13.3
Impact Factor
Theranostics 2016; 6(6):828-836. doi:10.7150/thno.14695 This issue Cite
Research Paper
Efficient Capture and Isolation of Tumor-Related Circulating Cell-Free DNA from Cancer Patients Using Electroactive Conducting Polymer Nanowire Platforms
1. New Experimental Therapeutic Branch, National Cancer Center, Goyang, Gyeonggi-do 410-769, South Korea;
2. Lung Cancer Branch, National Cancer Center, Goyang, Gyeonggi-do 410-769, South Korea;
3. Global Research Laboratory, Department of Biochemistry and Molecular Biology, Korea University College of Medicine;
4. Department of Medical Science, Yonsei University College of Medicine, 50 Yonsei-Ro, Seodaemun-Gu, Seoul 03722, Korea;
5. College of Veterinary Medicine, Konkuk University, Seoul, Korea.
* These authors contributed equally.
Received 2015-12-15; Accepted 2016-2-27; Published 2016-4-11
Abstract
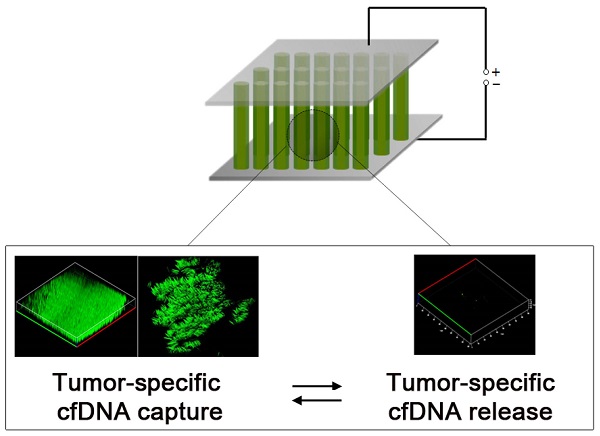
Circulating cell-free DNA (cfDNA) is currently recognized as a key non-invasive biomarker for cancer diagnosis and progression and therapeutic efficacy monitoring. Because cfDNA has been detected in patients with diverse types of cancers, the use of efficient strategies to isolate cfDNA not only provides valuable insights into tumour biology, but also offers the potential for developing new cancer-specific targets. However, the challenges associated with conventional cfDNA extraction methods prevent their further clinical applications. Here, we developed a nanostructured conductive polymer platform for the efficient capture and release of circulating cfDNA and demonstrated its potential clinical utility using unprocessed plasma samples from patients with breast and lung cancers. Our results confirmed that the platform's enhanced efficiency allows tumor-specific circulating cfDNA to be recovered at high yield and purity.
Keywords: cancer, circulating cell-free DNA, liquid biopsy, conducting polymer, nanowired structure, mutation detection.
Introduction
The substantial progress made in cancer nanotechnology has resulted in an extraordinary ability to detect early-stage cancers, assess disease progression, and guide therapeutic effectiveness with high sensitivity and specificity.[1-3] Recently, circulating cancer biomarkers such as circulating tumour cells (CTCs), cancer-related circulating cell-free DNA (cfDNA), and exosomes have emerged as powerful alternatives for diagnosing cancer and for predicting and evaluating the treatment responses that can assist in decision-making for patient-specific therapy.[4-9] Assessment of circulating biomarkers offers notable advantages over conventional tissue biopsy in that it can be used for noninvasively and simply analysing the clinical relevance of the biomarkers by monitoring tumour heterogeneity at multiple sites and tumour evolution at multiple time points; ultimately, this allows specific analysis of mutations and epigenetic modifications.[10-12] Currently, circulating cfDNA, in particular, is recognized as a valuable diagnostic and prognostic tool because detectable levels of cfDNA in peripheral blood are directly correlated with tumour burden.[13-15] cfDNA investigation allows not only for identification of diverse patterns of genomic alterations during tumour evolution and adaption, but also for direct exploration of potential challenges associated with genetic tumour heterogeneity. However, given the rarity and highly fragmented nature of tumour-specific cfDNA, cfDNA analysis is likely to be extremely challenging and thus requires highly efficient extraction procedures for successful implementation of various downstream applications. Conventional methods of isolating and extracting cfDNAs from blood plasma have limited practical applicability in clinical settings because they are well suited to processing, extraction, and analysis of cfDNA with medium and high molecular weight rather than short DNA fragments. Furthermore, they ensure optimal recovery efficiency and quality only when used with a sufficient amount of blood plasma (>1 mL) and a reasonable concentration of cfDNA in the sample. For further biomedical research applications, improved analysis methods for precise and sensitive quantification of mutant DNA present in a sample must be urgently developed.[16-18] Recently, we demonstrated the development of an electroactive conducting polymer polypyrrole (Ppy) platform for the efficient capture and release of CTCs.[19-21] Here, we expanded this concept to include the recovery of cfDNA from blood plasma. Our novel approach involves the use of free-standing, 3D arrays of Au nanowires coated with the conducting polymer Ppy (“Ppy/Au NWs”) (Figure 1a). The key strategies employed in this study were developed based on the following features: i) vertically aligned nanowire designs, including large surface areas and well-defined architecture, offer unique constructs that can readily serve as a reservoir for the efficient adsorption of circulating cfDNA, and ii) electrochemical performance attained through the interchange of charges along the Ppy backbones regulates the association/dissociation interactions between individual nanowires and negatively charged DNA molecules. In addition to possessing unique physical and chemical properties, Ppy exhibits reversible potential-induced transitions in two phases: i) in the oxidized state as a result of π-conjugated delocalized backbones, which ultimately allows positive charge distribution on the polymeric chains, and ii) in the reduced state, when anions are incorporated into the polymeric backbone and neutralize the positive charge of the polymer chains.[22-25] Therefore, the nanostructured electric field-responsive surface exhibits markedly enhanced performance in the isolation of cfDNA, particularly tumour-related highly fragmented DNA, with high yield and purity. Furthermore, it is important to note that the large surface area of nanowires promotes reliable recovery regardless of the experimental conditions, even at very low concentrations of cfDNA (≥10 pg/mL) or with small sample volume (≥100 µL), for example. Indeed, the Ppy/Au NW platform provides a convenient and efficient strategy to maximize the recovery of cfDNA through a highly sensitive, cost-effective, and simple process.
