cationic dendrimer G5/siRNA complex (N/P =10:1) showed comparable accumulation in the submandibular gland (Supplementary Figures S11 and S12). However, the amount of siRNA in the submandibular gland and pancreas was significantly elevated for the amphiphilic dendrimer (AD)/siRNA complex with an N/P ratio of 10:1 (Figures 4 and 5). Moreover, similar to the PECbD1/siRNA 15:1 complex, the AD/siRNA complex with an N/P ratio of 5:1 was also located around the nucleus in pancreatic cells (Figure 5b), showing a promising prospect for application. Here, the N/P ratio was calculated as the molar ratio of 'total end amines in dendrimer' to 'phosphates in siRNA'.
It seems that, in most cases, carrier-encapsulated siRNA accumulates less in glandular tissues than naked siRNA. This may be attributed to carriers deliver more siRNA to other tissues, such as liver and/or tumor. However, AD/siRNA with an N/P ratio of 10:1 and most siRNA formulations containing PDMAEMA-derived materials transported more siRNA into the submandibular gland and pancreas than naked siRNA. The distribution patterns varied among different carriers. The physicochemical properties, such as molecular specificity, particle size, and zeta potential, may be the important influence factors for their distribution patterns (supplementary Table S3). However, it is very difficult to identify the exact influence factor(s) since it is so complicated for the in vivo behaviors of carrier-encapsulated siRNA.
In vivo distribution of AD/siRNA complexes in C57BL/6 mice (2.5 mg/kg for siRNA). (a) Whole-body imaging at given time points after intravenous administration. (b) Fluorescence assessment of isolated organs of mice at the end observation point (5 days after injection). (c) Semi-quantitative analyses of (b) using a molecular imaging software package (Carestream Health, Toronto, Canada). The data were normalized to corresponding tissues from saline-treated animals. Each bar represents the mean ± SEM of 2-3 independent experiments. *P < 0.05 vs corresponding tissue(s) from mice treated with naked siRNA. n = 3 for saline, naked siRNA, and AD/siRNA 10:1 groups; n = 2 for AD/siRNA 5:1 group.
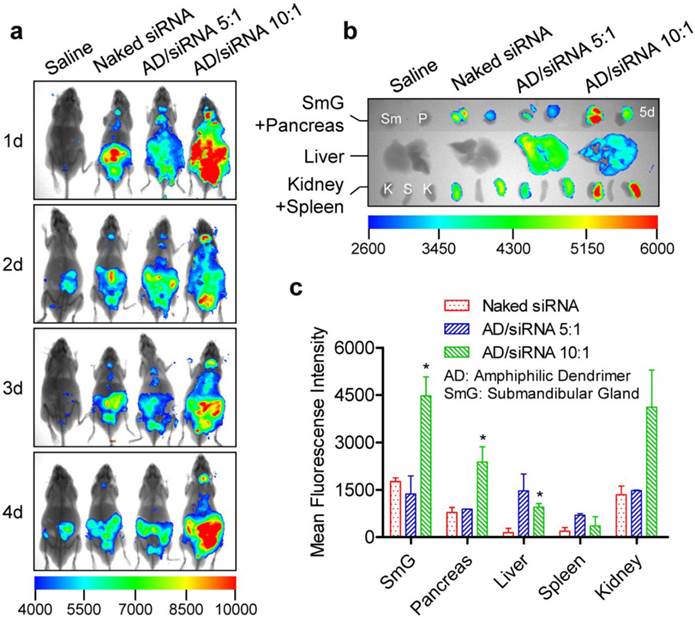
Collectively, the carriers described above cover many representative siRNA delivery systems, including liposomes, peptides, polymers, dendrimers, and inorganic materials. Although the issue of the presence or absence of siRNA accumulation in certain tissues (glands) was not affected by these carriers, the amount of siRNA might be remarkably influenced. No doubt, these findings provide a better understanding of the pharmacokinetic properties of siRNA and will help to optimize the therapeutic regimens of siRNA-based drugs.
