13.3
Impact Factor
Theranostics 2016; 6(9):1425-1439. doi:10.7150/thno.15359 This issue Cite
Review
Clinical Applications of NanoVelcro Rare-Cell Assays for Detection and Characterization of Circulating Tumor Cells
1. Urologic Oncology Program and Uro-Oncology Research Laboratories, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, California, USA;
2. Department of Molecular and Medical Pharmacology, California NanoSystems Institute, Crump Institute for Molecular Imaging, University of California, Los Angeles, Los Angeles, California, USA;
3. Department of Pathology, Guangdong Provincial Hospital of TCM, Guangzhou University of Chinese Medicine, Guangzhou, China.
4. Department of Surgery, University of California, Los Angeles, Los Angeles, California, USA;
5. Liver Transplantation and Hepatobiliary Surgery, University of California, Los Angeles, Los Angeles, California, USA;
6. Center for Pancreatic Disease, University of California, Los Angeles, Los Angeles, California, USA;
7. Department of Surgery Greater Los Angeles Veteran's Affairs Administration, Los Angeles, California, USA.
† These authors contribute equally to this work.
Received 2016-2-23; Accepted 2016-5-6; Published 2016-6-15
Abstract
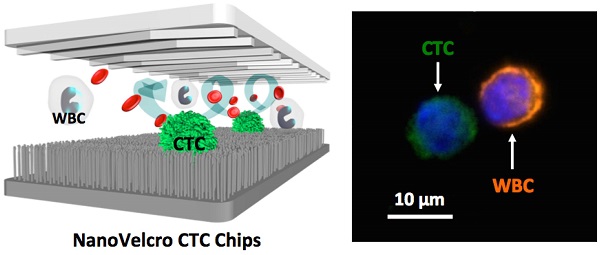
Liquid biopsy of tumor through isolation of circulating tumor cells (CTCs) allows non-invasive, repetitive, and systemic sampling of disease. Although detecting and enumerating CTCs is of prognostic significance in metastatic cancer, it is conceivable that performing molecular and functional characterization on CTCs will reveal unprecedented insight into the pathogenic mechanisms driving lethal disease. Nanomaterial-embedded cancer diagnostic platforms, i.e., NanoVelcro CTC Assays represent a unique rare-cell sorting method that enables detection isolation, and characterization of CTCs in peripheral blood, providing an opportunity to noninvasively monitor disease progression in individual cancer patients. Over the past decade, a series of NanoVelcro CTC Assays has been demonstrated for exploring the full potential of CTCs as a clinical biomarker, including CTC enumeration, phenotyping, genotyping and expression profiling. In this review article, the authors will briefly introduce the development of three generations of NanoVelcro CTC Assays, and highlight the clinical applications of each generation for various types of solid cancers, including prostate cancer, pancreatic cancer, lung cancer, and melanoma.
Keywords: Circulating tumor cell
Circulating tumor cell (CTC)
Pathologic evaluation remains the gold standard for diagnosis and prognosis in the care of cancer patients. This approach typically relies on the tissue specimens obtained by surgical excision or radiographically guided biopsy. While tremendous amounts of information can be obtained from tissues, including histopathology and molecular signatures, this approach has several disadvantages. First, the procedures to obtain tissues are both invasive and costly. The risk of morbidity and psychological stress on the patients largely limit the feasibility of invasive procedures. Moreover, it has been technically challenging to biopsy lesions of certain cancer types or at certain locations, for instance, the osteoblastic metastasis in prostate cancer. Finally, recent studies showing temporospatial heterogeneity [1-7] within a tumor raise serious concerns about how accurately a single biopsy represents a cancer that is spatially heterogeneous and evolves over time.
As an alternative to solid tumor biopsy, many propose the use of a “liquid biopsy” based on circulating tumor cell (CTC) sampling and are actively developing CTC capture techniques.[8] CTCs are rare tumor cells shed from all present disease sites that have active blood perfusion, including primary and metastatic tumors. Sampled through phlebotomy, CTCs can be obtained easily throughout the course of cancer; even during the late stages of metastatic disease without needing invasive and complex traditional biopsy procedures. The ability of serial CTC sampling performed over the course of disease offers the opportunity for real-time, dynamic monitoring of the disease evolution.[9, 10] Over the past decade, collaborative and interdisciplinary research groups including chemistry, material science, bioengineering, cancer biology, and oncology have been formed to focus their efforts upon CTC detection, isolation, and characterization.[11] These collaborative scientific endeavors have led to many important studies setting the foundation for the realization of CTCs to function as a liquid biopsy. Initial studies focused on enumeration [12-14] while recently, some groups have begun to show genomic[15-17] and transcriptomic[18, 19] similarities between CTCs and the traditional tumors biopsies. More and more evidence is supporting the use of CTCs for investigating the nature of cancer, guiding therapeutic interventions, and assessing emerging resistance.
Conventional CTC assays
The most widely used CTC detection assays include: (i) Immunomagnetic separation: these methods utilize capture agent-labeled magnetic beads to either positively select [13, 20, 21] CTCs targeting their surface markers (e.g., epithelial cell adhesion molecule [EpCAM]) or negatively deplete [22, 23] white blood cells (WBCs) using anti-CD45. The CellSearchTM Assay [12-14] is the only FDA-cleared CTC diagnostic technology for metastatic breast, prostate, and colorectal cancers. This assay harvests CTCs with anti-EpCAM-coated magnetic beads, and the subsequent immunocytochemistry (ICC) process helps to identify CTCs (DAPI+/cytokeratin, CK+/CD45-) from nonspecifically captured WBCs (DAPI+/CK-/CD45+). Recently, several new systems (e.g., MagSweeper[24], IsoFlux[25], Cynvenio,[26] magnetic sifters,[27] VerIFAST[28] and AdnaGen/Qiagen[29]) have been developed to further improve detection speed and efficiency. (ii) Flow cytometry: In conjunction with the use of fluorescent markers, flow cytometry [30, 31] is one of the most mature technologies for analyzing and sorting subpopulations of cells. However, this flow-based methodology often has limited detection power due to the low abundance of CTCs, and is unable to provide the CTCs' morphological information. An improved method, known as ensemble-decision aliquot ranking (eDAR),[32, 33] was developed to address this weakness. (iii) Microscopy imaging: Microscopy imaging [34-36] of ICC-treated blood samples allows for highly sensitive detection of CTCs, accompanied with their morphometric characteristics and protein expression. Currently, Epic Sciences is one of the leaders in the commercial sector, now providing Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory tests for both CTC enumeration and characterization. In contrast to the previous three approaches, which require the use of CTC markers, the following two approaches are recognized as label-free methods. (iv) CTC filters: Filter-based approaches [37-41] have been established to trap CTCs according to their sizes. A wide collection of commercial kits/systems from Clearbridge,[39, 40] Rarecells,[42] ScreenCell,[43] and Creatv MicroTech etc. are now available to support research utility. Nevertheless, concerns regarding overlooking small-sized CTCs have been raised. (v) Dielectrophoresis: CTCs can be sorted from WBCs in the presence of a dielectrophoretic field, since the CTC's dielectric properties (depending on their diameter, membrane area, density, conductivity and volume) are different from those of WBCs. ApoCell's technology[44] leverages these differences in a microfluidic flow channel to isolate CTCs. Silicon Biosystems' DEPArray™ combines the use of microscopy imaging and dielectrophoresis sorting[45] to identify and isolate pre-sorted CTCs, paving the way for downstream single-CTC molecular characterizations. (vi) Others: Several reviews [46-48] also summarized a wide collection of CTC detection technologies which may not be included in this article.