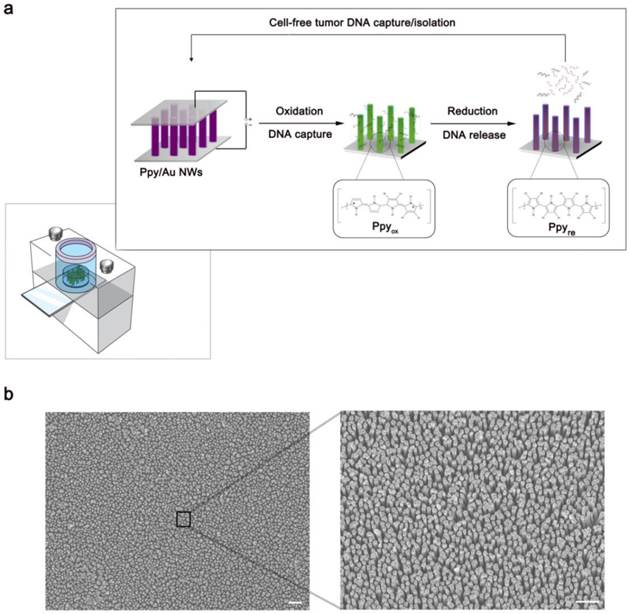
(a) Schematic representation of Ppy-coated Au nanowires (Ppy/Au NWs). The insets highlight the spontaneous redox behaviour of Ppy in response to the applied electric fields that is closely linked to the DNA recovery efficiency. (b) Left: top-view FE-SEM image of the Ppy/Au NWs (scale bar, 1 µm). Right: magnified FE-SEM image of individual nanowires (scale bar, 2 µm).
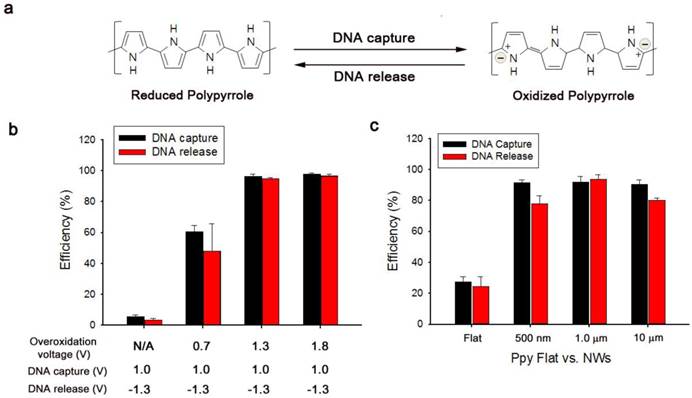
(a) Reversible adsorption and desorption of cfDNA on as-prepared platforms triggered by the electrochemical oxidation-reduction process in the Ppy layer. (b) Evaluation of the corresponding cfDNA capture/release performance at various overoxidation potentials applied to the resulting Ppy. (c) Comparison of the capture/release performance between a flat Ppy substrate and Ppy/Au NWs of various lengths. The data shown represent the means ± SD of 5 independent experiments.
