13.3
Impact Factor
Theranostics 2015; 5(4):399-417. doi:10.7150/thno.10351 This issue Cite
Research Paper
Targeted Antiangiogenesis Gene Therapy Using Targeted Cationic Microbubbles Conjugated with CD105 Antibody Compared with Untargeted Cationic and Neutral Microbubbles
1. Chongqing Key Laboratory of Ultrasound Molecular Imaging, Institute of Ultrasound Imaging, The Second Affiliated Hospital of Chongqing Medical University, Chongqing 400010, P. R. China.
2. Department of Ultrasound, The Children's Hospital of Chongqing Medical University, Chongqing 400014, P. R. China.
3. Department of Gastrointestinal & Anorectal Surgery, The Second Affiliated Hospital of Chongqing Medical University, Chongqing 400010, P. R. China.
4. Department of Endocrine & Breast Surgery, The First Affiliated Hospital of Chongqing Medical University, Chongqing 400016, P. R. China.
5. Department of Otolaryngology, The Third People's Hospital of Chongqing, Chongqing 400014, P. R. China.
6. Department of Ophthalmology, The Second Affiliated Hospital of Chongqing Medical University, Chongqing 400010, P. R. China.
7. Chongqing Jinxin Advertisement Company, Chongqing Daily News Group, Chongqing 400012, P. R. China.
Received 2014-8-17; Accepted 2015-1-2; Published 2015-2-1
Abstract
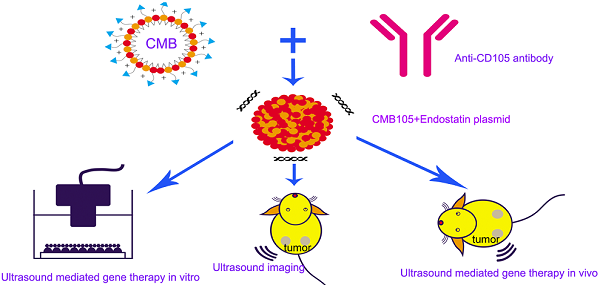
Objective This study aimed to develop targeted cationic microbubbles conjugated with a CD105 antibody (CMB105) for use in targeted vascular endothelial cell gene therapy and ultrasound imaging. We compared the results with untargeted cationic microbubbles (CMB) and neutral microbubbles (NMB).
Methods CMB105 were prepared and compared with untargeted CMB and NMB. First, the microbubbles were characterized in terms of size, zeta-potential, antibody binding ability and plasmid DNA loading capacity. A tumor model of subcutaneous breast cancer in nude mice was used for our experiments. The ability of different types of microbubbles to target HUVECs in vitro and tumor neovascularization in vivo was measured. The endostatin gene was selected for its outstanding antiangiogenesis effect. For in vitro experiments, the transfection efficiency and cell cycle were analyzed using flow cytometry, and the transcription and expression of endostatin were measured by qPCR and Western blotting, respectively. Vascular tube cavity formation and tumor cell invasion were used to evaluate the antiangiogenesis gene therapy efficiency in vitro. Tumors were exposed to ultrasound irradiation with different types of microbubbles, and the gene therapy effects were investigated by detecting apoptosis induction and changes in tumor volume.
Results CMB105 and CMB differed significantly from NMB in terms of zeta-potential, and the DNA loading capacities were 16.76±1.75 μg, 18.21±1.22 μg, and 0.48±0.04 μg per 5×108 microbubbles, respectively. The charge coupling of plasmid DNA to CMB105 was not affected by the presence of the CD105 antibody. Both CMB105 and CMB could target to HUVECs in vitro, whereas only CMB105 could target to tumor neovascularization in vivo. In in vitro experiments, the transfection efficiency of CMB105 was 24.7-fold higher than the transfection efficiency of NMB and 1.47-fold higher than the transfection efficiency of CMB (P<0.05). With ultrasound-targeted microbubble destruction (UTMD)-mediated gene therapy, the transcription and expression of endostatin were the highest in the CMB105 group (P<0.001); the antiangiogenesis effect and inhibition of tumor cells invasion was better with CMB105 than CMB or NMB in vitro (P<0.01). After gene therapy, the tumor volumes of CMB105 group were significantly smaller than that of CMB and NMB, and many tumor cells had begun apoptosis in the CMB105 group, which had the highest apoptosis index (P<0.001).
Conclusions As a contrast agent and plasmid carrier, CMB105 can be used not only for targeted ultrasound imaging but also for targeted gene therapy both in vitro and in vivo. The plasmid DNA binding ability of the CMB was not affected by conjugation of the CMB with the CD105 antibody, and because of its targeting ability, the gene transfection efficiency and therapeutic effect were better compared with the untargeted CMB and NMB. The advantages of targeted gene therapy with CMB105 in vivo were more prominent than with CMB or NMB because neither can target the endothelia in vivo.
Keywords: Ultrasound-mediated gene delivery (UMGD), Antiangiogenesis, Target, Cationic microbubbles
Introduction
Gene therapy offers an effective method to prevent and treat many refractory diseases; however, this method cannot currently be used in clinical therapy. Effective gene therapy requires high gene transfection efficiency and expression. Viral-mediated gene therapy has shown high gene transfer efficiency; however, its toxicity and immunity limit its application in clinical therapy (1). To overcome the problem of security, other physical and chemical methods have been reported to enhance gene transfection efficiency; one important method is ultrasound targeted microbubble destruction (UTMD)-mediated gene therapy. In 1996, Porter demonstrated the possibility of transferring DNA using ultrasound with microbubbles (2); since that time, this method has attracted the attention of many researchers. However, the primary problem of this method is that its low transfection efficiency limits its use; thus, most researchers have focused on how to improve the gene transfection efficiency.
In the process of UTMD-mediated gene therapy, microbubbles have always served as exogenous cavitation nuclei. They reduce the ultrasound energy threshold necessary for sonoporation to occur (3, 4) and can also serve as vectors. Ordinary microbubbles carry either a net neutral or slightly negative surface charge, which Nikolitsa et al (5) called neutral microbubbles (NMB) based on their surface potential characterization. This type of microbubble minimizes interactions with cellular or molecular components in plasma (6) because both nucleic acids and the cell surface are negatively charged. For use as a vector, it is better for the microbubbles to carry a positive surface charge, which could enhance their interactions with negatively charged nucleic acids and cells. Therefore, cationic microbubbles (CMB) were developed, and several studies have reported that the use of CMB could increase the gene carrying capacity and clearly enhance gene transfer. Wang et al (7) reported that when using CMB, the mean CBLuc expression was 20-fold higher than when using NMB. Sun et al (8) reported that CMB could bind 70% more plasmid DNA than the commercial definity microbubbles, and when delivering the therapeutic AKT gene to cure ischemic rat myocardium, the therapeutic effect was better compared with definity microbubbles. Cationic lipids have been reported to have cell toxicity, but many scholars have not reported that cationic microbubbles were toxiferous to cells (5, 6, 7, 8).