Microfluidics-enabled CTC assays
The microfluidic affinity-capture devices [49] developed by Toner et al. sparked the recent research efforts focused on the development of nanotechnology-enabled CTC assays. This 1st-generation (gen) device [49] (i.e., CTC-Chip) featured chemically etched microposts on a silicon substrate, on which anti-EpCAM antibodies were covalently functionalized. These embedded microposts were designed to maximize the contact between the device surfaces and the flow through cells. Following CTC capture, ICC was conducted to identify CTCs. The CTC-Chips demonstrated significantly more success in enumeration performance than most of the conventional CTC assays. Thereafter, similar device configurations were adapted to create new microfluidic chips (e.g., geometrically enhanced differential immunocapture, GEDI [50] approach and Biocept's CTC assay [51]), and different antibody capture agents were employed. Recently, a unique "Ephesia" approach [52] based on microposts of capture agent-coated magnetic beads self-assembled in a microchip demonstrated combined advantages of both microfluidic and immunomagnetic cell sorting. The 2nd-gen device [53] (i.e., herringbone-chip, HB-Chip) from the same group was made from an imprinted polydimethylsiloxane (PDMS) component on a glass slide. Microscale herringbone patterns were engineered into the PDMS component to introduce microvortices, leading to enhanced contact between the CTCs and the antibody-coated chip surfaces. In addition to the commonly used ICC technique, the transparent nature of the HB-Chip allowed for imaging of the captured CTCs by standard clinical histopathological stains (i.e., haematoxylin and eosin stain). Although the microfluidic setting improves CTC-capture performance, the majority of the microfluidic CTC assays suffers from depth of field issues when performing microscopy imaging due to the vertical depths of 3-dimensional device configurations. Time-consuming multiple cross-sectional imaging scans that generate large image files are required in order to avoid out-of-focus or superimposed micrographs. By coupling a pair of microelectrodes at the terminal of a plastic microfluidic chip[54], enzymatic release of the captured CTCs can be electrically counted without the issue of microscopy imaging. In contrast to their 1st and 2nd-gen devices, their 3rd-gen iChip[55] represents a groundbreaking label-free approach, which combines negative immunomagnetic depletion processes with an inertial focusing setting in an integrated microchip. Most importantly, this approach allows for the recovery of unmanipulated CTCs with desired molecular integrity and viability, allowing for downstream expressional profiling[18], as well as ex vivo culture and drug susceptibility testing[56]. The sorting mechanism of iChip, however, was recently reported to compromise the isolation of CTC clusters, which potentially contain CTCs with high metastatic potential.[57] “Cluster-Chip”, a microchip that can be used individually or in conjunction with CTC-iChips to isolate CTC clusters, was developed to address this issue.[58] Other microfluidic CTC assays based on unique principles, including micro-nuclear magnetic resonance (μNMR) platform[59], cell rolling[60], supported lipid bilayer (SLB)-coated microfluidic devices[61], and Vortex technology[62, 63] have also been developed and demonstrated. In addition to the microfluidic assays developed for the enumeration, molecular characterization, and ex vivo expansion of CTCs, a sectioned microfluidic device (known as the Velocity Valley Chip) that selectively captures CTCs in a manner dependent on the number of magnetic beads grafted on the surface of a given CTC [64] has been designed. This device was employed to separate CTCs into subpopulations by EpCAM expression of individual CTCs. Overall, microfluidic technology has shown its potential in enriching and isolating CTCs amenable for subsequent molecular and functional characterizations.
NanoVelcro CTC Assays: Three generations of development
Recent advances in the field of nanotechnology offer powerful solutions [65-67] resulting in a wide range of in-depth characterizations of CTCs while drastically reducing costs. Ultimately, deployment of these emerging advances will bring oncology closer to the goal of personalized care. It has long been recognized that there are nanoscale components present in the tissue microenvironment, including the extracellular matrix, and the cellular membrane. These provide structural and biochemical support that regulate cellular behavior and fate. Inspired by the nanoscale interactions observed in the tissue microenvironment, Dr. Tseng's research team at UCLA pioneered the development of “NanoVelcro” cell-affinity substrates [68, 69]. In this unique approach, capture agent-coated nanostructured substrates are utilized to immobilize CTCs with high efficiency. The working mechanism of NanoVelcro cell-affinity substrates mimics that of VelcroTM - when the two fabric strips of a Velcro fastener are pressed together, interactions between the hairy surfaces on two strips leads to strong affinity between cells and nanosubstrates. In addition to the silicon nanowire substrate (SiNS)[68], the general applicability of the NanoVelcro cell-affinity assay is supported by extensive research endeavors devoted to exploiting different nanomaterials, e.g., polymer dots[70]/nanopillars[71], TiO2 nanowires[72]/nanoparticles[73], layer-by-layer-assembled nanostructures[74], gold clusters on silicon nanowires[75], Fe3O4 nanoparticles[76], DNA networks[77], and graphene oxide nanosheets[78] to achieve high affinity capture of CTCs and other types of rare cells. In parallel, the team has also established a 3-color ICC protocol[79] using DAPI, anti-CD45, and anti-CK to identify nanosubstrate-immobilized CTCs. Single-cell image cytometry data covering DAPI staining, CK/CD45 expression and object size can be used to distinguish CTCs (DAPI+/CK+/CD45-, sizes>6 µm) from nonspecifically captured WBCs (DAPI+/CK-/CD45+, sizes<12 µm), and cellular debris. With the initial proof-of-concept demonstration of the NanoVelcro substrates and ICC protocol in place, three generations of NanoVelcro CTC Chips have been established[69] (Figure 1) to achieve different clinical utilities.
Conceptual illustration of three generations of the NanoVelcro CTC Assays developed by the UCLA team to achieve different clinical utilities. 1st-Gen NanoVelcro Chip [9, 80], composed of a silicon nanowire substrate (SiNS) and an overlaid microfluidic chaotic mixer, was created for CTC enumeration. In conjunction with the use of the laser capture microdissection (LCM) technique, 2nd-gen NanoVelcro-LMD technology [16, 81, 82], was developed for single-CTC isolation. The individually isolated CTCs can be subjected to single-CTC genotyping. By grafting thermoresponsive polymer brushes onto SiNS, 3rd-gen Thermoresponsive NanoVelcro CTC Chips [83, 84] were developed for purification of CTCs via capture and release of CTCs at 37°C and 4°C, respectively. The surface-grafted polymer brushes were responsible for altering the accessibility of the capture agent on NanoVelcro substrates, allowing for rapid CTC purification with desired viability and molecular integrity. (Reprinted with permission from Tseng 2014, Copyright, American Chemical Society). We compare the performance and differences of the three generations of NanoVelcro CTC Assays in a table.
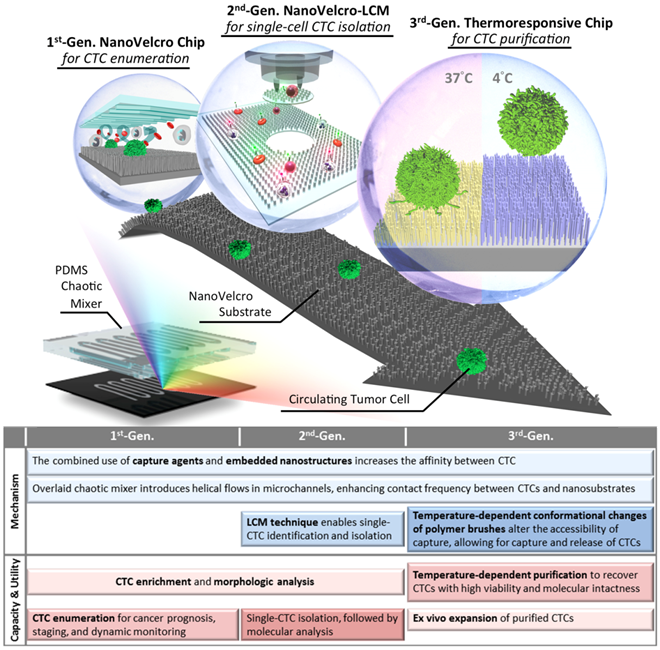
The 1st-gen NanoVelcro Chip [9, 80], composed of a SiNS and an overlaid microfluidic chaotic mixer, was created for CTC enumeration. The performance (>85% of CTC capture efficiency) of these NanoVelcro Chips was measured using artificial CTC samples. Side-by-side analytical validation studies using clinical blood samples show that the sensitivity of the 1st-gen NanoVelcro Chip exceeds [9] that of the FDA-approved CellSearchTM Assay. Notably, the NanoVelcro-like approach allows immobilization of CTCs onto a flat and small surface, thus facilitating the implementation of subsequent high-resolution immunofluorescence microscopy imaging of CTCs without multiple cross-sectional imaging scans required for the majority of the existing microfluidic CTC assays. Moving beyond CTC enumeration, the 2nd-gen NanoVelcro Chips[16, 81, 82], known as NanoVelcro-LCM approach were developed by replacing SiNS with a transparent substrate covered with nanofibers made of PLGA, i.e., poly(lactic-co-glycolic acid). The transparent PLGA NanoVelcro substrate retains the desired CTC capture performance, and allows for seamless integration with a laser capture microdissection (LCM) technique to isolate immobilized CTCs with single-cell resolution. The individually isolated CTCs can be subjected to single-CTC genotyping (both Sanger sequencing [82] and next-generation sequencing [NGS][16, 81]) to serve as liquid biopsies. In order to increase throughput, lower labor, and address the need for viable/unfixed CTCs, the UCLA team developed the 3rd-gen Thermoresponsive NanoVelcro Chips [83, 84] and demonstrated the ability to capture and release viable CTCs at 37°C and 4°C, respectively. By grafting thermoresponsive polymer brushes[83] (poly(N-isopropylacrylamide, PIPAAm) onto SiNS via atom transfer radical polymerization, the temperature-dependent conformational changes of polymer brushes can effectively alter the accessibility of capture agents on SiNS, allowing for rapid CTC purification with desired viability and molecular integrity. The advent of the 3rd-gen Thermoresponsive NanoVelcro Chips is expected to open up new opportunities to connect with a wider range of molecular and functional assays. The continuous research endeavors put together by the UCLA team and its clinical collaborators have demonstrated the use of NanoVelcro CTC assays in clinical settings to facilitate the concept of CTC-based liquid biopsy. These results are briefly summarized in this review article.