Results and Discussion
Characterization of the Ppy/Au NW platform by using DNA spiked into human plasma
First, we present field emission scanning electron microscopy (FE-SEM) images showing the detailed morphological characteristics of Ppy/Au NWs in Figure 1b. The magnified view clearly shows that nanowires synthesized using anodic alumina oxide (AAO) template-assisted electrochemical deposition were on average 1.0 µm in length and 100 nm in diameter. As a proof-of-concept study, we used Ppy/Au NW arrays to efficiently isolate circulating cfDNA directly from blood plasma (Figure 2). The principal advantage of Ppy is that it enables the tuning of electrochemical activities. By inducing reversible switching between the oxidized and reduced states along the polymeric backbone, the oxidized Ppy surfaces strongly influence the binding interactions with the corresponding negatively charged DNA molecules.[26, 27]
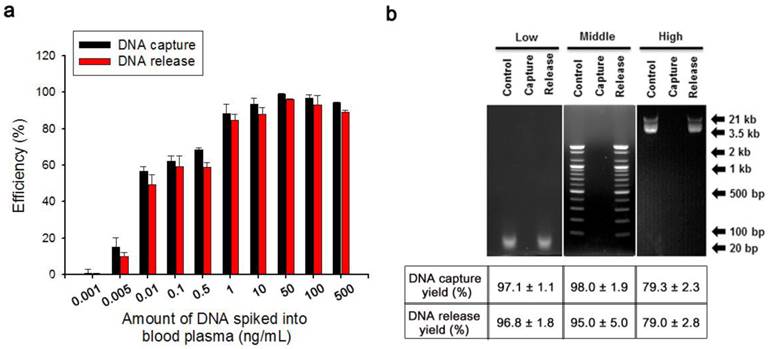
Based on the abovementioned property, we examined the efficiency of electrochemically assisted DNA capture and isolation using artificial blood samples prepared through ex vivo spiking of a reference 100-bp DNA ladder (50 ng) into 200 µL of blood plasma collected from healthy donors. The procedure of efficiently recovering DNA from blood plasma comprised three simple steps: i) overoxidation of Ppy/Au NWs at 1.8 V during 2 min of electrical stimulation, ii) DNA capture upon exposure to 1.0 V, and iii) DNA release from the nanowires with electrical stimulation at -1.3 V. First, we tested the effect of overoxidation on the induction of specific adsorption of the spiked DNA by applying various positive potentials (Figure 2b). Interestingly, the capture efficiency was greatly enhanced as the applied overoxidation potentials were gradually increased, whereas a negligible effect on DNA capture was observed in the absence of overoxidation. Therefore, overoxidation treatment at higher potentials clearly imparted additional functions to the nanostructured Ppy platform, inducing a high distribution of positive charges on the polymer backbone and thereby strengthening the intermolecular forces between DNA and overoxidized Ppy. However, DNA extraction performance of overoxidized Ppy/Au NWs can vary over the reaction time (Figure S1). When overoxidized at 1.8 V for 2 min, Ppy/Au NWs exhibited maximal recovery of cfDNA from blood plasma. By contrast, overoxidation of Ppy at 1.8 V for ≥5 min led to a drastic decrease in the DNA extraction yield as a consequence of a loss of positive charge in the Ppy backbone.[28-31] Next, we systematically explored optimal experimental conditions necessary to improve DNA recovery without DNA degradation. After 2 min of overoxidation at 1.8 V, DNA capture was examined as a function of the applied voltages and reaction times. Capture efficiency was maximized following the application of 1.0 V for ≥30 s (Figure S2a-b). Unbound DNA or non-specifically bound plasma constituents (proteins, lipids, hormones, etc.) were immediately removed through repetitive washing from Ppy/Au NW arrays. As shown in Figure S3, we evaluated the isolation of DNA from both blood plasma and PBS that were spiked with DNA ladders of three sizes: 10 bp (low range), 100 bp (middle range), and 3.5 kb (high range). Interestingly, DNA recovery was similar in both cases of extraction, implying that various components present in the plasma did not interfere with the performance of DNA capture/release. We further investigated the non-destructive release of the captured DNA from Ppy/Au NWs in response to electric fields. The results showed that positive charges on the polymeric chain could be readily neutralized by applying high negative potentials, and this was primarily responsible for the release of the captured DNA (Figure S2c). We also directly investigated the DNA release pattern at six time points, revealing that a maximal release rate of ~95% was achieved when -1.3 V was applied for 5 min (Figure S2d). However, prolonged electrical stimulation for ≥10 min adversely affected DNA release. This phenomenon might be associated with the mechanical instability of Ppy accompanied by the shrinkage and cracking of the film. We also attempted to elucidate the impact of the nanostructured surface geometry on DNA interactions by using a spiked DNA ladder. Vertically aligned nanowire structures featuring extremely high surface areas appeared likely to provide more binding sites compared with flat structures (Figure 2c). Notably, the unique advantages of 3D nanowire arrays in DNA recovery might be attributed to their large surface-to-volume ratio and high wire density, which ultimately lead to increased interactions with the target DNA. In addition, the large surface area of the nanowires provided the high sensitivity required to extract very low levels of tumour-related cfDNA present in blood plasma (Figure 3a). The results clearly show that particularly when the DNA concentration was ≥1 ng/mL, the proposed platform demonstrated high performance in DNA capture and release with >90% efficiency. Unexpectedly, this platform could also be successfully used to extract DNA at a concentration as low as 10 pg/mL with an efficiency of 50%. Subsequently, to obtain additional evidence of enhanced DNA recovery from nanowires, we performed assays in which we varied the molecular weights of the spiked DNA (Figure 3b).
To examine how DNA size affected recovery efficiency, we spiked 50 ng DNA ladders of three different sizes (10 bp (low range), 100 bp (middle range), and 3.5 kb (high range)) into 200 µL of healthy human blood plasma, and then performed gelelectrophoresis analysis (Figure 3b). After the capture experiments were conducted by applying dual potentials of 1.8/1.0 V, no DNA bands were visible in the supernatant, which indicated that overoxidized Ppy possessed sufficient positive charge to bind all of the DNA fragments present in blood plasma. However, DNA fragments were clearly detected in the solution after stimulation at -1.3 V, and this observation could be attributed to a massive release of the captured DNA from Ppy/Au NWs.