In ultrasound-mediated gene therapy, CMB can be used not only as a carrier to improve the gene carrying capacity but also to protect the gene from DNase degradation in serum. Due to the positively charged surfaces of CMB, the local gene concentration is increased at the therapy sites (7). Another method for increasing the local gene concentration is to produce microbubbles that target aggregations. This approach is not novel, and many studies have previously reported the various uses of this technique. For this technique, microbubbles are linked with special antibodies or ligands that bind to disease-associated molecular markers expressed on endothelial cells (9-11). Certain common endothelial markers, such as VEGFR2, endothelial adhesion molecules (P-selectin or intercellular adhesion molecule [ICAM]-1), and CD105 (endoglin), and several types of microbubbles targeted to those markers have been reported (12-14). These targeted microbubbles could aggregate to vessels that express a related antigen and could be used to detect small tumors at an earlier stage, evaluate the response to antiangiogenic therapy, and assess the malignancy and invasive risk.
Due to the advantages of CMB and targeted microbubbles, we attempted to develop a new targeted CMB that can not only target the endothelia but also carry plasmids efficaciously to achieve targeted therapy. We selected CD105 as a therapeutic target marker because of its security and specificity. This marker is a cell membrane glycoprotein and a specific marker of newly formed blood vessels. CD105 is highly expressed in endothelial cells in breast cancer. In addition, VEGFR2 levels were lower than CD105 in these cells (15). In a murine model bearing breast and colon carcinomas, the CD105 antibody caused no significant side effects (16). In addition, activation of the VEGFR2 signaling pathway causes unwanted adverse effects, such as cardiotoxicity, gastrointestinal perforation, poor wound healing, and hypertension (17, 18).
The endostatin gene was selected for use in antiangiogenesis therapy in our study because of its excellent antiangiogenic ability. In 1999, Feldman et al (19) reported that liposomes complexed to endostatin plasmids could inhibit the growth of human breast cancer implanted in a nude mice model. Calvo et al (20) reported that human endostatin could significantly delay tumor onset, decrease tumor multiplicity and prolong the survival of animals bearing mammary carcinoma. Although endostatin is outstanding for its excellent antiangiogenic ability, it is not used in clinical therapy because of its instability; thus, gene therapy may be the best method to use its advantages in antiangiogenesis.
Materials and methods
Plasmid preparation
A plasmid containing an expression cassette for human endostatin was constructed by GeneCopoeiaTM Co. (Guangzhou, China); the upstream and downstream primers were 5' atcgttcgaaccatggcgccgaggtgcccctggcc 3' and 5' ATCGTGCGGCCGCACTCGAGGTACTTGGAGGCAGTCATGAAGCTGT3', respectively. This sequence was directly amplified by polymerase chain reaction (PCR). The DNA segment and pEZ-M46 vector were digested and ligated to create the recombinant plasmid pEZ-M46-ES. Green fluorescent protein (GFP) was encoded in pEZ-M46-ES as the reporter gene and used to evaluate the efficiency of gene delivery to the HUVECs. The recombinant plasmid is abbreviated as ES-GFP in our paper. Plasmids were grown under kanamycin selection in the host strain DH5α and purified using alkaline lysis and chromatographic methods using an Endo Free kit (Qiagen, Valencia, CA). The concentration of plasmid DNA was determined by spectrophotometry (NanoDrop 2000/2000C, Thermo Scientific, Waltham, MA, USA) and adjusted to 500 ng/μl for the experiment.
Cells culture
The HUVECs and MDA-MB-231 cells (both obtained from ATCC) were maintained in RPMI 1640 medium supplemented with 10% volume of fetal bovine serum at 37℃ in a 5% CO2 humidified atmosphere. All cells were cultured to 70% to 90% confluence before passaging. When used for experiments, HUVECs were cultured with the culture supernatant of MDA-MB-231 cells. Before the microbubble targeting experiment, the HUVECs underwent immunohistochemical tests to ensure that their cell membranes expressed the special target marker CD105. The cell numbers for all the experiments were determined with a hemocytometer.
Tumor model
All animals were treated according to the guidelines for the Care and Use of Laboratory Animals. Female BALB/c nude mice (4 weeks old and weighing 18-20 g) were purchased from Slac Laboratory Animal Co., Ltd (Shanghai, China). For inoculation, MDA-MB-231 tumor cells were resuspended in PBS, and cell suspensions were injected subcutaneously in both hind limbs of 24 nude mice (1×106 cells/100 μl per limb). Xenograft tumors were allowed to grow for 14 days before the experimental studies began, and the tumor volumes were calculated using the following formula: [π/6 x length x (width)2]. The mean tumor volume was approximately 0.44 cm3. The mice were then randomly sorted into 4 groups, and no significant differences were observed in mean tumor volume between these groups (P=0.579). The left hind limbs of each mouse were underwent ultrasound imaging and gene therapy.
CMB105, Control-CMB105, CMB, and NMB preparation and characterization
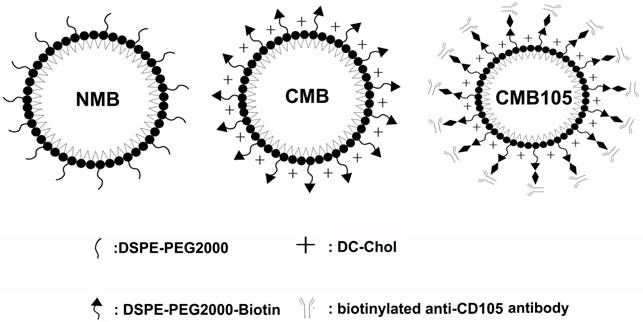
CMB and NMB were prepared by sonication of the relevant lipid mixtures in the presence of octafluoropropane (C3F8) as described by Sun et al (8). The lipid shell compositions of the microbubbles are summarized in Table 1. Diagrams of each type of microbubble are shown in Figure 1. After preparation, PBS was used to adjust the concentration of the microbubbles to 1×109 microbubbles/ml.
Next, 0.3 ml of CMB (approximately 3.0×108 microbubbles) was combined with streptavidin (90 μg, Sigma) for 30 min at 4℃, centrifuged at 400 g×4 min and washed with PBS twice. Then, either 20 μg of biotinylated CD105 antibody or a biotinylated immunoglobulin G (IgG) isotype control (both purchased from Abcam, Cambridge, UK) was added per vial of CMB to produce either a CD105-targeted or a control CMB.
Schematics of NMB, CMB and CMB105. The three types of microbubbles all consisted of a lipid shell surrounding a C3F8 gas core. The lipid shell of the NMB in this study consisted of DPPC and DSPE-PEG2000, whereas the shell of CMB and CMB105 consisted of DPPC, DSPE-PEG2000-Biotin and DC-Chol. DSPE-PEG2000-Biotin was used to increase the stability of the CMB, and DC-Chol was used to positively charge the surface. CMB105 were constructed through the conjugation of a biotinylated CD105 antibody with CMB using a biotin-avidin reaction.
All four types of microbubbles were stable in suspension for up to one month at 4℃. The size distribution and zeta-potential were analyzed in optiMEM using a laser particle size analyzer system (Zeta SIZER 3000 HS; Malvern, USA).