Enumerating CTCs using 1st-gen NanoVelcro CTC Assay
Given the CTC detection performance observed for the 1st-gen NanoVelcro CTC Assay [80], continuous efforts were devoted to test its utility for CTC detection in different solid tumors in conjunction with the use of combined capture and ICC antibodies. The initial clinical studies focused on prostate cancer with the intention to address the issue that CellSearchTM assay is unable to detect CTCs in a large portion of late stage prostate cancer patients [14]. These clinical validation studies [9] were jointly conducted by Urologic Oncology teams at Ronald Reagan UCLA Medical Center and Cedars-Sinai Medical Center (CSMC). Forty prostate cancer patients (32 with metastatic disease and 8 with localized disease) were recruited. CTCs were identified in all 40 patients, indicating a consistent efficiency of 1st-gen NanoVelcro Assay for CTC enumeration in prostate cancer patients across different stages of disease. The team also performed serial enumeration allowing the comparison of CTC number changes after 4-10 weeks of therapy, and observed a statistically significant reduction in CTC counts in the clinical responders. Further, long-term follow ups were also performed for 460 days with serial CTC collection and enumeration. In this case, CTC numbers faithfully represented the initial response and subsequent failures during nilutamide and sipuleucel-t treatment. This study demonstrates the consistency of the 1st-gen NanoVelcro CTC Assay over time for CTC enumeration, and shows that continuous monitoring of CTC numbers can be employed to follow responses to different treatments and monitor disease progression.
In addition to the prostate cancer application described above, the pancreatic cancer research team at Ronald Reagan UCLA Medical Center investigated [85] the feasibility of applying NanoVelcro Enumeration Assay to detect CTCs in patients with pancreatic ductal adenocarcinoma (PDAC). The goal is to explore the use of CTCs as an adjunctive biomarker at the time of PDAC presentation. Venous blood was collected prospectively from 100 consecutive, pre-treatment PDAC patients. Utilizing the 1st-gen NanoVelcro CTC Chips, samples were evaluated for the presence and number of CTCs. CTC enumeration data was then evaluated as a diagnostic and staging biomarker for PDAC. Evaluation of CTCs as a diagnostic revealed the presence of CTCs in 54/72 patients with confirmed PDAC (sensitivity = 75.0%, specificity = 96.4%). Furthermore, a cut-off of >3 CTCs in 4-mL blood was able to discriminate between local/regional and metastatic disease. Together, our results highlight the utility of CTCs as a liquid biopsy to better inform diagnosis and staging of PDAC, importantly, at the time of disease presentation.
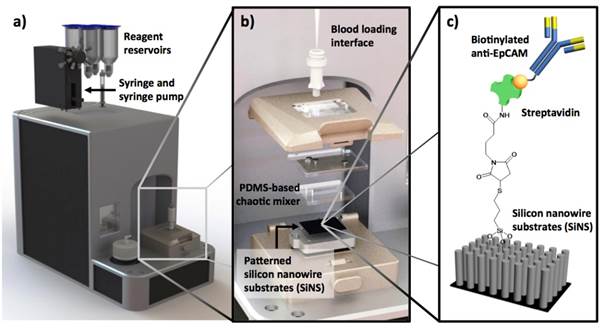
Incremental improvement of the 1st-gen NanoVelcro CTC Assays has led to a user-friendly system and protocol allowing convenient CTC enumeration studies at different facilities. In preparation for its FDA 510K clinical study, the latest version of NanoVelcro CTC Enumeration Assay (Figure 2) has been subjected to a series of studies, including calibration/interference tests using two different cancer cell lines (MCF7 and PC3), duplicated studies using clinical samples from pancreatic cancer and liver cancer patients, and side-by-side comparison with CellSearchTM Assay using clinical samples from more than 100 prostate cancer patients. These data have shown that the NanoVelcro Assay is sufficiently reproducible for 510K trials.
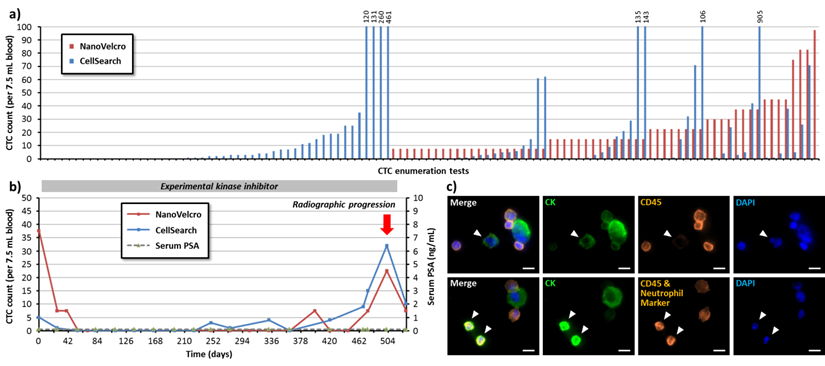
With continuous accumulation of CTC enumeration data, the UCLA/CSMC team has amassed a cohort of prostate cancer patients followed with serial CTC counts by NanoVelcro and CellSearchTM assays that are annotated with patient clinical data. These studies show parallels and differences in the output of these two assays. Figure 3a shows a comparison of the enumeration results of CellSearchTM assay and NanoVelcro assay from more than 100 samples collected from metastatic castration-resistant prostate cancer (mCRPC) patients. In this patient cohort, the NanoVelcro CTC Assay detected counts ranging from 0-98 cells per 7.5 mL of blood, in comparison to CellSearchTM that yielded 0-905 counts per 7.5 mL of blood. In most patients, changes in both NanoVelcro and CellSearchTM CTC counts reflected disease progression - counts decreased immediately after the initiation of anti-cancer therapies. Similarly, counts rose at the time of disease progression (Figure 3b). Both CellSearchTM and NanoVelcro assays capture cells based on EpCAM expression and identify putative CTCs based on CK and CD45 fluorescence signals. However, the differences in each system, such as the use of nanostructured substrate and different imaging modality may contribute to the variation in the dynamic range of the assay. Higher resolution fluorescence microscopy in CTC detection suggests that certain white blood cells may be falsely counted as CTCs given potential CK false-positivity. This would result in unusually high CTC counts, as many research teams have observed (Figure 3c).
The latest version of NanoVelcro CTC Enumeration Assay is performed using (a) a fluidic handler that has been designed and fabricated to introduce processed blood samples into NanoVelcro Chips. (b) Engineering design of NanoVelcro Chips, in which the mechanical click-on approach allows instant assembly of the device. (c) Silanation reaction and NHS chemistry were employed to covalently link streptavidin onto the SiNW substrate, allowing conjugation of biotinylated anti-EpCAM prior to CTC detection studies.
Comparison of NanoVelcro and CellSearchTM Assays in CTC enumeration for metastatic castration-resistant prostate cancer (mCRPC) patients. More than 100 contemporary CTC enumeration results from NanoVelcro and CellSearchTM Assays are shown in (a). The data demonstrated parallels and differences in the output of these two assays. The NanoVelcro CTC assay detected counts ranging from 0-98 cells per 7.5 mL of blood, in comparison to CellSearchTM that yielded 0-905 counts per 7.5 mL of blood. (b) A typical case showing the NanoVelcro and CellSearchTM counts in reflection of patient's disease progression. Cell counts decreased immediately after the initiation of anti-cancer therapies, and rose at the time of disease progression. (c) (Upper row) some of cells present with strong CK fluorescence signal and weak CD45 signal (arrowheads), but could be identified as WBCs with fluorescence staining neutrophil markers (lower row), suggesting the existence of false positive events that may contribute to the unusually high cell counts.
Applying conventional cytopathology standards to morphologic analysis may help eliminate the false-positive events. To further address this issue, more studies are needed to understand the assay and the nature of CTCs. Meanwhile, larger scale clinical trials with parallel tests and head-to-head comparison have to be conducted to validate the performance of the CTC assay. Careful subgroup analysis must be performed to identify the factors that may have affected the results. The UCLA/CSMC team thus explored the use of the NanoVelcro CTC Assay for studying CTC subpopulations in prostate cancer patients.