Gene silencing in the submandibular gland
It is both interesting and important to know whether the siRNA accumulating in glands can mediate gene silencing. Therefore, siSCD1, an RNase-resistant siRNA targeting SCD1, a key enzyme in fatty-acid metabolism, was used to evaluate the knockdown efficiency in vivo. Elevated expression levels of SCD1 are correlated with obesity [37, 38] and tumor malignancy [39, 40]. It is also involved in germ-cell determination [41], liver cell differentiation [42], and cardiac development [43]. Previously we confirmed that SCD1 is moderately expressed in the submandibular glands of normal mice (data not shown). Here, each of five groups of mice (6/group) were given one of the following formulations: (1) normal saline, (2) naked siRNA-1, (3) naked siSCD1, (4) IVF3.0/siRNA-1, or (5) IVF3.0/siSCD1. siRNA-1 was a negative control because it has no targeting gene in mouse, rat, and human. IVF3.0 is a lipid-based in vivo delivery system. The siRNA dose was 2.5 mg/kg for groups 2-5, and the mice were sacrificed at 72 h after administration.
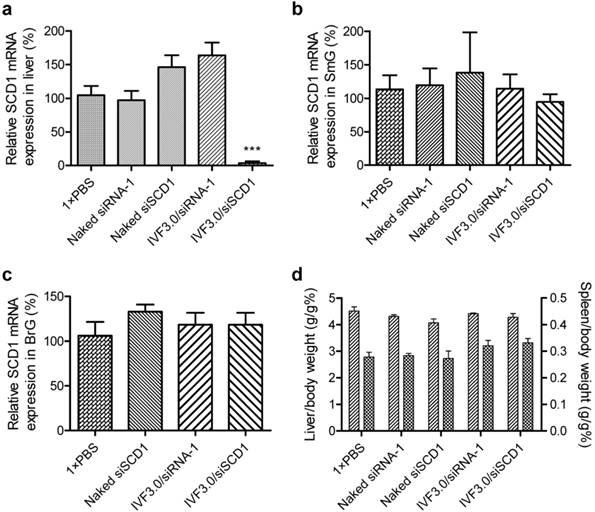
The data revealed that the SCD1 expression in liver was significantly repressed by IVF3.0/siSCD1 with high knockdown efficiency (>90%; Figure 6a), demonstrating that IVF3.0 can effectively deliver siRNA into hepatocytes and siSCD1 shows high potency in vivo. However, naked siSCD1 did not inhibit SCD1 expression in the submandibular gland, bulbourethral gland, and liver (Figure 6a-c). IVF3.0/siSCD1 also displayed equivocal gene silencing in the submandibular and bulbourethral glands (Figure 6b and c). These phenomena suggested that although siRNA strongly accumulates in these glands, neither naked nor carrier-formulated siRNA enter the cytoplasm to mediate gene silencing. CLSM (confocal laser scanning microscopy) examination of cryosections of the submandibular gland also showed that siRNA was barely detectable in the cytoplasm even when encapsulated by delivery carriers (Figures 5, S7, S8, S10, and S12). No significant difference was found between the organ coefficients of liver and spleen (Figure 6d), indicating that the treatments are safe and well-tolerated.