Percent molar ration of lipid-based components of CMB105, CMB and NMB
| Microbubbles | Surface nature | DPPC | DSPE-PEG2000-Biotin | DSPE-PEG2000 | Dc-Chol |
|---|---|---|---|---|---|
| NMB | Neutral | 5 | 0 | 2 | 0 |
| CMB | Biotinylated cationic | 5 | 2 | 0 | 0.5 |
| CMB105 | Biotinylated cationic | 5 | 2 | 0 | 0.5 |
DPPC: Dipalmitoyl phosphatidylcholine; DSPE-PEG2000-Biotin: 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[biotinyl (polyethylene glycol)-2000]; DSPE-PEG2000: 1,2-dipalmitoyl-sn-glycero-3-phosphoethanol-amineN-[methoxy(polyethylene glycol)-2000]; Dc-Chol: 3-(N-(N',N'-Dimethylaminoethane) carbamoyl)cholesterol. All of these chemicals were obtained from Avanti Polar Lipids, Alabaster, AL.
Assessment of antibody binding to microbubbles
To assess whether the antibody combined with the CMB successfully, a secondary antibody labeled by FITC was combined with the CMB105 or Control-CMB105 (C-CMB105) using the same steps described for the first antibody. Confocal fluorescence microscopy (Zeiss, German) was used to obtain the images.
Assessment of microbubbles targeting an in vitro cell monolayer
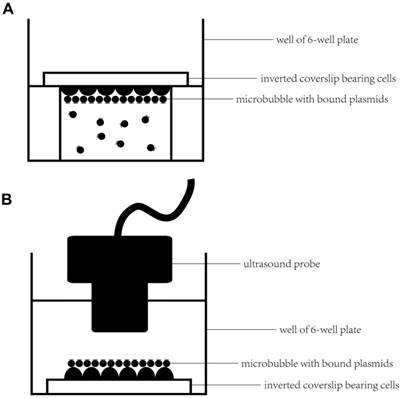
An inverted method was selected to assess the microbubbles targeting the cell monolayer in vitro (21, 22). In brief, HUVECs were grown on 4.84 cm2 square coverslips (22×22 mm), which were placed in a 6-well plate in advance. When cells were cultured to approximately 70% confluence, the inverted method (Figure 2A) was selected to promote microbubbles binding to HUVECs. First, an aseptic plastic ring was inserted in one well of a 6-well plate, and then, approximately 4 ml of optiMEM was added to fill the well to slightly above the top of the ring. Next, 100 μl of diluted microbubbles (approximately 5×107 microbubbles) was added to the medium. Five minutes later, when nearly all of the microbubbles had floated to the top of the medium, a coverslip of HUVECs was placed on the platform with the cells facing the microbubbles to ensure that the cells were in contact with the microbubbles. After 15 min at RT, the coverslips were gently washed twice with PBS and examined under a microscope using a 20× or 40× objective.
Blocking studies were performed by preincubating the cells bearing coverslips with the anti-CD105 antibody (30 μg/ml) or the isotype control antibody IgG (30 μg/ml) for 10 min; then, the CMB105 were incubated with HUVECs as described above.
Images were acquired from five random fields using a microscope. The number of bound microbubbles in each field was determined using the Image Plus software.
Assessment of microbubbles targeting endothelial cells in vivo
1. Ultrasound imaging
Both healthy and tumor-bearing nude mice were anesthetized with 1% pentobarbital and placed in a heated environment to maintain their body temperature at 37℃. Then, 100 μl (approximately 5×107) of microbubbles was injected into the tail vein within 2 s, followed by 200 μl of physiological saline. Contrast-enhanced ultrasound imaging was performed using a MyLab 70 (Esaote, MyLab70, X-vision, Italy) system in the CnTI mode; a linear transducer with a transmission frequency of 10 MHz, a mechanical index (MI) of 0.08 and a depth of 20 mm were used, and all mice were imaged with the same instrument under the conditions by a professional operator. Conventional ultrasound imaging using NMB, untargeted CMB, C-CMB105 and CMB105 were performed in healthy mice first to obtain their circulation characteristics. The transducer was placed on side of the mice to ensure that the heart and liver could be depicted. Characteristics in blood were obtained by assessments at the heart level.
Before ultrasound imaging, immunohistochemistry of neovascular endothelial cells in tumor was performed (21, 22) to assess CD105 expression.
Experimental setup for microbubbles targeted to the cell monolayer and ultrasound-mediated gene delivery in vitro. (A) First, an aseptic plastic ring was inserted in one well of a 6-well plate; a coverslip bearing HUVECs was placed on the platform with the cells facing the microbubbles to ensure that the cells were in contact with the microbubbles for 15 min at RT. (B) A coverslip with HUVECs and bound microbubbles was placed upright in the well of the 6-well plate after gently washing the coverslip twice with PBS. Serum-free optiMEM was added to the well; then, the targeted bound microbubbles were observed under a microscope or ultrasound-mediated gene transfer was performed by submerging the sterile ultrasound probe into the medium.
2. Molecular ultrasound imaging of tumor xenografts
After a rapid bolus injection of microbubbles and a 10-minute period to enable the accumulation of microbubbles at the target site and clearance of unbound microbubbles, the ultrasound parameters used was in the same manner as described above. A 10-s cine-loop imaging sequence was acquired, followed by a destructive pulse (MI=3.0, transmission frequency=10 MHz) to destroy all the microbubbles within the slice; 30 seconds later, a second 10-s cine-loop imaging sequence was acquired. The signal intensity before the destructive pulse represented the echoes from both the targeted binding microbubbles and the circulating microbubbles, and the signal intensity after the destructive pulse represented the echoes from the vascular replenishment of residual circulating microbubbles. Thus, the former intensity subtracted from the latter intensity represented the relative amount of targeted binding microbubbles (13, 23). All animals recovered after imaging, and no acute toxicity was been observed with these microbubbles.
3. In vivo competition experiment and imaging
For the competition experiment, 150 μg of CD105 antibody was injected into the tail vein; 10 min later, CMB105 were injected to obtain a contrast ultrasound image with the same parameters as described above. In total, 6 tumor-bearing nude mice were used in this test.
4. Image analysis
The images were analyzed using self-made DFY-II software designed by Professor Yuan-Y Zheng at our research institute. Through many studies, this software to been shown to be able to precisely analyze the signal intensity of ultrasound imaging (24, 25). The regions of interest were defined, consisting the entire tumor at maximum; we were then able to obtain the time-intensity curves and calculate the signal intensity of the bound microbubbles.
Assessment of plasmids binding to microbubbles
Through assessing the ability of microbubbles to target endothelial cells in vitro and in vivo, we found that CMB105 had the ability to target to endothelial cells in vivo, whereas C-CMB105 could not target endothelia; thus, C-CMB105 were not used in our further plasmid binding and gene transfer experiments.