Subclassification of Prostate Cancer CTCs
Over the past 3 years, the UCLA/CSMC team has performed CTC analysis using NanoVelcro CTC Assay in conjunction with fluorescence microscopy[79, 86] that enabled visualization of cellular features of the captured CTCs and pathologic review for cellular morphology and nuclear size. In their initial observational study, the team analyzed serial blood specimens collected from men with various disease states ranging from localized prostate cancer to advanced metastatic castration-resistant disease. A mathematical modeling and unsupervised clustering of CTC nuclear size distribution identified 3 distinct subsets of CTCs, i.e., very-small-nuclear CTCs (vsnCTCs, nuclear size < 8.54 µm), small-nuclear CTCs (snCTCs, nuclear size between 8.54 µm and 14.99 µm) and large-nuclear CTCs (lnCTCs, nuclear size > 14.99 µm). snCTCs and vsnCTCs seem to appear in metastatic prostate cancer patients, while vsnCTCs occurred predominantly in patients with metastasis in visceral organs such as the liver or lungs, and vsnCTC counts were significantly higher in patients with visceral lesions compared to those without. The UCLA/CSMC team also found that vsnCTCs emerge prior to appearance of visceral metastasis on clinical imaging.[87]
As the UCLA/CSMC team further expand the investigation for this phenomenon in a bigger patient database, they identified 28 metastatic prostate cancer patients who had progressed through next generation hormonal maneuvers such as abiraterone, enzalutamide, or an equivalent drug. Serial blood specimens were used retrospectively for CTC enumeration and subgroup analysis (Figure 4). Fifteen out of 28 patients presented with visceral lesions and 13 had bone-only disease at their first CTC enumeration. Six out of 13 non-visceral metastatic patients developed visceral lesions during follow-up, and vsnCTCs were detected 86-196 days prior to radiographic detection of the visceral lesions. Four patients had vsnCTCs detected without the presence of visceral lesions by the time of this analysis, but the UCLA/CSMC team is still following some of them for visceral progression in the future. Overall, vsnCTCs were detected in all the patients with visceral metastasis, and none of the patients without vsnCTCs developed visceral metastasis in this study.
Aside from the potential predictive utility of vsnCTCs, the UCLA/CSMC team also analyzes the relationship between vsnCTCs and patients' response to therapeutic interventions. The team observed reduction of vsnCTC count occurred at initiation of anti-cancer treatment. Conversion from vsnCTC(-) to vsnCTC(+) was seen prior to progression of visceral lesions under the treatment.
The data summarized above point toward a potential benefit in adding simple morphologic categorization (i.e., nuclear size) to CTC enumeration, and prompt the group to further enhance the understanding of the association between vsnCTCs and visceral metastasis in prostate cancer patients.
Detecting NSCLC CTCs using aptamer-grafted NanoVelcro Chips
Antibody capture agents (e.g., anti-EpCAM) are commonly used with marker-based CTC enrichment platforms. Considering the high production cost and poor storage stability known for antibodies, researchers have been exploiting alternative CTC capture agents, e.g., aptamers. Aptamers are single-stranded oligonucleotides with function similar to antibodies because they are both able to differentiate between other molecules and cells. These molecules can be produced by the process called in vitro cell-SELEX (systematic evolution of ligands by exponential enrichment)[88-90], allowing for custom productions of aptamer-based CTC capture agents. In collaboration with the Chinese Academy of Science, a group of aptamers specific to non-small cell lung cancer (NSCLC) cells was generated through in vitro cell-SELEX process. By coating these aptamers onto the NanoVelcro Chips, the study team was not only able to capture NSCLC CTCs with high efficiency, but also recover the nanosubstrate-immobilized CTCs upon treatment with a nuclease solution. On the basis of the dual aptamer capture agents, the joint team recently showed a rational design of several aptamer cocktails based on a panel of existing aptamers. By grafting these aptamer cocktails onto NanoVelcro Chips, the devices exhibited [91] enhanced and differential capture performances using blood samples collected from a cohort of NSCLC patients. This study also shows the feasibility of exploiting the aptamer-grafted NanoVelcro Chips to dissect CTC heterogeneity and to monitor treatment responses.
1st-Gen. NanoVelcro CTC Assay identified very-small-nuclear CTCs (vsnCTCs) in prostate cancer patients with visceral metastasis. The presence of vsnCTCs has been associated with the presence of visceral metastasis. Serial CTC enumerations also suggested that vsnCTCs emerge before the development/detection of visceral lesions and thus may be a predictive biomarker for visceral metastasis in prostate cancer patients.

Molecular Analysis of CTCs using 2nd-gen NanoVelcro Chip
Mutational Analysis of Circulating Melanoma and Pancreatic Cancer Cells
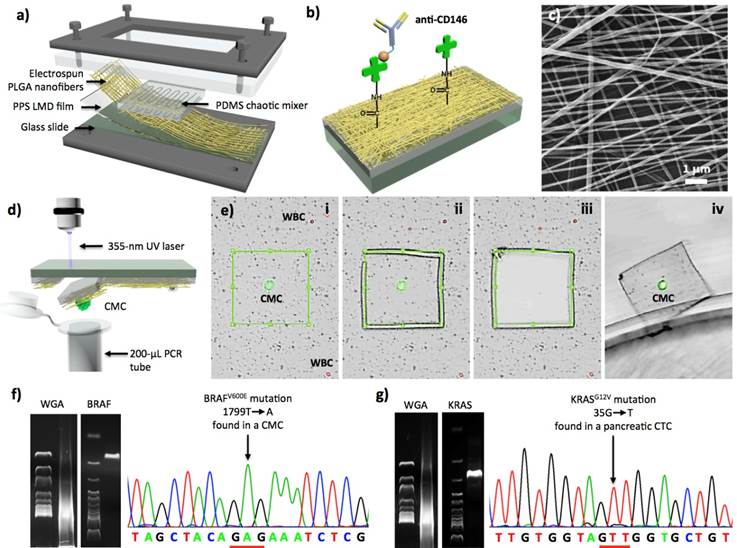
Although 1st-gen NanoVelcro Chips allow efficient and reproducible detection and subclassification of CTCs in patient blood, challenges remain in 1) broadening its general applicability for detecting other types of solid-tumor CTCs that exhibit surface markers other than EpCAM, and 2) enabling the isolation of single CTCs for subsequent molecular analyses. To test the general applicability of the NanoVelcro-based cell-affinity assay, the UCLA team explored a melanoma-specific capture agent[92] (i.e., anti-CD146) to capture circulating melanoma cells (CMCs; a subcategory of solid-tumor CTCs). On the foundation of the 1st-gen NanoVelcro Chips, 2nd-gen NanoVelcro-LCM technology (Figure 5) was demonstrated to not only capture CMCs with high efficiency, but also enable highly specific isolation of single CMCs immobilized on the nanosubstrates without contamination by background WBCs. The non-transparent SiNS in 1st-gen NanoVelcro Chips was replaced with a transparent PLGA-nanofiber-embedded substrate, prepared by depositing electrospun PLGA nanofibers onto a commercial laser microdissection (LMD) slide (Figure 5a), followed by streptavidin-mediated conjugation of anti-CD146 (Figure 5b and c). Both artificial and patient blood samples were obtained to optimize and validate the performance of the PLGA-NanoVelcro Chips. Here, a new 4-color ICC protocol for parallel staining of FITC-labeled anti-Mart1, TRITC-labeled anti-HMW-MAA, DAPI, and Cy5-labeled anti-CD45 was established to identify CMCs (DAPI+/Mart1+/HMW-MAA+/CD45-, and 40 mm>diameter>10 mm) among nonspecifically captured WBCs (DAPI+/Mart1-/HMWMAA-/CD45+ and diameter<10 mm) and cellular debris, immobilized on the PLGA NanoVelcro substrates. These PLGA NanoVelcro Chips exhibit similar performance to capture CMCs compared to those observed for 1st-gen NanoVelcro Chips. Most importantly, the transparent PLGA NanoVelcro substrate allows for seamless integration with a LCM technique to isolate immobilized CMCs with single-cell resolution (Figure 5d and e). After CMC capture and ICC melanoma lineage validation, a LMD microscope (Leica) or a LCM microscope (ArcturusTM Life Technology) was used to cut out and harvest single CMCs. After conducting whole genome amplification (WGA) and targeted PCR amplification on the isolated CMCs, the amplified DNA materials were subjected to Sanger sequence analysis (Figure 5f). To examine the clinical utility of the optimized 2nd-gen NanoVelcro-LCM technology, the UCLA team then performed single-CMC isolation and genotyping using peripheral blood samples collected from multiple stage-IV melanoma patients, whose melanomas have been previously characterized by conventional PCR-based techniques (cobas 4800, Roche) to contain a signature oncogenic mutation, i.e., BRAFV600E. Over the past five years the oncology field has experienced a paradigm shift in treatment of metastatic melanoma. BRAF inhibitors (e.g., vemurafenib), designed for treating melanomas harboring the BRAFV600E oncogenic mutation, have had unprecedented response rates in excess of 60-80%.[93, 94] The BRAFV600E mutation (present in 60% of melanomas) is an indispensable requisite for response to this agent. Thus, detecting the BRAFV600E mutation can be employed as a companion diagnostic solution that can guide the implementation of vemurafenib treatment, as well as facilitate the clinical development of the new BRAF inhibitors. The ability to detect BRAFV600E mutation in CMCs could evolve into a companion diagnostic that would avoid invasive tissue sampling.