(a) DNA capture/release efficiencies of Ppy/Au NWs as a function of the amount of DNA (100 bp) spiked into blood plasma. (b) (Top) Agarose gel electrophoresis of DNA ladders of 3 sizes, 10 bp (low range), 100 bp (middle range), and 3.5 kb (high range). Control (left lanes), DNA markers; Capture (middle lanes), unreacted DNA solution after capture experiment was performed using Ppy/Au NWs; Release (right lanes), the solution containing DNA fragments released from Ppy/Au NWs. (Bottom) Capture and release efficiencies of Ppy/Au NW platforms measured using various DNA ladders. The data shown represent the means ± SD of 5 independent experiments.
(a) Concentrations of the eluted cfDNA from blood plasma donated by healthy volunteers and patients with breast cancer and lung cancer, respectively. (b) Integrity of the cfDNAs extracted using Ppy/Au NWs and Qiagen kit was assessed using 2% agarose gels. In both the top and bottom images, the left panel shows 100-bp and 3.5-kb DNA markers; the middle and right panel represents cfDNAs extracted using Qiagen kit and Ppy/Au NWs from the plasma of patients with breast (B1-B5) and lung (L1-L5) cancers. (c) Confocal fluorescence images of the captured and released cfDNA from the plasma of lung cancer patient. Ppy/Au NWs that were overoxidized using dual stimulations of 1.8/1.0 V strongly adsorbed cfDNAs, and this was detected under the confocal microscope after staining with PicoGreen (left and middle figures). Immediately after an electric field of -1.3 V was applied, the captured cfDNAs were completely released from Ppy/Au NWs (right figure).
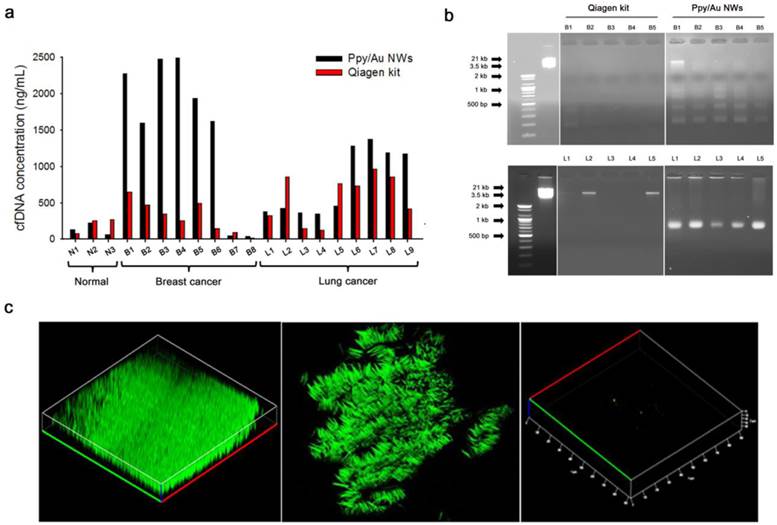
We present the capture and release yields of various DNA ladders quantified for Ppy/Au NWs. Interestingly, DNA adsorption is likely to be size-dependent. Whereas low- and middle-range DNA ladders exhibited high recovery rates, rates declined slightly for high-range DNA ladders. This could be attributed to the densely packed architecture of the nanowire arrays: large DNA fragments would be trapped and prevented from penetrating deeply into the individual nanowires, and therefore electrostatic recognition could occur at or near the top surface of the nanowires. Consequently, the binding ability of large DNA molecules to cationic Ppy NW arrays appeared to be slightly lower than that of small DNA molecules. Given that cfDNAs in the bloodstream are generally 180-1000 bp in size, which is characteristic of the apoptotic process[6, 7], our platform is sufficiently suitable for the extraction of tumour-specific small fragmented DNA.