Evaluations of the amount of plasmid DNA that can bind to each type of microbubble were performed as described by Panje et al (21), with some variations: 5×108 NMB, CMB or CMB105 were incubated with varying doses of ES-GFP plasmids (10, 20, 40, and 80 μg) in optiMEM for 15 min to allow spontaneous electrostatic charge-coupling between the DNA and microbubbles. Then, the samples were centrifuged for 4 min at 400 g to form two phases: the upper layer appeared milky white and contained microbubbles with bound DNA, and the lower clear layer contained unbound DNA. The lower clear layer was collected and centrifuged for 5 min at 14,000 g using 0.45-μm centrifugation filters to remove MB shell contaminants. The concentration of unbound DNA was determined using spectrophotometry (NanoDrop 2000/2000C, Thermo Scientific, Waltham, MA, USA), and by multiplying by the total sample volume, we obtained the total amount of unbound DNA. The amount of bound DNA was calculated by subtracting the amount of unbound DNA from the total amount of DNA added initially. By increasing the amount of initial DNA, the maximum amount of bound DNA was determined and regarded as the loading capacity for 5×108 microbubbles. All experiments were performed in triplicate.
The DNA-microbubble interaction was also demonstrated by laser confocal fluorescence microscopy. Ten micrograms of ES-GFP plasmid was added to 5×108 microbubbles, and the first and second steps described above were repeated. Then, the upper milky white layer was collected, incubated with propidium iodide (PI, Sigma, USA) for 15 min in PBS, and centrifuged at 400 g×4 min to form two phases. The lower layer was discarded, the upper layer was resuspended in PBS, and the previous step was repeated twice. Finally, the samples were diluted and imaged at 1000× magnification using a confocal laser scanning microscope. All experiments were performed in triplicate.
To assess whether binding with DNA affected the ability of the target to attach to HUVECs, three types of microbubbles were first conjugated with DNA and then incubated with HUVECs as described before.
Assessment of HUVEC membrane permeability and viability after UTMD with NMB, CMB or CMB105
First, we selected the inverted method (Figure 2A) to target the microbubbles to HUVECs; the details were the same as described previously. After washing the unbound microbubbles, coverslips bearing HUVECs with bound microbubbles were placed in the wells of a 6-well plate upright (Figure 2B), 6 ml of serum-free optiMEM was added into the well, and a sterile ultrasound probe was submerged in the optiMEM; the distance between the ultrasound probe and HUVECs was approximately 1 cm. After preparation, ultrasound was applied at 1 MHz, 1 W/cm2, and 50% duty cycle (DC) for 30 s using the ultrasound gene transfer machine (UGT 1025, CQMU, Chongqing, China), which was produced by our own institute (Ultrasonographic Image Research Institute of Chongqing Medical University, China) and used for gene transfer in vitro and in vivo; the diameter of the probe was approximately 2 cm.
After sonication, the effects on permeabilization of the plasma membrane and cell proliferation were measured as we previously reported (26, 27). In brief, the permeabilization was detected by staining with 3,6-diacetoxyfluoran Di-O-acetylfluorescein (FDA, Sigma, USA) and PI; proliferation was detected using a 3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyl-tetrazolium bromide (MTT) assay.
UTMD-mediated gene transfer in vitro
Forty micrograms of ES-GFP plasmid was added to 5×108 microbubbles (NMB, CMB, and CMB105, respectively); the procedure of plasmid binding to microbubbles was the same as described before. Both inverted and upright set-ups (Figure 2A and 2B) were used to ensure that the microbubbles in the transfer step were bound to HUVECs. Assessments of the effect of targeted gene transfer were performed as described in the “Assessment of HUVEC membrane permeability and viability” section. Then, 5×107 microbubbles of NMB, CMB, or CMB105 was added into each well; after binding with the ES-GFP plasmid, ultrasound (UGT 1025, CQMU, Chongqing, China) was applied at 1 MHz, 1 W/cm2, and 50% duty cycle (DC) for 30 seconds (28). Within the transfer procedure, serum-free optiMEM was used, and the plates were then placed in a humidified 5% CO2 atmosphere at 37℃ for 2 h. The plates were then washed once with RPMI 1640 serum-free medium, and finally, 2 ml of RPMI 1640 medium supplemented with 10% volume of fetal bovine serum was added to each well (5). The plates were then incubated in a humidified 5% CO2 atmosphere before analysis for gene expression.
Flow cytometry
Transfection efficiency analysis
After transfection for 24 h or 48 h, both the control and treated HUVECs were collected to measure the expression of the plasmid-encoded GFP gene. Flow cytometry (FACS-Calibur; Becton Dickinson, San Jose, CA, USA) with the excitation setting at 488 nm was used to analyze the transfection efficiency. At least 10,000 cells were acquired for each test measurement. All experiments were performed in triplicate.
Cell cycle analysis
Both the control and treated HUVECs were collected before being transferred or were collected 24 h, 48 h, or 72 h after being transferred. In brief, the HUVECs were digested with 0.25% trypsin and washed 3 times in 1 ml of pre-cooled PBS; then, 1 ml of pre-cooled 70% ethanol was added to the cell pellet, and the cells were fixed overnight at 4℃. The following day, the cells were collected by centrifugation and washed twice with PBS; finally, the cells were resuspended in 0.5 ml of PI solution (50 mg/ml PI, 100 mg/ml RNase A and 0.2% Triton X-100). The cells were then incubated at 4℃ for 30 min in the dark, and flow cytometry (FACS-Calibur; Becton Dickinson, San Jose, CA, USA) was used to measure the cell cycle distribution. At least 10,000 cells were acquired for each test measurement. All experiments were performed in triplicate.
Quantitative real-time polymerase chain reaction (qPCR)
The total RNA was extracted by phenol-chloroform extraction and ethanol precipitation using TRIZOL (Invitrogen, Life Technologies, Gaithersburg, MD, USA). The integrity of the RNA was determined using 1.5% agarose gels containing ethidium bromide. The reverse transcription to cDNA was performed using the Takara PrimeScript RT Reagent kit (AMV; Takara, Tokyo, Japan) with Bio-Rad iQ™ 5. The reaction conditions were as follows: 25℃ for 10 min, 42℃ for 50 min, and 85℃ for 5 min. The qPCR amplification conditions were as follows: 94℃ for 4 min; 35 cycles of 94℃ for 20 sec, 60℃ for 30 sec, and 72℃ for 30 sec; finally, the signal was measured at 72℃. The primer sequences are shown in Table 2.