As a second model, the 2nd-gen NanoVelcro-LCM technology was utilized to pursue characterization of KRAS mutation in pancreatic cancer CTCs as more than 95% of patients have activating mutations.[95],[96] Using a similar approach to that described above, both KRASG12V (Figure 5g) and KRASG12D mutations[97] were detected in the pancreatic CTCs immobilized on the PLGA NanoVelcro substrates.
Whole Exome Sequencing of CTCs
Knowing that identification of genetic alteration was possible, it was hypothesized that the NanoVelcro-LCM approach could be used to monitor the dynamic tumor biology of a cancer by profiling CTCs. As a result, a streamlined process [81], for whole exome sequencing (WES) of CTCs based on the 2nd-gen platform, was established. Single CTCs were isolated from a prostate cancer patient, and WBCs were utilized here as control representing germline DNA. After whole genome amplification (WGA), DNA was sequenced by standard exon-capture targeted sequencing. The results indicated that 25 to 80% of the targeted exome regions were sequenced with a mean coverage of 29 to 48X, and that no chromosomal loss occurred during the isolation and sequencing processes. In looking across the genomic information from the samples, the study team concluded that there were more shared mutations among individual CTCs than there were between CTCs and WBCs. The similarity of CTCs and differences between CTCs and the WBC control verified the feasibility of using the NanoVelcro-LCM platform to capture pure CTCs for WES.
Whole Genome Sequencing (WGS) of CTCs
Given the success with WES on single-CTCs isolated by the 2nd-gen NanoVelcro Assay, the team further refined the protocol for high-quality WGS on single-CTCs. The major technological breakthroughs included (i) the utilization of ethanol instead of paraformaldehyde fixation to better preserve single-CTC DNA integrity [98]; (ii) the utilization of multiple displacement amplification (MDA) to decrease the loss of unamplified segments [81] as well as bias introduced by polymer chain reaction (PCR)-based amplification [99]; and (iii) the rigorous quality check to select high-quality CTC samples by a multiplexed PCR based on eight housekeeping genes. Utilizing a streamlined protocol, the mutational landscapes of serially-collected single CTCs were compared with the primary and metastatic tumor tissues in a mCRPC patient at the whole genome level.
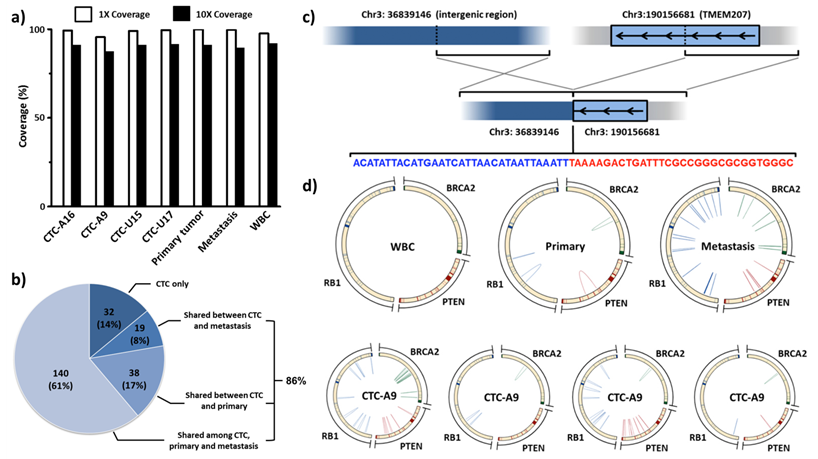
In 4 sequenced individual CTCs, 30X sequencing depth (data not shown) and above 95% sequencing coverage in WGS (Figure 6a) were achieved. These single-CTC WGS data allow the subsequent comparisons. Twenty nine percent of founder single nucleotide variants (SSNVs) in tumor tissues were found in CTCs. In addition, 86% of clonal mutations in CTCs were traced back to either the primary or metastatic tumor (Figure 6b). The research team found and validated an intrachromosomal rearrangement in chr3 (Figure 6c) and an interchromosomal rearrangement between chr13 and chr15. These tumor specific rearrangements were shared between both tumor tissues and most of the CTCs (Figure 6d), but not identified in WBCs and normal adjacent tissue. At the same time, highly heterogeneous short rearrangements were discovered in important tumor suppressor genes, including PTEN, RB1 and BRCA2, in all tumor and CTC samples [16].
2nd-Gen NanoVelcro-LCM technology for single-CTC isolation, followed by mutational analyses. (a) The PDMS chaotic mixer is layered on top of a NanoVelcro chip that contains PLGA nanofibers [82]. (b) For the binding of biotinylated capture agents (i.e., anti-CD146 for CMC and anti-EpCAM for pancreatic cancer), streptavidin is conjugated to PLGA nanofibers. (c) An image of the electrospun PLGA nanofibers by using SEM. (d) The graphic illustration of LMD-based single-CMC isolation. (e) The process to isolate single CMCs consists of (i) identification of CMC, (ii) isolation of the selected CMC using laser dissection, followed by (iii), and (iv) discharge of CMC from the silicon substrate into a 200 μl PCR tube. (f) Results of single-CMC WGA and gel electrophoresis after amplification in PCR with BRAF-specific primer. Through Sanger sequencing, further affirmation is gained because of the display of CMCs exhibiting the unique BRAFV600E mutation. (g) Pancreatic CTCs and the KRASG12V mutation present [97].
2nd-Gen NanoVelcro CTC Chips for single-CTC isolation, followed by whole genome sequencing (WGS). (a) Single-CTC whole genome sequencing successfully demonstrated [16] above 90% coverage. (b) Clonal single nucleotide variants (SSNVs) shared by more than three CTCs were identified. These SSNVs were repeatedly detected in more than three single-cell sequencing runs and can be considered high confident mutations. It is interesting to note that 86.0% of these clonal SSNVs in CTCs can be traced back to either the primary or metastatic tissues. (c) Interchromosomal rearrangement involving TMEM207 was found in CTCs and tumor tissues. (d) Complex rearrangements involving tumor suppressor genes, including PTEN, RB1 and BRCA2, were found in CTCs and tumor tissues but not in WBCs.
Molecular Analysis of CTCs using 3rd-gen Thermoresponsive NanoVelcro Chip
Though the 2nd-gen NanoVelcro-LCM technology [16, 81, 82] exhibited exceptional precision in single-CTC isolation, this approach suffered from its limited throughput due to the labor-intensive procedure of LCM. Since fixation is needed for ICC, this approach does not yield viable CTCs. There have been significant research endeavors devoted to developing new CTC purification methods that overcome this technical challenge. For example, the MGH CTC-iChips[55] are capable of sequential depletions of normal blood cells, to recover unmanipulated CTCs which can be cultured ex vivo for drug susceptibility testing. Several issues, e.g., lack of control over CTC purity after their recovery, and high operation costs associated with complicated device design, need to be resolved in order to translate these new methods into research and clinical settings. The 3rd-gen Thermoresponsive NanoVelcro Chips [83, 84] will have the potential to overcome all of the challenges encountered in the field.
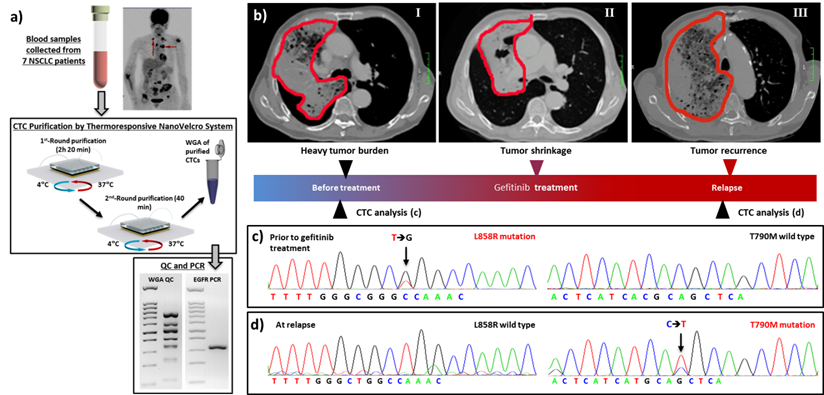
The move toward molecular characterization and functional analysis of CTCs creates an urgent need for (i) efficient isolation, (ii) improved cell quality, and (iii) lower technical demand on the end user. The 3rd-gen thermoresponsive NanoVelcro Chip[83, 84] was engineered with these issues in mind. This chip can successfully capture and release CTCs at 37 ºC and 4 °C, respectively, via grafting thermoresponsive polymer brushes onto the SiNS. This unique idea takes advantage of the fact that the polymer brushes undergo temperature-dependent conformational changes. This alters the accessibility of the CTC surface to capture agents on the NanoVelcro Chips. Ultimately, this allows for rapid, viable CTC purification with intact nucleic acid content. To optimize the CTC purification parameters for the 3rd-gen Thermoresponsive NanoVelcro Chips, we used H1975 anti-EpCAM-positive non-small cell lung cancer (NSCLC) cells as a model system. We found that by performing two rounds of capture/release CTCs can be obtained with higher purity, allowing for EGFR point mutation analysis (Figure 7a). Sanger sequencing of purified CTCs from the 7 NSCLC patients' blood samples showed strong molecular correlation with tumor tissues. We also observed the emergence of secondary T790M mutation in serial CTC analyses from a NSCLC patient who received EGFR inhibitor (Figure 7b-d). This patient developed resistance to EGFR inhibitor soon afterwards. These results indicated the potential utility of CTCs as a tool for detecting the emerging resistance to targeted therapies.