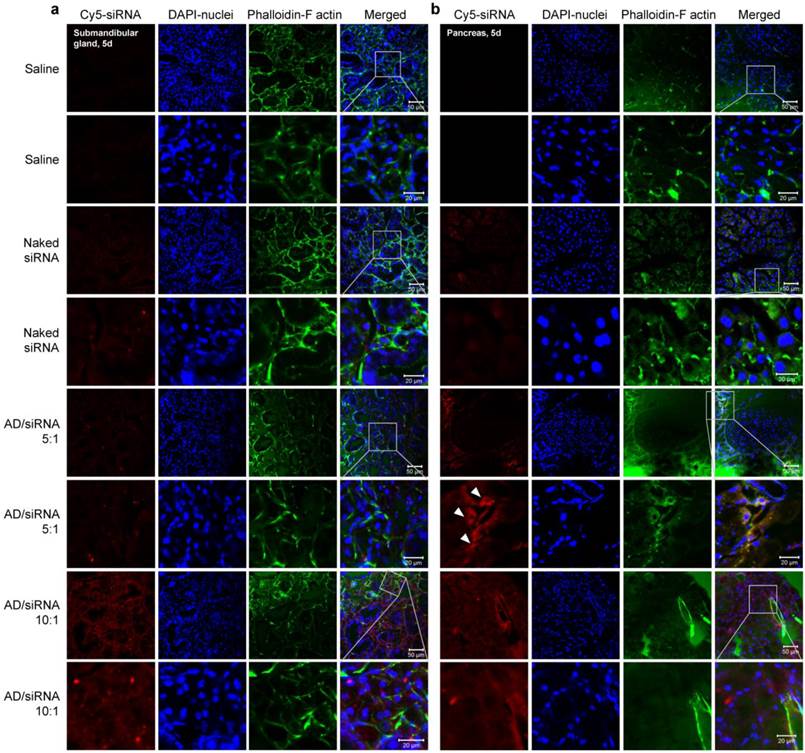
Confocal laser scanning microscopy images of cryosections of submandibular gland (a) and pancreas (b). For all four formulations (saline, naked siRNA, AD/siRNA 5:1, and AD/siRNA 10:1), magnified images (lower panels) were acquired by zooming in on the indicated areas of the parental images (upper panels). Scale bars are shown in the lower right corners of the merged images.
Silencing efficiency and organ coefficients of mice treated with various formulations. (a-c) Relative SCD1 mRNA expression in liver, submandibular gland, and bulbourethral gland at 72 h after treatment. (d) Organ coefficients of liver and spleen indicating the general safety of the various treatments. Data are shown as mean ± SEM, n = 6; ***p <0.001 vs SCD1 expression in mice treated with 1×PBS. SmG, submandibular gland; BrG, bulbourethral gland.
Gene-silencing mediated by direct injection of siRNA into the submandibular gland. Data are shown as mean ± SEM, n = 6; **p <0.01 vs SCD1 expression in untreated mice. SmG, submandibular gland.
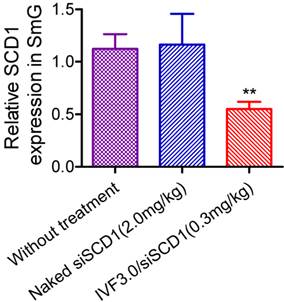
We are very concerned with the feasibility of realization of RNAi in glandular cells. It seems that systemically-administered IVF3.0/siRNA could not enter the cytoplasm of submandibular cells. Benefiting from the submandibular gland is just located in the neck in mice, we can readily perform gene silencing assay by local injection of IVF3.0/siRNA into the gland. As a result, we found that IVF3.0/siSCD1 significantly suppressed SCD1 expression as ~45% knockdown efficiency was achieved at 0.3 mg/kg (Figure 7). However, no inhibition occurred with naked siSCD1, even at 2.0 mg/kg (Figure 7). These results demonstrated carrier-encapsulated siRNA could cross cell membrane of the submandibular gland and effective gene repression indeed could be achieved. It open a window for the treatment of salivary gland-related diseases (such as xerostomia) using siRNA-based therapeutics.
In summary, intravenously administered siRNA does not mediate gene silencing in the submandibular gland, indicating that the gland only plays the role of a reservoir, allowing sustained release of siRNA into the circulation. However, local injection of carrier-encapsulated siRNA does achieve satisfactory gene knockdown efficiency, permitting potential drug development by changing local skin-penetration injection to retroductal administration [44].
Conclusions
In this study, we comprehensively investigated the pharmacokinetics of intravenously-administered siRNA in mice. In accord with previous studies [21], systemically-injected naked siRNAs were eliminated into the kidneys and bladder, processed in the liver, and subsequently secreted into the gallbladder and intestine. More importantly, benefiting from the structural properties of fenestrated capillaries, a large proportion of siRNAs accumulated in the submandibular gland, bulbourethral gland and pancreas, where they remained for a long time. Given that the siRNAs themselves did not mediate gene knockdown in the glands, we deduced that glands play a pivotal role as natural siRNA reservoirs, allowing the slow release of siRNA in vivo. Delivery carriers may markedly influence the in vivo behavior of siRNA. Successful gene silencing was achieved by injecting IVF3.0/siRNA directly into the submandibular gland, providing an opportunity for salivary gland-related siRNA drug development. In conclusion, this study has expanded our understanding of the pharmacokinetic properties of systemically-administered siRNAs and will facilitate the therapeutic development of siRNAs.