The primer sequences of endostatin, VEGF and caspase-3 detected by qPCR
| Detected gene | Primer sequences | |
|---|---|---|
| COL18A1 Endostatin | hCOL18A1F | GATTTGGCATGAGGGGAATG |
| hCOL18A1R | AAGAAAGTCAAACGGAAACTGC | |
| VEGF | hVEGFF | AATCGAGACCCTGGTGGACA |
| hVEGFR | TGTTGGACTCCTCAGTGGGC | |
| Caspase-3 | hcasepase3F | AGAACTGGACTGTGGCATTGAG |
| hcasepase3R | GCACAAAGCGACTGGATGAA | |
Western blot analysis
Both the control and treated HUVECs were collected 48 h after being transferred. Briefly, the proteins were separated by SDS-PAGE on 12% (w/v) polyacrylamide gels and electrotransferred onto Millipore polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA). The completion of protein transfer from the gels to the membranes was verified by staining the gels with Coomassie Blue R-250. The membranes were blocked in Tris-buffered saline containing 0.05% Tween 20 and 5% nonfat dry milk for 60 min at room temperature. The blots were then incubated with 1:1000 rabbit-anti-human endostatin antibody (Abcam, Cambridge, UK), rabbit-anti-human VEGF antibody (Abcam, Cambridge, UK) or rabbit-anti-human caspase-3 antibody (Abcam, Cambridge, UK) for 1.5 h at room temperature in blocking solution. Then, the membranes were washed in Tris-buffered saline and incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:3000) in blocking solution for 1.5 h. The blots were examined using a chemiluminescent substrate (Santa Cruz Biotechnology, Santa Cruz, CA, USA) according to the manufacturer's instructions. The protein bands were normalized to GAPDH, and all blots were quantified using the software Quantity One (Bio-Rad, Hercules, CA, USA).
Effect on angiogenesis in vitro
The 96-well plate, Matrigel, and serum-free RPMI-1640 culture medium were first pre-cooled; then, the Matrigel was diluted by the serum-free RPMI-1640 culture medium (1:2). Next, 50 μl of the diluted Matrigel was added to each well and incubated at 37℃ for 2 h. Then, 24 h after transfection with different types of microbubbles, the control and transferred HUVECs were resuspended in RPMI 1640 medium supplemented with 10% volume of fetal bovine serum and incubated at 37℃ in a 5% CO2 humidified atmosphere for 8 h; approximately 0.2×104 cells were added to each well. Then tubular structures were examined under a microscope using 10× or 20× objectives. Images were acquired from five random fields, and the number of tubular structures in each field was determined by direct counting. Each treatment was performed in triplicate.
Effect on the invasion ability of MDA-MB-231 cell in vitro
The 24-well plate, Matrigel, and serum-free RPMI-1640 culture medium were first pre-cooled; then, the Matrigel was diluted as described previously. Then, 100 μl of the diluted Matrigel was added to the upper chamber of the 24-well transwell and incubated at 37℃ for at least 4 h for gelling. The gelled Matrigel was then gently washed with warm serum-free RPMI-1640 culture medium, and the MDA-MB-231 cells were harvested using 0.25% trypsin. The cells were washed 3 times with RPMI-1640 medium containing 1% volume of FBS, and the cells were resuspended in medium containing 1% volume of FBS at a density of 5×105 cells/ml. Next, 200 μl of the cell suspension was placed into the Matrigel, and the lower chamber of the transwell was filled with 600 μl of culture medium from the control or treated HUVECs after being transferred 24 h with the 3 different types of microbubbles. The MDA-MB-231 cells were incubated at 37℃ for 24 h, and the noninvasive cells on top of the transwell were scraped off with a cotton swab. The transwells were removed from the 24-well plates, stained with 0.1% crystal violet at 37℃ for 30 min and examined under an inverted microscope using 20× or 40× objectives. Images were acquired from five random fields, and the number of invasive cells in each field was determined using Image Plus software. Each treatment was performed in triplicate.
For in vivo UTMD-mediated gene transfer
For in vivo UTMD-mediated gene transfer to the left hind limb tumor xenografts, mice were prepared as described before, and the right hind limb tumor xenografts were used as control. First, 400 μg of ES-GFP was incubated with 5×109 microbubbles (NMB, CMB, and CMB105, respectively); the plasmid binding and centrifuging were performed as described before. Then, the concentration of different types of microbubbles with plasmid binding was adjusted to 5×108 microbubbles/ml; 500 μl microbubbles-plasmid solutions were administered intravenously through tail vein within 30 seconds per mouse, and 10 min after injection, ultrasound-mediated gene transfer was performed. Ultrasound (UGT 1025, CQMU, Chongqing, China) was applied at 1 MHz, 2 W/cm2, and 50% duty cycle (DC) for 30 seconds (29). The probe was positioned directly on top of the tumor xenografts with ultrasonographic gel in between. The size of tumor xenografts was recorded every 3 days. The tumor growth curve was delineated, and the tumor growth inhibition rate was calculated as follows: inhibition rate = (mean tumor volume control - mean tumor volume treatment)/mean tumor volume control × 100. Fifteen days after transfer, the mice were sacrificed for proliferating cell nuclear antigen (PCNA) and terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling assay (TUNEL) immunohistochemisty staining (25, 30) to evaluate the gene therapy effect.
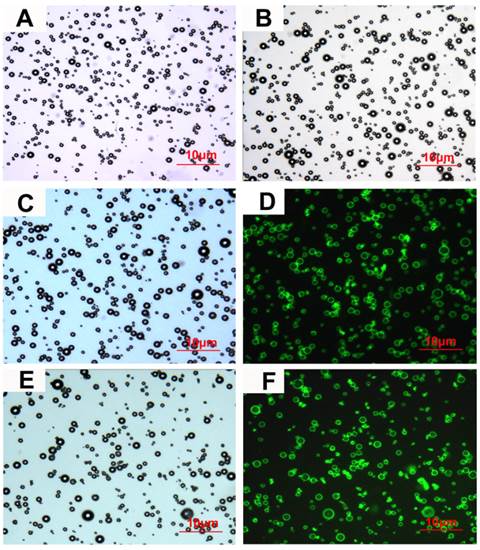
Characterization of microbubbles under microscope. (A), (B), (C) and (E) Bright-field images of NMB, CMB, CMB105 and C-CMB105, respectively. (D) and (F) Successful binding of the biotinylated CD105 antibody and isotype control (IgG) antibody, respectively, assessed using laser confocal microscopy (excited at 488 nm, emitted at 583 nm). The images were all acquired at 1000x magnification.
Statistical analysis
All the data are expressed as the mean ± standard deviation and were analyzed using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). Differences between means with P<0.05 were considered significantly different. Comparisons between groups were performed using a one-way ANOVA. The least significant difference (LSD) and paired t-test with two-sided were used for further comparisons.
Results
Characterization of microbubbles
The characteristics of the different types of microbubbles are listed in Table 3. No significant differences were observed in the mean diameters or concentrations between NMB, CMB and CMB105; however, the zeta-potentials of CMB (26.44±2.13 mV) and CMB105 (26.62±2.48 mV) were significantly higher than for NMB (-2.38±0.56 mV; P<0.001), and no significant difference was observed between CMB and CMB105 (P>0.05). Inverted microscopy images of NMB, CMB, CMB105 and C-CMB105 are shown in Figures 3A, 3B, 3C and 3E, respectively. The conjugation between the antibody and microbubbles was confirmed visually by laser confocal microscopy (Figures 3D and 3F), demonstrating the successful binding of CD105 antibody and isotype control (IgG) antibody, respectively.