Proof-of-concept studies on Ppy/Au NWs conducted using plasma from cancer patients
To evaluate the technical and clinical utility of our Ppy/Au NW platform, we examined its capacity to capture and release cfDNA from plasma samples collected from healthy donors and patients with breast cancer and lung cancer (Figure 4). The Qiagen kit was chosen for this study because it resulted in the highest yield of DNA from plasma samples of breast and lung cancer patients (Figure S4).[32-35] Figure 4a shows the quantification of the cfDNA extracted using Ppy/Au NWs (black bars) and the Qiagen kit (red bars) from plasma samples. Notably, compared with the Qiagen kit, Ppy/Au NWs yielded considerably higher amounts of cfDNA from the samples of the majority of patients with breast and lung cancers. In the case of breast cancer patients in particular, the cfDNA yields obtained using Ppy/Au NWs were 4-fold higher than those obtained using the Qiagen kit. By contrast, the mean concentrations of cfDNA extracted from normal samples did not differ significantly between Ppy/Au NWs and the Qiagen kit. In agreement with previous studies, cfDNA levels were observed to be considerably higher in cancer patients than in healthy donors. [7, 13]
Here, we also found that the mean purity of the cfDNA obtained from the Ppy/Au NW platform was very high at 1.97 ± 0.02, as determined based on the A260/A280 ratio. Thus, our platform enables efficient extraction of cfDNA with sufficiently high quality and purity to allow further molecular analysis. Furthermore, we monitored the occurrence of isolated cfDNA fragments using agarose gel electrophoresis. Typically, cfDNAs are highly likely to form a ladder pattern, which is a key signature of the apoptotic process.[6] The cfDNAs extracted using our Ppy/Au NW platform exhibited an apoptotic ladder of bands in agarose gels, highlighting the distinctive characteristic of circulating cfDNA (Figure 4b). By comparison, cfDNAs isolated using the Qiagen kit showed low or no prominent bands, which might be associated with the relatively low concentrations of the eluted cfDNA. Next, we performed confocal microscopy and examined how the electrochemical behaviours of Ppy manipulated the intermolecular interaction with circulating cfDNAs (Figure 4c). Because the PicoGreen dye binds selectively to double-stranded DNA and enhances the fluorescence signal, we used it as a reliable indicator to detect the adsorption-desorption behaviours of cfDNA on Ppy/Au NWs. Notably, nanowires that were overoxidized using combined 1.8/1.0 V stimulation were abundantly and specifically stained with PicoGreen, which indicated the direct attachment of negatively charged cfDNAs. The reconstructed 3D confocal images revealed a strong distribution of green fluorescence along the surface of individually isolated nanowires (Fig. 4c, left and middle). However, as per our hypothesis, the green fluorescence emitted from Ppy/Au NWs disappeared immediately upon exposure to a field of -1.3 V for 5 min, which confirmed that most of the attached cfDNAs were completely released from the nanowires (Fig. 4c, right).
Integrity of cfDNAs assessed by amplifying cancer-related genes
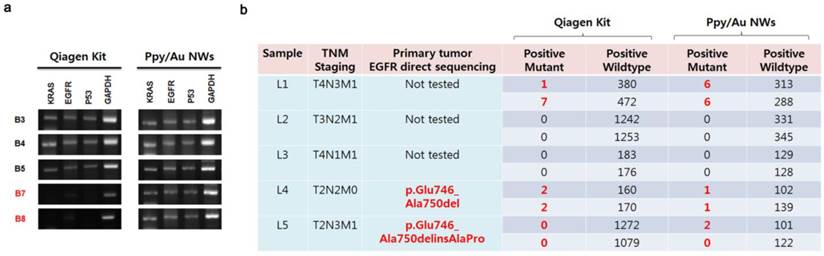
Lastly, we attempted to quantitatively detect cancer-related mutations in the released cfDNAs by directly amplifying target sequences, and we used GAPDH as a control gene in these assays (Fig. 5a). Our results showed that most of the cfDNAs extracted using Ppy/Au NWs and the Qiagen kit were closely related to specific PCR bands corresponding to the EGFR, KRAS, and P53 genes. However, in the case of samples B7 and B8, the KRAS, EGFR, and P53 genes were not amplified from the cfDNAs collected using the Qiagen kit, but all three genes were clearly amplified when cfDNAs were extracted using Ppy/Au NWs. Because the levels of eluted cfDNA can strongly influence the detection and quantification of certain target genes, we explored the feasibility of analysing EGFR mutations in the recovered cfDNAs using droplet digital PCR (ddPCR). We determined the relative numbers of EGFR mutants in the eluted cfDNAs and compared them with the confirmed EGFR status in the corresponding tumour tissue (Fig. 5b). In the case of sample L4, we identified EGFR mutations (Exon 19 deletion) in cfDNAs extracted using both Ppy/Au NWs and the Qiagen kit at mutant-to-WT ratios of 0.98% and 1.25%, respectively, which agreed with the mutation data from their tumour tissues. In the case of L5, mutation was detected in the tissue, but the EGFR gene was not amplified from the cfDNA extracted using the Qiagen kit. However, EGFR mutations were verified in the cfDNAs isolated using Ppy/Au NWs, and their abundance was 1.98%. Conversely, in the case of L1, although testing for primary tumour tissue was not available, the assessment of the sample led to the identification of EGFR mutation (Exon 19 deletion) through its detection in cfDNAs isolated using both Ppy/Au NWs and the Qiagen kit at mutant-to-WT ratios of 2.1% and 1.5%, respectively.
(a) Electrophoresis results for the amplified fragments of KRAS, EGFR, P53, and GAPDH genes. The patterns of gene amplification might be strongly correlated with the concentrations of the cfDNAs extracted from the plasma samples. (b) EGFR gene alterations in the cfDNAs extracted from plasma samples of lung cancer patients, determined using droplet digital PCR. In our results, a close concordance in EGFR mutation was noted between the eluted cfDNAs and tumour tissues.