Supplementary Material
Supplementary tables and figures.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81402863), the Postdoctoral Science Foundation of China (2014M550008 and 2015T80016), the National High-tech R&D Program of China (2014AA021103 and 2012AA022501), the National Drug Program of China (2015ZX09102-023-002 and 2014ZX09304313-001). We appreciate the technical support of Ms. Ning Hou (Institute of Molecular Medicine, Peking University) in cryosection preparation. We thank Dr. Rupei Tang (School of Pharmaceutical Science, Jiangnan University and Engineering Research Center for Biomedical Materials, School of Life Science, Anhui University) for providing the PEA and PEAG polymers, Dr. Anjie Dong (Department of Polymer Science and Technology, School of Chemical Engineering and Technology, Tianjin University) for supplying all the PDMAEMA-based materials, and Dr. Ling Peng (Aix-Marseille Université, CNRS, Centre Interdisciplinaire de Nanoscience de Marseille) for providing the dendrimers (G5 and AD). We also thank Dr. Iain C. Bruce (Peking University) for critically reading this manuscript.
Competing Interests
The authors declare no competing financial interests.
References
1. Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711-9
2. Lares MR, Rossi JJ, Ouellet DL. RNAi and small interfering RNAs in human disease therapeutic applications. Trends Biotechnol. 2010;28:570-9
3. Coelho T, Adams D, Silva A, Lozeron P, Hawkins PN, Mant T. et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N Engl J Med. 2013;369:819-29
4. Suanprasert N, Berk JL, Benson MD, Dyck PJ, Klein CJ, Gollob JA. et al. Retrospective study of a TTR FAP cohort to modify NIS+7 for therapeutic trials. J Neurol Sci. 2014;344:121-8
5. Fitzgerald K, Frank-Kamenetsky M, Shulga-Morskaya S, Liebow A, Bettencourt BR, Sutherland JE. et al. Effect of an RNA interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum LDL cholesterol in healthy volunteers: a randomised, single-blind, placebo-controlled, phase 1 trial. Lancet. 2014;383:60-8
6. Tabernero J, Shapiro GI, LoRusso PM, Cervantes A, Schwartz GK, Weiss GJ. et al. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 2013;3:406-17
7. Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA. et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067-70
8. Geisbert TW, Lee AC, Robbins M, Geisbert JB, Honko AN, Sood V. et al. Postexposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: a proof-of-concept study. Lancet. 2010;375:1896-905
9. Wooddell CI, Rozema DB, Hossbach M, John M, Hamilton HL, Chu Q. et al. Hepatocyte-targeted RNAi therapeutics for the treatment of chronic hepatitis B virus infection. Mol Ther. 2013;21:973-85
10. Thompson JD, Kornbrust DJ, Foy JW, Solano EC, Schneider DJ, Feinstein E. et al. Toxicological and pharmacokinetic properties of chemically modified siRNAs targeting p53 RNA following intravenous administration. Nucleic Acid Ther. 2012;22:255-64
11. Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W. et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002-7
12. Li SD, Chen YC, Hackett MJ, Huang L. Tumor-targeted delivery of siRNA by self-assembled nanoparticles. Mol Ther. 2008;16:163-9
13. Viel T, Boisgard R, Kuhnast B, Jego B, Siquier-Pernet K, Hinnen F. et al. Molecular imaging study on in vivo distribution and pharmacokinetics of modified small interfering RNAs (siRNAs). Oligonucleotides. 2008;18:201-12