Summary of microbubble characteristics
| Zeta-potential (mV) | Diameter (µm) | Concentration (×109/ml) | |
|---|---|---|---|
| NMB | -2.38±0.56 | 1.42±0.1 | 3.64±0.24 |
| CMB | 26.44±2.13* | 1.42±0.15 | 4.26±0.61 |
| CMB105 | 26.62±2.48* | 1.44±0.21 | 4.13±0.17 |
*P<0.001 relative to NMB, n=3 per group
Microbubble binding to HUVECs in vitro
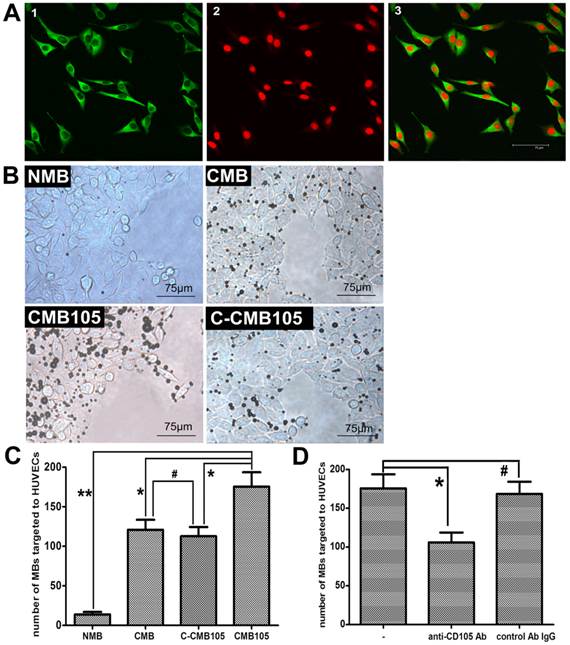
CD105 expression on HUVEC membranes was confirmed by laser confocal microscopy (Figure 4A). High expression of green fluorescence was observed on the HUVEC membranes, confirming CD105 expression. Therefore, HUVECs expressing CD105 were used for the microbubble target binding test, and the results are presented in Figure 4B. For NMB, only a slight non-specificity binding to HUVECs was observed; for CMB, we observed increased amounts of targeted bound microbubbles because of the surface of the target cells usually bears a net negative charge, although the CMB had no targeting ability. Thus, the CMB could attach to HUVECs through electrostatic interaction, which was also reported by Nikolitsa et al (5). For CMB105, more targeted bound microbubbles were observed compared with CMB, C-CMB105 and NMB (Figure 4C, P<0.01 vs. CMB and C-CMB105; P<0.001 vs. NMB). The difference between CMB and C-CMB105 was not significant (P>0.05), indicating that the mechanism of C-CMB105 to target to HUVECs was the same as CMB but through electrostatic interaction, not the reaction between antibody and antigen, and that C-CMB105 did not have target ability, different from CMB105. For blocking studies, HUVECs were pre-incubated with various concentrations of anti-CD105 antibody (Supplementary Material: Figure S1), an obvious inhibition was observed when HUVECs were preincubated with anti-CD105 antibody (30 μg/ml), whereas no inhibition was found when preincubated with isotype control antibody (Figure 4D).
Interaction between microbubbles and HUVECs in vitro. (A) Green fluorescence represents the expression of endoglin on the membrane of HUVECs; red fluorescence represents nuclear staining by PI (40× objective). (B) Different types of microbubbles targeted to HUVECs observed under a microscope (40× objective). (C) Direct counting of microbubbles targeted to HUVECs using Image Plus software. The ability of NMB, CMB and C-CMB105 to target HUVECs was different than CMB105 (*P<0.01, **P<0.001), but the difference between CMB and C-CMB105 was not significant (#P>0.05), n=3 per group. (D) Binding of CMB105 onto HUVECs with and without preincubation of the cells with anti-CD105 antibody or an isotype control antibody IgG (*P<0.01, #P>0.05), n=3 per group.
Microbubbles binding to endothelial cells in vivo
a) Circulation characteristics of NMB, CMB, C-CMB105 and CMB105 in healthy mice
The circulation characteristics of each type of microbubbles are shown in Supplementary Material: Figure S2. For peak enhancement and time-to-peak, no significant differences were observed between these 4 types of microbubbles. The half-life of CMB, C-CMB105 and CMB105 was 8.35±1.27, 7.63±1.61 and 7.27±1.33 min, respectively. The blood levels of these microbubbles decreased rapidly, and the signal intensity in liver was higher than in blood (Figure S2B, S2C, S2D), likely because the reticuloendothelial system had a higher phagocytic of cationic substances. Different from microbubbles bearing cationic potential, the signal intensity in blood levels of NMB did not decrease below 50% of the peak enhancement, and no retention was observed in liver (Figure S2A).
b) Evaluation of targeted bound microbubbles using ultrasound molecular imaging
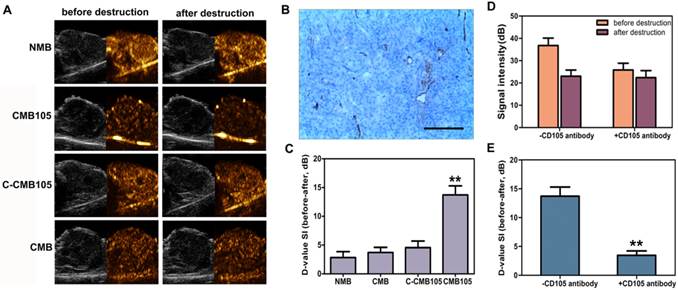
CD105 expression in neovascular endothelial cells in tumor was confirmed by immunohistochemistry (Figure 5B). Typical ultrasound images for NMB, CMB, C-CMB105 and CMB105 before and after a destructive pulse are presented in Figure 5A and were analyzed using self-made DFY-II software as described in Methods. The difference value (D-value) was calculated as the signal intensity (before) minus the signal intensity (after), which represents the amount of targeted bound microbubbles. The D-value of CMB105 (13.71±1.58) was significantly different from the D-value of NMB (2.84±1.03), CMB (3.71±0.89) and C-CMB105 (4.56±1.14, Figure 5C, P<0.001). For in vivo competition experiments, various doses of anti-CD105 antibody was pre-injected (Supplementary Material: Figure S3), and an injection of about 150 μg free anti-CD105 antibody significantly decreased the signal intensity of targeted bound CMB105 (Figure 5D and 5E). These results confirmed that only CMB105 could successfully target neovascular endothelial cells that expressed endoglin.
Plasmid binding ability of microbubbles
The binding of plasmids and microbubbles was also observed by laser confocal microscopy (Figure 6A). Red fluorescence represents successful plasmid binding because only PI could stain the plasmid. After the plasmids were bound, when the microbubbles were excited by laser confocal microscopy at 488 nm, red fluorescence was observed at approximately 630 nm. No red fluorescence was observed in NMB, but it could be observed in CMB and CMB105, indicating that only the CMB could bind plasmids successfully through charge-coupling, and the binding was not altered after the CMB were conjugated with the CD105 antibody.