Conclusions
In this study, we developed a simple, fast, and reliable strategy to extract tumor-specific circulating cfDNA from unprocessed plasma using an electroactive Ppy/Au NW platform. The nanostructured electric field-mediated Ppy surface exhibited excellent performance in the isolation of cfDNA at high yield and purity. Ppy appears to be well suited as a unique electroactive interface for modulating the surface charge in response to external electrical stimuli, and thus for repetitively manipulating DNA-Ppy surface adsorption/desorption interactions. Our findings have demonstrated the platform's enhanced performance in cfDNA recovery and verified tumour-specific genetic alterations in the eluted cfDNA. Overall, the Ppy/Au NW platform can be used as an efficient and flexible tool in both basic and clinical cancer-related fields and will ultimately provide highly accurate diagnostic and prognostic information, thus yielding improved outcomes for cancer patients.
Experimental Section
Fabrication of Ppy-coated Au NWs (Ppy/Au NWs)
An approximately 150-nm-thick Au layer was deposited onto one surface of the AAO template (Whatman; pore diameter, 100 nm) by employing the conventional thermal evaporation technique. Au nanowires were electrochemically grown within the pores of the Au-backed AAO membrane by using a gold-plating solution (Orotemps 24 RTU Rack) and applying cyclic voltammetry in the -1.1 - 0 V potential range at a scan rate of 100 mV/s. All electrochemical experiments were conducted using a potentiostat/galvanostat (BioLogic SP-150) in 3-electrode cells, in which Ag/AgCl, platinum wire, and the designed platform were employed as reference, counter, and working electrodes, respectively. After fixing on the indium tin oxide (ITO) surface by using a conductive carbon paste, the generated membrane was dissolved in an aqueous NaOH solution (2 M) for 4 h to remove the AAO template; this ultimately produced vertically aligned arrays of Au NWs of distinct lengths. To prepare Ppy-coated Au NWs, we performed electrochemical deposition of Ppy on freestanding Au NWs in an aqueous mixture of 0.1 M pyrrole and 0.01 M poly(sodium 4-styrenesulfonate) (PSS) by applying chronoamperometry (CA) at 0.8 V (vs. Ag/AgCl) for 20 s. The resulting Ppy/Au NWs were rinsed several times with water and incubated in Tris-HCl (pH 7.5) for the electrochemical overoxidation of the thin layer of Ppy by applying 1.8 V (vs. Ag/AgCl) for 2 min.
In order to evaluate the chemical composition of the Ppy/Au NWs, survey and high-resolution X-ray photoelectron spectroscopy (XPS) scans were performed with a K-alpha (Thermo Scientific Inc., U.K.), equipped with a monochromatic Al Kα X-ray source with variable spot size (30-400µm in 5µm steps).
Sample collection and preparation
Whole blood was collected in Vacutainer tubes containing the anti-coagulant EDTA by using procedures approved by the NCC Institutional Review Board (IRB # NCCCTS08-112, NCCNCS13-717). Plasma was separated using a refrigerated centrifuge (3000 × g, 10 min). To evaluate the capture and release efficiency of our Ppy/Au NW platforms, we tested them using artificial blood samples that were prepared by the ex vivo spiking of DNA ladders (50 ng) into 200 µL of blood plasma obtained from healthy donors. For clinical applications, we collected blood samples from 3 healthy volunteers and 17 patients with breast and lung cancers.
DNA capture and release
Before evaluating their DNA-capture efficiency, we immersed the overoxidized Ppy/Au NWs in blood samples for 30 min at room temperature in order to promote DNA interaction with individual nanowires; the samples were i) artificial samples containing spiked 50 ng of DNA molecules in 200 µL of human plasma or ii) unprocessed plasma samples (0.2-1 mL) obtained from 3 healthy donors and 17 cancer patients. Next, immediate DNA capture was performed by applying 1.0 V (vs. Ag/AgCl) for 30 s. Subsequently, Ppy/Au NWs were washed twice by 500 µL of nuclease free water (NFW) to remove unbound DNA or non-specifically bound plasma constituents. Then, the captured DNA was eluted in NFW (20-100 µL) by electrical stimulation of Ppy/Au NWs at -1.3 V (vs. Ag/AgCl) for 5 min. The captured and released DNA was quantified by using the PicoGreen assay according to the manufacturer's protocol. The obtained DNA was measured spectrophotometrically at 260 and 280 nm, and the A260/A280 ratio was used to assess the purity of the captured and released DNA. The interactions between Ppy/Au NWs and DNA were visualized by examining PicoGreen fluorescence under a Zeiss LSM 710 confocal microscope.
PCR and gel electrophoresis
All primers were synthesized by Macrogen, Korea. DNA ladders (low: 10-100 bp; middle: 100 bp to 2 kb; high: 3.5-21 kb) were purchased from Bioneer, Korea. All PCR amplifications were performed in a GeneAmp PCR system 9600 (Perkin Elmer, Norwalk, CT, USA) in a final reaction volume of 20 µL that contained template DNA, 2 oligonucleotide primers (5 pM), 200 mM of each dNTP, 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 1.5 mM MgCl2, 0.1% (w/v) gelatin, and 1 U of Taq DNA polymerase (Perkin Elmer). PCR amplification consisted of 45 cycles of denaturation (94°C, 30 s), annealing (64°C, 30 s), and elongation (72°C, 1 min). PCR products were electrophoresed on 2% agarose gels and stained with ethidium bromide to visualize their presence under a UV transilluminator.