3rd-Gen NanoVelcro CTC Chips for non-small cell lung cancer (NSCLC) CTC purification, followed by mutational analysis. (a) The workflow demonstrating [83] the use of 3rd-gen NanoVelcro Chip for non-small cell lung cancer CTC purification followed by detection of EGFR mutations. (b-d) Longitudinal data showed the mutational analysis of CTCs from a NSCLC patient. (c) L858R mutation was detected in CTCs and tumor tissues before gefitinib (1st-Gen EGFR inhibitor) treatment. Tumor regression was observed soon after initiation of gefitinib, as shown in (b). (d) The secondary T790M mutation was detected later in CTCs when the patient's disease progressed.
Significant progress is currently being made in the area of rapid CTC purification from whole blood samples. Through the development of a user-friendly interface for the 3rd generation Thermoresponsive NanoVelcro Chips we will be able to facilitate CTC characterization. The TR-NanoVelcro assay will produce a number of exciting opportunities for clinical applications. Using Thermoresponsive NanoVelcro Chips one could create patient-specific, CTC-derived cancer lines via ex vivo expansion. Such a tool could be used to conduct ex vivo testing for sensitivity and resistance to therapy that is patient specific, bringing the field of oncology closer to the goal of personalized care.
Summary of NanoVelcro CTC Assays
The unique advantages of NanoVelcro CTC Assays in contrast to other existing systems are briefly summarized below. First of all, the three generations of NanoVelcro CTC Assays share two common working mechanisms - (i) the combined use of capture agents and embedded nanostructures leads to enhanced affinity between CTCs and NanoVelcro substrates, and (ii) the overlaid PDMS chaotic mixer increases contact frequency between CTCs and NanoVelcro substrates - to achieve desired CTC capture performances. Furthermore, the use of contemporary micro-fabrication techniques in the fabrication of the two key functional components (i.e., nanostructured substrate and PDMS chaotic mixer) guarantees the scalability, reproducibility, and cost-effectiveness to produce these NanoVelcro CTC Assays. Additional advantages include (i) Flexibility: these assays can be employed for both detection and isolation of CTCs with viability and molecular intactness; (ii) Speed and precision: Runtime with semi-automated microscopy allows for optimal pathologic review of all CTCs avoiding contamination. The combined use of 1st-gen NanoVelcro Enumeration Assay and 2nd gen NanoVelcro-LCM technique, takes approximately 4 h to complete the enumeration and isolation of individual CTCs. This is less time than is required for the CellSearchTM assay (4h for enumeration only), and CTC-(HB)Chip (ca. 6-8 h for enumeration only); (iii) Cost efficiency. The current cost of the CellSearchTM assay is approximately $1,200. Performing a NanoVelcro enumeration or isolation assay (including the materials, devices fabrication, surface coating and antibodies) is less than $50; (iv) Sample processing capacity: Given the improved sensitivity of NanoVelcro Assays, we are able to recover CTCs from only 2-mL blood samples. If required, we are able to process up to 5-mL blood in one assay. Multiple units and rounds operating in parallel and in sequence will allow even higher capacity. (v) Simple user interface: The computerized interface for operation and simplified slide-in & click-on chip holder and fluid handler facilitate setup and user-to-user variation making processing and analysis simple. Over the continuous evolution process of the past decade, the research team has accumulated significant research experience and technical knowhow conferring several unique capacities to each generation of NanoVelcro Assay. These capacities include high-resolution fluorescent imaging for morphological analysis of CTCs, LCM technique for single-cell isolation, and temperature-dependent CTC purification. With these unique capacities, NanoVelcro Assays are able to address unmet needs in oncology practice, such as stratification of heterogeneous CTC population (1st-Gen), molecular profiling of CTCs (2nd-Gen and 3rd-Gen), and rapid CTC purification for liquid biopsy (3rd-Gen).
Future scientific and clinical developments
Moving forward, future research endeavors with nanotechnology-enabled CTC assays will be driven by particular needs: (i) acquiring a fundamental understanding of the nanointerfaces between CTCs (e.g., how the underlying physical/chemical properties of any given nanosubstrate affect their CTC-capture performance, as well as the viability and molecular integrity of captured CTCs); (ii) developing new CTC-capture/release mechanisms governed by physiologically compatible stimulations for instant isolation/purification of CTCs with desired viability and molecular integrity to allow for downstream ex vivo molecular characterization; (iii) exploiting the growing number of multi-omic analytical technologies (that could result from other research initiatives within NCI Nanotechnology Alliance Program) with single-cell resolution to characterize the heterogeneous CTC pool; (iv) exploring the use of rare-cell culture techniques that will enable ex vivo expansion of purified CTCs for in-depth studies (e.g., xenograft models and drug susceptibility testing); (v) studying other types of circulating rare cells (e.g., tumor associated macrophage and stromal cells) and non-cellular particles (e.g., exosomes), which also carry information about the tumor microenvironment.
Following development of these technologic advances, challenges will remain in utilizing these new assays to address unmet needs in the areas of cancer biology and, most importantly, clinical oncology. Research endeavors should be devoted to: (i) performing multi-omic molecular characterizations on CTCs in parallel with tumor tissues from the same patients (including primary and metastatic sites when available) to further refine the CTC-tumor relationship. Such efforts are crucial to the development of CTC-based liquid biopsies. It is conceivable that CTCs may be used as surrogate tumor tissue that will provide relevant information for personalization of cancer treatment; (ii) dissecting CTC subpopulations according to their distinct phenotypes (e.g., molecular fingerprints, morphological characteristics, and behaviors) in order to address the issue of heterogeneity in tumor/CTC pool. For instance, a subpopulation of CTCs with defined small nuclei was discovered to strongly correlate with the presence of visceral metastasis in prostate cancer, offering a new way to detect the onset of the most lethal disease progression events; (iii) conducting analyses on serial CTC samples through monitoring the dynamic change of CTC subpopulations and their multi-omic molecular signatures to better understand the evolution of cancer, which is currently limited by the difficulty of obtaining tumor tissues; (iv) effectively generating and applying CTC-derived cell lines as well as xenograft models to better understand the oncogenic/resistant mechanism, and evaluate a wide range of treatment options that can potentially benefit individual patients. Validation in appropriately powered studies will be needed as these ideas translate directly into the clinical setting. Ultimately, regulatory and commercial efforts will be required to bring these tools to the population at large.
Conclusion and outlook
Early successes in the field of nanotechnology have shown great promise for addressing existing, urgent, and unmet needs in clinical oncology. As the scientific understanding of the dynamic and complex biology of cancer evolves, it has become clear to clinical scientists and cancer biologists that characterizing this dynamic biology will add an important dimension to clinical data. Oncologists practicing cancer care in this evolving biologic environment are already accustomed to handling temporal variation of data. Monitoring the dynamic alterations of biological variables, which themselves follow a distinct and biologically relevant rhythm, is a fundamental part of clinical medicine. Given the limitations of performing serial biopsies or the limited data obtainable in single biomarker panels, to date, this type of dynamic characterization has been possible only in animal models or in limited biomarker panels. The promise that analysis of CTCs and other circulating entities holds is the ability to study the dynamic biology in the system that bears the greatest relevance: the individual patient. In this era of molecular medicine that has brought us beyond the cell to the level of DNA, RNA, and proteins, it has become exceedingly clear that no two patients are identical and no two cancers are identical. Having a non-invasive means of dissecting these differences bridges the gap between the laboratory and the clinic. While these ideas are young, the successes seen in this field provide ample cause for continued work and fuel the enthusiasm for launching integrated transdisciplinary research in this transformative field.