Evaluation of targeted bound microbubbles in vivo. (A) Ultrasound images using NMB, CMB, C-CMB105 and CMB105 before and after the destructive pulse. (B) CD105 expression in neovascular endothelial cells in tumors was confirmed by immunohistochemistry, bar: 100 μm. (C) D-value of NMB, CMB, C-CMB105 and CMB105. The D-value of CMB105 was highest (**P<0.001). n=6 per group. (D) In vivo competitive binding experiments. Binding of CMB105 with and without pre-injection of CD105 antibody. (E) Mean D-value of CMB105 with and without pre-injection of CD105 antibody. Pre-injection of CD105 antibody led to significantly decreased binding of CMB105 in vivo (**P<0.001). n=6 per group.
Plasmid DNA interactions with NMB, CMB and CMB105. (A) Red fluorescence represents plasmid DNA successfully bound to microbubbles; green fluorescence represents anti-CD105 antibody successfully bound to CMB; and yellow fluorescence represents the superposition of red fluorescence and green fluorescence. For NMB, almost no red fluorescence was observed; for CMB and CMB105, red fluorescence could be observed. (B) Quantitative assessment of DNA binding to NMB, CMB and CMB105 (n=3 per group). The DNA loading capacity of CMB and CMB105 was significantly higher than for NMB, regardless of how many plasmids were added to the microbubble solution (**P<0.0001). The difference in DNA loading capacity between CMB and CMB105 was not significant. *P<0.01 compared with 10, 40, and 80 μg plasmid DNA, within-group comparisons; #P>0.05 compared with 80 μg plasmid DNA, within-group comparisons. (C) Binding with DNA had a slight effect on the ability to target HUVECs for CMB (*P<0.05), whereas no effect was observed for CMB105 and NMB (#P>0.05), n=3 per group.
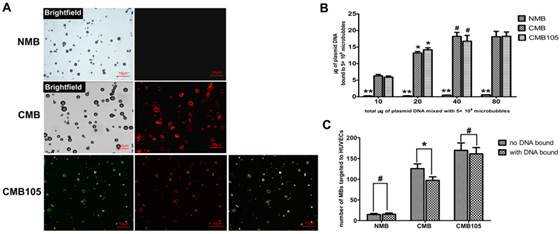
Quantitative assessment of the DNA loading capacity of the different types of microbubbles revealed significantly higher plasmid binding abilities for CMB and CMB105 compared with NMB (Figure 6B, P<0.0001) at every dose. The differences in DNA binding ability between CMB and CMB105 were not significant, which confirmed our previous conclusion that plasmid binding to CMB was not altered after CMB was conjugated with the CD105 antibody. As the amount of added plasmid increased, the amount of plasmid bound to CMB and CMB105 also increased. However, when 40 μg plasmid was added to the microbubble solution, no further increases in the amount of plasmids bound to CMB and CMB105 were observed (P>0.05), indicating that the binding capacity of the microbubbles had reached saturation. Therefore, when 40 μg plasmid was added to the microbubble solution, the amount of plasmid bound to microbubbles was regarded as the saturated DNA loading capacity for 5×108 CMB and CMB105. The saturated loading capacities for CMB and CMB105 were 18.21±1.22 μg and 16.76±1.75 μg, respectively.
Binding with DNA exerted little effect on the interaction between CMB and HUVECs (Figure 6C, P<0.05) and no effects on NMB or CMB105 (Figure 6C, P>0.05).
Evaluation of cell membrane permeability and viability
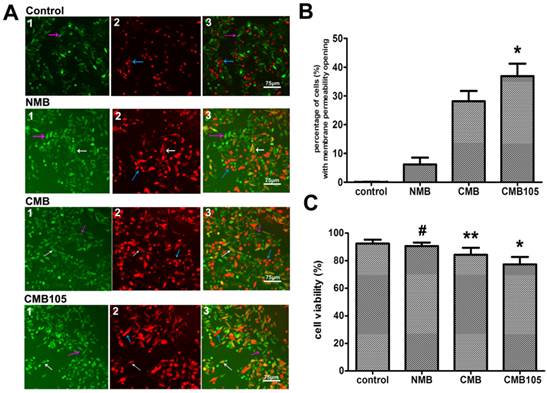
The effect of UTMD on HUVEC membrane permeability is illustrated in Figure 7A. PI and FDA are a common combination in studies of mammalian cell membrane permeability. FDA is a type of lipid molecule without charge that does not express any fluorescence itself but can be enzymatic hydrolyzed to express green fluorescence after free diffusion into cells, where it can be retained due to polarity. After UTMD, the cell membrane permeability increased, and thus, the FDA leaked to the extracellular region and the intracellular fluorescence decreased. If the cell membrane was reversibly open, part of the FDA would remain in the intracellular region, and PI would be allowed to enter into the cells and express red fluorescence; thus, those cells would express yellow fluorescence (white arrow, Figure 7A, “3”) because of the combination of green and red fluorescence. If the cell membrane was irreversibly open, all the FDA would overflow extracellularly, and only the red fluorescence stained by PI could be detected, representing dead cells (blue arrow, Figure 7A, “3”). If permeability of the cell membrane did not change, only FDA staining positive with a bright green fluorescence would be observed (purple arrow, Figure 7A, “3”). Because the experimental procedure was somewhat long and complicated, the cell death rate increased; thus, even in the control group, a few dead cells were observed. Figure 7 shows that the ratio of cells with reversibly open membrane permeability in the CMB105 group (36.89±4.28%) and CMB group (28.12±3.58%) was significantly higher than in the NMB (6.19±2.35%) and control groups (nearly 0); these differences were significant (Figure 7B, P<0.01). The effects of microbubbles and ultrasound on cell viability were evaluated by an MTT assay (Figure 7C); the cell viability measurements of the HUVECs in the CMB and CMB105 groups were lower than the NMB and control groups (P<0.01). Cell viability is affected by many factors, such as the concentration of microbubbles, ultrasound power, and blast time; thus, the use of correct parameters is very important in the cell viability and transfer experiments.
Evaluation of cell membrane permeability and viability. (A) Effect on HUVEC membrane permeability after blasting with NMB, CMB or CMB105. White arrows represent cells with increased membrane permeability, purple arrows represent cells with no change in the permeability, and blue arrows represent dead cells. (B) The percentage of cells with increased membrane permeability was highest after treatment using ultrasound with CMB105 (*P<0.01 compared with control, NMB and CMB). n=3 per group. (C) MTT analysis of cell viability. After ultrasound with CMB or CMB105, the cell viability decreased (*P<0.01 compared with control, NMB and CMB groups, **P<0.01 compared with control, NMB and CMB105, #P>0.05 compared with control). n=3 per group.
UTMD-mediated ES-GFP gene transfection in cell culture. (A) GFP expression in HUVECs observed using a laser confocal microscope 24 h post-transfection (20× objective). (B) Typical flow cytometry images for quantitative analysis of transfection efficiency 24 h and 48 h post-transfection. The percentage of GFP-positive cells was determined as the gene transfection efficiency. (C) Analysis of gene transfection efficiency. The transfection efficiency of CMB105 after 24 h was 24.7-fold higher compared with NMB and 1.47-fold higher compared with CMB (*P<0.01 compared with CMB, **P<0.001 compared with NMB). The difference between 24 h and 48 h in each group was not significant (#P>0.05). n=3 per group.