14. Solano EC, Kornbrust DJ, Beaudry A, Foy JW, Schneider DJ, Thompson JD. Toxicological and pharmacokinetic properties of QPI-1007, a chemically modified synthetic siRNA targeting caspase 2 mRNA, following intravitreal injection. Nucleic Acid Ther. 2014;24:258-66
15. Heidel JD, Yu Z, Liu JY, Rele SM, Liang Y, Zeidan RK. et al. Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. Proc Natl Acad Sci U S A. 2007;104:5715-21
16. van de Water FM, Boerman OC, Wouterse AC, Peters JG, Russel FG, Masereeuw R. Intravenously administered short interfering RNA accumulates in the kidney and selectively suppresses gene function in renal proximal tubules. Drug Metab Dispos. 2006;34:1393-7
17. Merkel OM, Librizzi D, Pfestroff A, Schurrat T, Behe M, Kissel T. In Vivo SPECT and Real-Time Gamma Camera Imaging of Biodistribution and Pharmacokinetics of siRNA Delivery Using an Optimized Radiolabeling and Purification Procedure. Bioconjugate Chemistry. 2009;20:174-82
18. Merkel OM, Librizzi D, Pfestroff A, Schurrat T, Buyens K, Sanders NN. et al. Stability of siRNA polyplexes from poly(ethylenimine) and poly(ethylenimine)-g-poly(ethylene glycol) under in vivo conditions: effects on pharmacokinetics and biodistribution measured by Fluorescence Fluctuation Spectroscopy and Single Photon Emission Computed Tomography (SPECT) imaging. J Control Release. 2009;138:148-59
19. Gao S, Dagnaes-Hansen F, Nielsen EJ, Wengel J, Besenbacher F, Howard KA. et al. The effect of chemical modification and nanoparticle formulation on stability and biodistribution of siRNA in mice. Mol Ther. 2009;17:1225-33
20. Abrams MT, Koser ML, Seitzer J, Williams SC, DiPietro MA, Wang W. et al. Evaluation of efficacy, biodistribution, and inflammation for a potent siRNA nanoparticle: effect of dexamethasone co-treatment. Mol Ther. 2010;18:171-80
21. Huang Y, Hong J, Zheng S, Ding Y, Guo S, Zhang H. et al. Elimination pathways of systemically delivered siRNA. Mol Ther. 2011;19:381-5
22. Eguchi A, De Mollerat Du Jeu X, Johnson CD, Nektaria A, Feldstein AE. Liver Bid suppression for treatment of fibrosis associated with non-alcoholic steatohepatitis. J Hepatol. 2016;64:699-707
23. Rozema DB, Blokhin AV, Wakefield DH, Benson JD, Carlson JC, Klein JJ. et al. Protease-triggered siRNA delivery vehicles. J Control Release. 2015;209:57-66
24. Nair JK, Willoughby JL, Chan A, Charisse K, Alam MR, Wang Q. et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J Am Chem Soc. 2014;136:16958-61
25. Huang Y, Lin D, Jiang Q, Zhang W, Guo S, Xiao P. et al. Binary and ternary complexes based on polycaprolactone-graft-poly (N, N-dimethylaminoethyl methacrylate) for targeted siRNA delivery. Biomaterials. 2012;33:4653-64
26. Lin D, Huang Y, Jiang Q, Zhang W, Yue X, Guo S. et al. Structural contributions of blocked or grafted poly(2-dimethylaminoethyl methacrylate) on PEGylated polycaprolactone nanoparticles in siRNA delivery. Biomaterials. 2011;32:8730-42
27. Gary DJ, Lee H, Sharma R, Lee JS, Kim Y, Cui ZY. et al. Influence of nano-carrier architecture on in vitro siRNA delivery performance and in vivo biodistribution: polyplexes vs micelleplexes. ACS Nano. 2011;5:3493-505
28. Crombez L, Morris MC, Dufort S, Aldrian-Herrada G, Nguyen Q, Mc Master G. et al. Targeting cyclin B1 through peptide-based delivery of siRNA prevents tumour growth. Nucleic Acids Res. 2009;37:4559-69