Acknowledgements
This work was supported by National Institutes of Health (R21CA151159, R33CA157396, P01CA168585, R33CA174562, P01CA098912, U01CA198900, and R44CA180482), Department of Defense (Idea Award W81XWH-11-1-0422 and Postdoctoral Training Award PC151088), Prostate Cancer Foundation (Young Investigator Award and Creativity Award), UCLA Prostate Cancer SPORE Program, the Steven Spielberg Discovery Fund in Prostate Cancer Research, the St. Anthony Prostate Cancer Research Fund, the CD McKinnon Memorial Fund for Neuroendocrine Prostate Cancer, and the Berns Family Fund.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J. et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90-4
2. Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883-92
3. Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E. et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353-7
4. Ross-Innes CS, Becq J, Warren A, Cheetham RK, Northen H, O'Donovan M. et al. Whole-genome sequencing provides new insights into the clonal architecture of Barrett's esophagus and esophageal adenocarcinoma. Nat Genet. 2015;47:1038-46
5. Boutros PC, Fraser M, Harding NJ, de Borja R, Trudel D, Lalonde E. et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet. 2015;47:736-45
6. Kreso A, O'Brien CA, van Galen P, Gan OI, Notta F, Brown AM. et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339:543-8
7. Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J. et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256-9
8. Alix-Panabieres C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem. 2013;59:110-8
9. Lu YT, Zhao L, Shen Q, Garcia MA, Wu D, Hou S. et al. NanoVelcro Chip for CTC enumeration in prostate cancer patients. Methods. 2013;64:144-52
10. Magbanua MJ, Sosa EV, Roy R, Eisenbud LE, Scott JH, Olshen A. et al. Genomic profiling of isolated circulating tumor cells from metastatic breast cancer patients. Cancer Res. 2013;73:30-40
11. Green BJ, Saberi Safaei T, Mepham A, Labib M, Mohamadi RM, Kelley SO. Beyond the capture of circulating tumor cells: next-generation devices and materials. Angew Chem Int Ed Engl. 2016;55:1252-65
12. de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H. et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302-9
13. Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC. et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781-91
14. Scher HI, Jia X, de Bono JS, Fleisher M, Pienta KJ, Raghavan D. et al. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 2009;10:233-9
15. Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P. et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014;32:479-84
16. Jiang R, Lu YT, Ho H, Li B, Chen JF, Lin M. et al. A comparison of isolated circulating tumor cells and tissue biopsies using whole-genome sequencing in prostate cancer. Oncotarget. 2015;6:44781-93
17. Ni X, Zhuo M, Su Z, Duan J, Gao Y, Wang Z. et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc Natl Acad Sci U S A. 2013;110:21083-8
18. Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S. et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012;487:510-3
19. Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT. et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351-6
20. Riethdorf S, Fritsche H, Muller V, Rau T, Schindlbeck C, Rack B. et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res. 2007;13:920-8
21. Shaffer DR, Leversha MA, Danila DC, Lin O, Gonzalez-Espinoza R, Gu B. et al. Circulating tumor cell analysis in patients with progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:2023-9
22. Jacob K, Sollier C, Jabado N. Circulating tumor cells: detection, molecular profiling and future prospects. Expert Rev Proteomics. 2007;4:741-56
23. Yang L, Lang JC, Balasubramanian P, Jatana KR, Schuller D, Agrawal A. et al. Optimization of an enrichment process for circulating tumor cells from the blood of head and neck cancer patients through depletion of normal cells. Biotechnol Bioeng. 2009;102:521-34
24. Talasaz AH, Powell AA, Huber DE, Berbee JG, Roh KH, Yu W. et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci U S A. 2009;106:3970-5
25. Harb W, Fan A, Tran T, Danila DC, Keys D, Schwartz M. et al. Mutational Analysis of Circulating Tumor Cells Using a Novel Microfluidic Collection Device and qPCR Assay. Transl Oncol. 2013;6:528-38
26. Winer-Jones JP, Vahidi B, Arquilevich N, Fang C, Ferguson S, Harkins D. et al. Circulating tumor cells: clinically relevant molecular access based on a novel CTC flow cell. PLoS One. 2014;9:e86717
27. Earhart CM, Hughes CE, Gaster RS, Ooi CC, Wilson RJ, Zhou LY. et al. Isolation and mutational analysis of circulating tumor cells from lung cancer patients with magnetic sifters and biochips. Lab Chip. 2014;14:78-88
28. Casavant BP, Guckenberger DJ, Berry SM, Tokar JT, Lang JM, Beebe DJ. The VerIFAST: an integrated method for cell isolation and extracellular/intracellular staining. Lab Chip. 2013;13:391-6
29. Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC. et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028-38
30. Allan AL, Vantyghem SA, Tuck AB, Chambers AF, Chin-Yee IH, Keeney M. Detection and quantification of circulating tumor cells in mouse models of human breast cancer using immunomagnetic enrichment and multiparameter flow cytometry. Cytometry A. 2005;65:4-14
31. He W, Wang H, Hartmann LC, Cheng JX, Low PS. In vivo quantitation of rare circulating tumor cells by multiphoton intravital flow cytometry. Proc Natl Acad Sci U S A. 2007;104:11760-5
32. Schiro PG, Zhao M, Kuo JS, Koehler KM, Sabath DE, Chiu DT. Sensitive and high-throughput isolation of rare cells from peripheral blood with ensemble-decision aliquot ranking. Angew Chem Int Ed Engl. 2012;51:4618-22
33. Zhao M, Schiro PG, Kuo JS, Koehler KM, Sabath DE, Popov V. et al. An automated high-throughput counting method for screening circulating tumor cells in peripheral blood. Anal Chem. 2013;85:2465-71
34. Krivacic RT, Ladanyi A, Curry DN, Hsieh HB, Kuhn P, Bergsrud DE. et al. A rare-cell detector for cancer. Proc Natl Acad Sci U S A. 2004;101:10501-4
35. Marrinucci D, Bethel K, Luttgen M, Bruce RH, Nieva J, Kuhn P. Circulating tumor cells from well-differentiated lung adenocarcinoma retain cytomorphologic features of primary tumor type. Arch Pathol Lab Med. 2009;133:1468-71
36. Marrinucci D, Bethel K, Kolatkar A, Luttgen MS, Malchiodi M, Baehring F. et al. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys Biol. 2012;9:016003
37. Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schutze K. et al. Isolation by size of epithelial tumor cells: a new method for the immunomorphological and molecular characterization of circulatingtumor cells. Am J Pathol. 2000;156:57-63
38. Zheng S, Lin H, Liu JQ, Balic M, Datar R, Cote RJ. et al. Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. J Chromatogr A. 2007;1162:154-61
39. Tan SJ, Yobas L, Lee GY, Ong CN, Lim CT. Microdevice for the isolation and enumeration of cancer cells from blood. Biomed Microdevices. 2009;11:883-92
40. Tan SJ, Lakshmi RL, Chen P, Lim WT, Yobas L, Lim CT. Versatile label free biochip for the detection of circulating tumor cells from peripheral blood in cancer patients. Biosens Bioelectron. 2010;26:1701-5
41. Lecharpentier A, Vielh P, Perez-Moreno P, Planchard D, Soria JC, Farace F. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br J Cancer. 2011;105:1338-41
42. Farace F, Massard C, Vimond N, Drusch F, Jacques N, Billiot F. et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br J Cancer. 2011;105:847-53
43. Desitter I, Guerrouahen BS, Benali-Furet N, Wechsler J, Janne PA, Kuang Y. et al. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer Res. 2011;31:427-41
44. Gupta V, Jafferji I, Garza M, Melnikova VO, Hasegawa DK, Pethig R. et al. ApoStream(TM), a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics. 2012;6:24133
45. Gagnon ZR. Cellular dielectrophoresis: applications to the characterization, manipulation, separation and patterning of cells. Electrophoresis. 2011;32:2466-87
46. Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8:329-40
47. Riethdorf S, Pantel K. Advancing personalized cancer therapy by detection and characterization of circulating carcinoma cells. Ann N Y Acad Sci. 2010;1210:66-77
48. Pantel K, Alix-Panabieres C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. 2010;16:398-406
49. Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L. et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235-9
50. Gleghorn JP, Pratt ED, Denning D, Liu H, Bander NH, Tagawa ST. et al. Capture of circulating tumor cells from whole blood of prostate cancer patients using geometrically enhanced differential immunocapture (GEDI) and a prostate-specific antibody. Lab Chip. 2010;10:27-9