Primers
| Primer name | Primer sequence (5' → 3') |
|---|---|
| KRAS forward primer | ACTGAATATAAACTTGTGGTAGTTGGACCT |
| KRAS reverse primer | ACTCATGAAAATGGTCAGAGAAACCTTTAT |
| EGFR (Exon 19) forward primer | GCACCATCTCACAATTGCCAGTTA |
| EGFR (Exon 19) reverse primer | AAAAGGTGGGCCTGAGGTTCA |
| P53 forward primer | CAGCACATGACGGAGGTTG |
| P53 reverse primer | TCATCCAAATACTCCACACGC |
| GAPDH forward primer | GGAGCGAGATCCCTCCAAAAT |
| GAPDH reverse primer | GGCTGTTGTCATACTTCTCA |
QIAamp circulating nucleic acid kit
We extracted cfDNA from 1 mL of plasma by using the QIAamp circulating nucleic acid kit (Qiagen) according to the manufacturer's protocol. Briefly, plasma samples were lysed using proteinase K and a lysis buffer and then the circulating nucleic acids were bound to the silica membrane by applying vacuum pressure. DNA fragments were recovered from the membrane after several washing steps.
Digital PCR
EGFR mutation in cfDNAs was examined by using PrimePCR ddPCR mutation assay kits for EGFR L858R and Exon 19 deletion (p.E746-A750del) on a QX200 ddPCR system according to the manufacturer's protocol (BioRad, Hercules, CA, USA). Water-oil emulsion droplets were generated from PCR mixtures that contained 8 μL of cfDNA, 10 μL of ddPCR supermix, and 2 μL of target primers and probes. After generating the droplets, PCR was performed in a thermal cycler. Positive droplets, which contained at least one copy of the amplified DNA, could be detected by the droplet reader for fluorescence analysis.
Mutation test for EGFR gene
The EGFR gene status in the primary tumour tissue was examined using the PCR-direct sequencing method. Genomic DNA was extracted and amplified using specific primers for Exons 19 and 21 of the EGFR gene. PCR products were sequenced using the BigDye terminator cycle sequencing kit and analyzed using an ABI PRISM 3100 DNA analyzer (Applied Biosystems, Foster City, CA, USA).
Supplementary Material
Supplementary figures.
Acknowledgements
This work was supported by a National Cancer Center grant from the Republic of Korea (1510070-1).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Schroeder A, Heller DA, Winslow MM, Dahlman JE, Pratt GW, Langer R, Jacks R, Anderson DG. Treating metastatic cancer with nanotechnology. Nat. Rev. Cancer. 2012;12:39-50
2. Ferrari M. Cancer nanotechnology: opportunities and challenges. Nat. Rev. Cancer. 2005;5:161-171
3. Chauhan VP, Jain RK. Strategies for advancing cancer nanomedicine. Nat. Mater. 2013;12:958-962
4. Bidard FC, Weigelt B, Reis-Filho JS. Going with the flow: from circulating tumor cells to DNA. Sci. Transl. Med. 2013;5:207
5. De Mattos-Arruda L, Cortes J, Santarpia L, Vivancos A, Tabernero J, Reis-Filho JS, Seoane J. Circulating tumour cells and cell-free DNA as tools for managing breast cancer. Nat. Rev. Clin. Oncol. 2013;10:377-389
6. Diaz LA, Bardelli A, Clin J. Liquid biopsies: genotyping circulating tumor DNA. Oncol. 2014;32:579-586
7. Elshimali YI, Khaddour H, Sarkissyan M, Wu Y, Vadgama JV. The clinical utilization of circulating cell free DNA (CCFDNA) in blood of cancer patients. Int. J. Mol. Sci. 2013;14:18925-18958
8. Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer.Clin. Chem. 2015;61:112-123
9. Yoshioka Y, Kosaka N, Konishi Y, Ohta H, Okamoto H, Sonoda H, Nonaka R, Yamamoto H, Ishii H, Mori M, Furuta K, Nakajima T, Hayashi H, Sugisaki H, Higashimoto H, Kato T, Takeshita F, Ochiya T. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat. Commun. 2014;5:3591
10. Benesova L, Belsanova B, Suchanek S, Kopeckova M, Minarikova P, Lipska L, Levy M, Visokai V, Zavoral M, Minariket M. Mutation-based detection and monitoring of cell-free tumor DNA in peripheral blood of cancer patients. Anal. Biochem. 2013;433:227-234
11. McLarty JL, Yhf CH. Circulating Cell-Free DNA: The Blood Biopsy in Cancer Management. Cell. Sci. Rep. 2015;2:00021
12. Speicher M, Pantel K. Tumor signatures in the blood. Nat. Biotechnol. 2014;32:441-443
13. Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer. 2011;11:426-437
14. Spellman PT, Gray JW. Detecting cancer by monitoring circulating tumor DNA. Nat. Med. 2014;20:474-475
15. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, Giuntoli RL, Goggins M, Hogarty MD, Holdhoff M, Hong SM, Jiao Y, Juhl HH, Kim JJ, Siravegna G, Laheru DA, Lauricella C, Lim M, Lipson EJ, Marie SKN, Netto GJ, Oliner KS, Olivi A, Olsson L, Riggins GJ, Bianchi AS, Schmidt K, Shih le-M, Shinjo SMO, Siena S, Theodorescu D, Tie J, Harkins TT, Veronese S, Wang TL, Weingart JD, Wolfgang CL, Wood LD, Xing D, Hruban RH, Wu J, Allen PJ, Schmidt CM, Choti MA, Velculescu VE, Kinzler KW, Vogelstein B, Papadopoulos N, Diaz LA Jr. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014;6:224ra24
16. Board RE, Williams VS, Knight L, Shaw J, Greystoke A, Ranson M, Dive C, Blackhall FH, Hughes A. Isolation and extraction of circulating tumor DNA from patients with small cell lung cancer. Ann. N. Y. Acad. Sci. 2008;1137:98-107
17. Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, Shrager JB, Loo BW Jr, Alizadeh AA, Diehn M. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014;20:548-554
18. Taniguchi K, Uchida J, Nishino K, Kumagai T, Okuyama T, Okami J, Higashiyama M, Kodama K, Imamura F, Kato K. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin. Cancer Res. 2011;17:7808-7815
19. Jeon S, Moon J, Lee E, Kim Y, Cho NY. An electroactive biotin-doped polypyrrole substrate that immobilizes and releases EpCAM-positive cancer cells. Angew. Chem. Int. Ed. 2014;53:4597-4602
20. Hong WY, Jeon S, Lee E, Cho YN. An integrated multifunctional platform based on biotin-doped conducting polymer nanowires for cell capture, release, and electrochemical sensing. Biomaterials. 2014;35:9573-9580
21. Jeon S, Hong WY, Lee E, Cho YN. High-purity isolation and recovery of circulating tumor cells using conducting polymer-deposited microfluidic device. Theranostics. 2014;4:1123-1132
22. Pernaut JM, Reynolds JR. Use of Conducting Electroactive Polymers for Drug Delivery and Sensing of Bioactive Molecules. A Redox Chemistry Approach. J Phys Chem. B. 2000;104:4080-4090
23. Wallace G, Spinks G. Conducting polymers - bridging the bionic interface. Soft Matter. 2007;3:665-671
24. Otero TF, Martinez JG. Biomimetic intracellular matrix (ICM) materials, properties and functions. Full integration of actuators and sensors. J Mater Chem. B. 2013;1:26-38
25. Dai Y, Blaisten-Barojas E. Energetics, structure, and charge distribution of reduced and oxidized n-pyrrole oligomers: a density functional approach. J. Chem. Phys. 2008;129:164903
26. Tsai YT, Choi CH, Gao N, Yang EH. Tunable wetting mechanism of polypyrrole surfaces and low-voltage droplet manipulation via redox. Langmuir. 2011;27:4249-4256
27. Xiaohua J, Xiangqin L. Overoxidized polypyrrole film directed DNA immobilization for construction of electrochemical micro-biosensors and simultaneous determination of serotonin and dopamine. Anal. Chim. Acta. 2005;537:145-154
28. Beck F, Braun O, Oberst M. Organic Electrochemistry in the Solid State-Overoxidation of Polypyrrole. Phys. Chem. 1987;91:967-974
29. Schlenoff JB, Xu H. Evolution of Physical and Electrochemical Properties of Polypyrrole during Extended Oxidation. J. Electrochem. Soc. 1992;139:2397-2401
30. Lewis TW, Wallace GG, Kim CY, Kim DY. Studies of the overoxidation of polypyrrole. Synthetic Met. 1997;84:403-404
31. Fermin DJ, Teruel H, Scharifker BR. Changes in the population of neutral species and charge carriers during electrochemical oxidation of polypyrrole. J. Electroanal. Chem. 1996;401:207-214
32. Page K, Hava N, Ward B, Brown J, Guttery DS, Ruangpratheep C, Blighe K, Sharma A, Walker RA, Coombes RC, Shaw JA. Detection of HER2 amplification in circulating free DNA in patients with breast cancer. Br. J. Cancer. 2011;104:1342-1348
33. Newman AM, Bratman SV, To J, Wynne J, Eclov NCW, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, Shrager JB, Loo BW Jr, Alizadeh AA, Diehn M. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014;20:548-556
34. Thress KS, Paweletz CP, Felip E, Cho BC, Stetson D, Dougherty B, Lai Z, Markovets A, Vivancos A, Kuang Y, Ercan D, Matthews SE, Cantarini M, Barrett JC, Jänne PA, Oxnard GR. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat. Med. 2015;21:560-564
35. Lebofsky R, Decraene C, Bernard V, Kamal M, Blin A, Leroy Q, Rio Frio T, Pierron G, Callens C, Bieche I, Saliou A, Madic J, Rouleau E, Bidard FC, Lantz O, Stern MH, Le Tourneau C, Pierga JY. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol. Oncol. 2015;9:783-790
Author contact
![]() Corresponding author: ynchore.kr
Corresponding author: ynchore.kr
 Global reach, higher impact
Global reach, higher impact