29. Kanasty RL, Whitehead KA, Vegas AJ, Anderson DG. Action and reaction: the biological response to siRNA and its delivery vehicles. Molecular therapy: the journal of the American Society of Gene Therapy. 2012;20:513-24
30. Cleaver O, Melton DA. Endothelial signaling during development. Nat Med. 2003;9:661-8
31. Huang Y, Wang X, Huang W, Cheng Q, Zheng S, Guo S. et al. Systemic Administration of siRNA via cRGD-containing Peptide. Sci Rep. 2015;5:12458
32. Huang Y, Cheng Q, Jin X, Ji JL, Guo S, Zheng S. et al. Systemic and tumor-targeted delivery of siRNA by cyclic NGR and isoDGR motif-containing peptides. Biomater Sci. 2016;4:494-510
33. Cheng Q, Huang Y, Zheng H, Wei T, Zheng S, Huo S. et al. The effect of guanidinylation of PEGylated poly(2-aminoethyl methacrylate) on the systemic delivery of siRNA. Biomaterials. 2013;34:3120-31
34. Lin D, Jiang Q, Cheng Q, Huang Y, Huang P, Han S. et al. Polycation-detachable nanoparticles self-assembled from mPEG-PCL-g-SS-PDMAEMA for in vitro and in vivo siRNA delivery. Acta Biomater. 2013;9:7746-57
35. Han S, Cheng Q, Wu Y, Zhou J, Long X, Wei T. et al. Effects of hydrophobic core components in amphiphilic PDMAEMA nanoparticles on siRNA delivery. Biomaterials. 2015;48:45-55
36. Lin D, Cheng Q, Jiang Q, Huang Y, Yang Z, Han S. et al. Intracellular cleavable poly(2-dimethylaminoethyl methacrylate) functionalized mesoporous silica nanoparticles for efficient siRNA delivery in vitro and in vivo. Nanoscale. 2013;5:4291-301
37. Flowers MT, Ntambi JM. Role of stearoyl-coenzyme A desaturase in regulating lipid metabolism. Curr Opin Lipidol. 2008;19:248-56
38. Hulver MW, Berggren JR, Carper MJ, Miyazaki M, Ntambi JM, Hoffman EP. et al. Elevated stearoyl-CoA desaturase-1 expression in skeletal muscle contributes to abnormal fatty acid partitioning in obese humans. Cell Metab. 2005;2:251-61
39. Ide Y, Waki M, Hayasaka T, Nishio T, Morita Y, Tanaka H. et al. Human breast cancer tissues contain abundant phosphatidylcholine(36ratio1) with high stearoyl-CoA desaturase-1 expression. PLoS One. 2013;8:e61204
40. Mohammadzadeh F, Mosayebi G, Montazeri V, Darabi M, Fayezi S, Shaaker M. et al. Fatty Acid Composition of Tissue Cultured Breast Carcinoma and the Effect of Stearoyl-CoA Desaturase 1 Inhibition. J Breast Cancer. 2014;17:136-42
41. Ben-David U, Gan QF, Golan-Lev T, Arora P, Yanuka O, Oren YS. et al. Selective elimination of human pluripotent stem cells by an oleate synthesis inhibitor discovered in a high-throughput screen. Cell Stem Cell. 2013;12:167-79
42. Rahimi Y, Mehdizadeh A, Nozad Charoudeh H, Nouri M, Valaei K, Fayezi S. et al. Hepatocyte differentiation of human induced pluripotent stem cells is modulated by stearoyl-CoA desaturase 1 activity. Dev Growth Differ. 2015;57:667-74
43. Zhang L, Pan Y, Qin G, Chen L, Chatterjee TK, Weintraub NL. et al. Inhibition of stearoyl-coA desaturase selectively eliminates tumorigenic Nanog-positive cells: improving the safety of iPS cell transplantation to myocardium. Cell Cycle. 2014;13:762-71
44. Arany S, Benoit DS, Dewhurst S, Ovitt CE. Nanoparticle-mediated gene silencing confers radioprotection to salivary glands in vivo. Mol Ther. 2013;21:1182-94
Author contact
![]() Corresponding authors: Y. Huang (yyhuangedu.cn, Tel: +86-10-62750683) or Z. Liang (liangzedu.cn, Tel./fax: +86-10-62769862).
Corresponding authors: Y. Huang (yyhuangedu.cn, Tel: +86-10-62750683) or Z. Liang (liangzedu.cn, Tel./fax: +86-10-62769862).
 Global reach, higher impact
Global reach, higher impact