51. Pecot CV, Bischoff FZ, Mayer JA, Wong KL, Pham T, Bottsford-Miller J. et al. A novel platform for detection of CK+ and CK- CTCs. Cancer Discov. 2011;1:580-6
52. Saliba AE, Saias L, Psychari E, Minc N, Simon D, Bidard FC. et al. Microfluidic sorting and multimodal typing of cancer cells in self-assembled magnetic arrays. Proc Natl Acad Sci U S A. 2010;107:14524-9
53. Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA. et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A. 2010;107:18392-7
54. Adams AA, Okagbare PI, Feng J, Hupert ML, Patterson D, Gottert J. et al. Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. J Am Chem Soc. 2008;130:8633-41
55. Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E. et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179ra47
56. Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC. et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216-20
57. Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA. et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110-22
58. Sarioglu AF, Aceto N, Kojic N, Donaldson MC, Zeinali M, Hamza B. et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat Methods. 2015;12:685-91
59. Castro CM, Ghazani AA, Chung J, Shao H, Issadore D, Yoon TJ. et al. Miniaturized nuclear magnetic resonance platform for detection and profiling of circulating tumor cells. Lab Chip. 2014;14:14-23
60. Myung JH, Gajjar KA, Saric J, Eddington DT, Hong S. Dendrimer-mediated multivalent binding for the enhanced capture of tumor cells. Angew Chem Int Ed Engl. 2011;50:11769-72
61. Chen JY, Tsai WS, Shao HJ, Wu JC, Lai JM, Lu SH. et al. Sensitive and specific biomimetic lipid coated microfluidics to isolate viable circulating tumor cells and microemboli for cancer detection. PLoS One. 2016;11:e0149633
62. Mach AJ, Kim JH, Arshi A, Hur SC, Di Carlo D. Automated cellular sample preparation using a Centrifuge-on-a-Chip. Lab Chip. 2011;11:2827-34
63. Dhar M, Wong J, Karimi A, Che J, Renier C, Matsumoto M. et al. High efficiency vortex trapping of circulating tumor cells. Biomicrofluidics. 2015;9:064116
64. Mohamadi RM, Besant JD, Mepham A, Green B, Mahmoudian L, Gibbs T. et al. Nanoparticle-mediated binning and profiling of heterogeneous circulating tumor cell subpopulations. Angew Chem Int Ed Engl. 2015;54:139-43
65. Yoon HJ, Kozminsky M, Nagrath S. Emerging role of nanomaterials in circulating tumor cell isolation and analysis. ACS Nano. 2014;8:1995-2017
66. Wang L, Asghar W, Demirci U, Wan Y. Nanostructured substrates for isolation of circulating tumor cells. Nano Today. 2013;8:347-87
67. Zhang P, Wang S. Designing fractal nanostructured biointerfaces for biomedical applications. Chemphyschem. 2014;15:1550-61
68. Wang S, Wang H, Jiao J, Chen KJ, Owens GE, Kamei K. et al. Three-dimensional nanostructured substrates toward efficient capture of circulating tumor cells. Angew Chem Int Ed Engl. 2009;48:8970-3
69. Lin M, Chen JF, Lu YT, Zhang Y, Song J, Hou S. et al. Nanostructure embedded microchips for detection, isolation, and characterization of circulating tumor cells. Acc Chem Res. 2014;47:2941-50
70. Sekine J, Luo SC, Wang S, Zhu B, Tseng HR, Yu HH. Functionalized conducting polymer nanodots for enhanced cell capturing: the synergistic effect of capture agents and nanostructures. Adv Mater. 2011;23:4788-92
71. Hsiao YS, Luo SC, Hou S, Zhu B, Sekine J, Kuo CW. et al. 3D bioelectronic interface: capturing circulating tumor cells onto conducting polymer-based micro/nanorod arrays with chemical and topographical control. Small. 2014;10:3012-7
72. Zhang N, Deng Y, Tai Q, Cheng B, Zhao L, Shen Q. et al. Electrospun TiO2 nanofiber-based cell capture assay for detecting circulating tumor cells from colorectal and gastric cancer patients. Adv Mater. 2012;24:2756-60
73. He R, Zhao L, Liu Y, Zhang N, Cheng B, He Z. et al. Biocompatible TiO2 nanoparticle-based cell immunoassay for circulating tumor cells capture and identification from cancer patients. Biomed Microdevices. 2013;15:617-26
74. Lee H, Jang Y, Seo J, Nam J-M, Char K. Nanoparticle-functionalized polymer platform for controlling metastatic cancer cell adhesion, shape, and motility. ACS Nano. 2011;5:5444-56
75. Park GS, Kwon H, Kwak DW, Park SY, Kim M, Lee JH. et al. Full surface embedding of gold clusters on silicon nanowires for efficient capture and photothermal therapy of circulating tumor cells. Nano Lett. 2012;12:1638-42
76. Banerjee SS, Paul D, Bhansali SG, Aher ND, Jalota-Badhwar A, Khandare J. Enhancing surface interactions with colon cancer cells on a transferrin-conjugated 3D nanostructured substrate. Small. 2012;8:1657-63
77. Zhao W, Cui CH, Bose S, Guo D, Shen C, Wong WP. et al. Bioinspired multivalent DNA network for capture and release of cells. Proc Natl Acad Sci U S A. 2012;109:19626-31
78. Yoon HJ, Kim TH, Zhang Z, Azizi E, Pham TM, Paoletti C. et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat Nano. 2013;8:735-41
79. Sun J, Masterman-Smith MD, Graham NA, Jiao J, Mottahedeh J, Laks DR. et al. A microfluidic platform for systems pathology: multiparameter single-cell signaling measurements of clinical brain tumor specimens. Cancer Res. 2010;70:6128-38
80. Wang S, Liu K, Liu J, Yu ZT, Xu X, Zhao L. et al. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew Chem Int Ed Engl. 2011;50:3084-8
81. Zhao L, Lu YT, Li F, Wu K, Hou S, Yu J. et al. High-purity prostate circulating tumor cell isolation by a polymer nanofiber-embedded microchip for whole exome sequencing. Adv Mater. 2013;25:2897-902
82. Hou S, Zhao L, Shen Q, Yu J, Ng C, Kong X. et al. Polymer nanofiber-embedded microchips for detection, isolation, and molecular analysis of single circulating melanoma cells. Angew Chem Int Ed Engl. 2013;52:3379-83
83. Hou S, Zhao H, Zhao L, Shen Q, Wei KS, Suh DY. et al. Capture and stimulated release of circulating tumor cells on polymer-grafted silicon nanostructures. Adv Mater. 2013;25:1547-51
84. Ke Z, Lin M, Chen JF, Choi JS, Zhang Y, Fong A. et al. Programming thermoresponsiveness of NanoVelcro substrates enables effective purification of circulating tumor cells in lung cancer patients. ACS Nano. 2015;9:62-70
85. Ankeny JS, Court CM, Hou S, Li Q, Song M, Wu D. et al. Circulating tumour cells as a biomarker for diagnosis and staging in pancreatic cancer. Br J Cancer. 2016 In press
86. Kamei K, Ohashi M, Gschweng E, Ho Q, Suh J, Tang J. et al. Microfluidic image cytometry for quantitative single-cell profiling of human pluripotent stem cells in chemically defined conditions. Lab Chip. 2010;10:1113-9
87. Chen JF, Ho H, Lichterman J, Lu YT, Zhang Y, Garcia MA. et al. Subclassification of prostate cancer circulating tumor cells by nuclear size reveals very small nuclear circulating tumor cells in patients with visceral metastases. Cancer. 2015;121:3240-51
88. Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818-22
89. Fang X, Tan W. Aptamers generated from cell-SELEX for molecular medicine: a chemical biology approach. Acc Chem Res. 2010;43:48-57
90. Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505-10
91. Zhao L, Tang C, Xu L, Zhang Z, Li X, Hu H. et al. Enhanced and differential capture of circulating tumor cells from lung cancer patients by microfluidic assays using aptamer cocktail. Small. 2016;12:1072-81
92. Rao C, Bui T, Connelly M, Doyle G, Karydis I, Middleton MR. et al. Circulating melanoma cells and survival in metastatic melanoma. Int J Oncol. 2011;38:755-60
93. Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA. et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809-19
94. Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H. et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596-9
95. Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549-54
96. Bryant KL, Mancias JD, Kimmelman AC, Der CJ. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. 2014;39:91-100
97. Court CM, Ankeny JS, Sho S, Hou S, Li Q, Hsieh C. et al. Reality of Single Circulating Tumor Cell Sequencing for Molecular Diagnostics in Pancreatic Cancer. J Mol Diagn. 2016 In press
98. Wang G, Brennan C, Rook M, Wolfe JL, Leo C, Chin L. et al. Balanced-PCR amplification allows unbiased identification of genomic copy changes in minute cell and tissue samples. Nucleic Acids Res. 2004;32:e76
99. Lovmar L, Fredriksson M, Liljedahl U, Sigurdsson S, Syvanen AC. Quantitative evaluation by minisequencing and microarrays reveals accurate multiplexed SNP genotyping of whole genome amplified DNA. Nucleic Acids Res. 2003;31:e129
Author contact
![]() Corresponding authors: Dr. Hsian-Rong Tseng and Edwin M. Posadas.
Corresponding authors: Dr. Hsian-Rong Tseng and Edwin M. Posadas.
 Global reach, higher impact
Global reach, higher impact