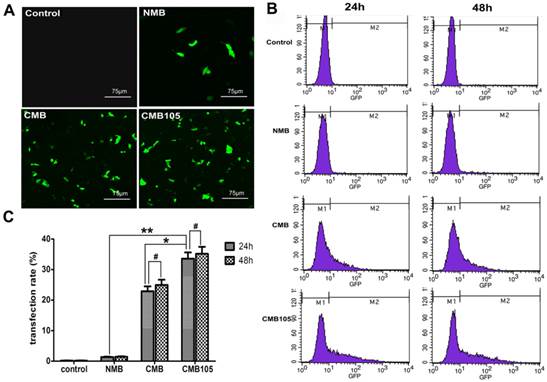
Quantitative assessment of UTMD-mediated gene transfection in cell culture
GFP expression was used to determine the efficiency of UTMD-mediated transfer of ES-GFP plasmid to HUVECs. A laser confocal microscope was used to observe the expression of GFP 24 h post-transfection (Figure 8A). The CMB and CMB105 images show numerous cells expressing GFP. In contrast, the NMB image indicates that few cells were expressing GFP, and nearly no GFP expression can be observed in the control image (Figure 8A). Quantitative analysis of ultrasound-mediated ES-GFP transfection was undertaken through flow cytometry 24 h and 48 h post-transfection, and the percentage of cells that expressed GFP was used to measure the transfection efficiency (Figures 8B and 8C). The transfection efficiency of CMB105 (33.60±2.02%) 24 h post-transfection was 24.7-fold higher than NMB (1.36±0.13%) and 1.47-fold higher than CMB (22.87±1.64%); these differences were significant (P<0.05). The differences in the different types of microbubble transfection after 24 h and 48 h were not significant (P>0.05).
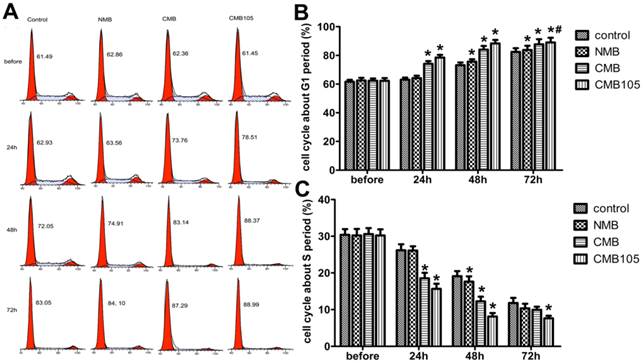
Effect on cell cycle after gene transfection
To evaluate whether the transfection of ES-GFP plasmid would exert a therapeutic effect, the cell cycle was quantitatively analyzed through flow cytometry because endostatin could inhibit the cell cycle of endothelial cells in the G1 phase (Figure 9). The cell cycle of HUVECs in the CMB105 group and CMB group was clearly inhibited in the G1 phase 24 h and 48 h post-transfection (Figure 9B). In addition, the percentage of cells that were inhibited in the G1 phase increased with time; this phenomenon was also observed in the control group due to the contact inhibition, especially 72 h post-transfection. However, even at 72 h, the percentage of cells that were inhibited in the G1 phase of the CMB105 group and CMB group was higher than control and NMB groups (Figure 9B, P<0.05). Cells in the S phase began to decrease in the CMB105 and CMB groups 24 h post-transfection, whereas the number of cells in the S phase in the control and NMB groups did not decrease; the trends concerning the cell cycle change correspond to the change in the G1 phase (Figure 9C).
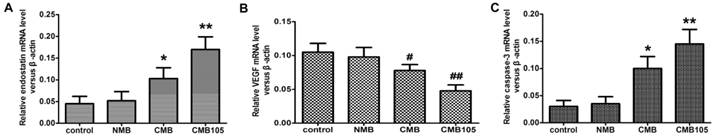
qPCR: transcription of ES-GFP plasmid and effect on VEGF or caspase-3
To evaluate the transcription of the ES-GFP gene after being transfected into HUVECs in vitro, the relative ratio of ES RNA in the treated and control HUVECs was determined through qPCR 48 h post-UTMD-mediated gene transfer (Figure 10). To evaluate the effect of endostatin on VEGF and caspase-3 expression, the relative ratio of VEGF and caspase-3 RNA was also determined (Figure 10). After being transferred with the 3 different types of microbubbles, the transcription level of the ES gene was highest for CMB105 (P<0.01 vs. CMB, NMB and control), followed by CMB (P<0.01 vs. CMB105, NMB and control), and no difference between NMB and the control was observed (P>0.05). With the increasing level of ES RNA, the VEGF RNA level decreased, whereas the caspase-3 RNA level increased (Figure 10), which may be due to the therapeutic effect induced by endostatin.
ES gene transfer induces cell cycle inhibition in the G1 phase. (A) DNA content flow cytometric profiles after transfection with ES gene by ultrasound with NMB, CMB and CMB105. (B) Analysis of the cell cycle in the G1 phase. The cell cycle was clearly inhibited in the G1 phase in the CMB105 and CMB groups transfected for 24 h. Comparisons between groups were performed simultaneously (*P<0.01 relative compared with control, #P>0.05 compared with CMB). n=3 per group. (C) Analysis of the cell cycle in the S phase. The percentage of HUVECs in the S phase was significantly decreased in the CMB and CMB105 groups after transfection for 24 h (*P<0.01 compared with control). n=3 per group.
qPCR analysis of the transcription of the ES-GFP plasmid and the effect on VEGF or caspase-3. The endostatin (A), VEGF (B) and caspase-3 (C) mRNA levels were measured by qPCR 48 h after UTMD-mediated ES gene delivery. Endostatin mRNA and caspase-3 mRNA levels were significantly higher with CMB delivery of the ES gene compared with NMB and the control group, and the highest levels were observed for CMB105 delivery. For VEGF mRNA, the levels were significantly lower for CMB delivery of the ES gene compared with NMB and the control group, and the lowest levels were observed with CMB105 delivery (*P<0.01 and #P<0.05 compared with control, NMB and CMB105, **P<0.001 and ##P<0.01 compared with control and NMB, n=6 per group).
Western blot analyses for the expression of endostatin protein and the effect on VEGF or caspase-3. The endostatin (A), VEGF (B) and caspase-3 (C) protein levels were measured by Western blotting 48 h after UTMD-mediated ES gene delivery. Endostatin protein and caspase-3 protein levels were significantly higher with CMB delivery of the ES gene compared with the NMB and control, and the highest levels were observed with CMB105 delivery. VEGF protein levels were significantly lower with CMB delivery of the ES gene compared with NMB and control, and the lowest levels were observed with CMB105 delivery (*P<0.05 compared with the control, NMB and CMB105 groups, **P<0.001 compared with the control and NMB groups, n=6 per group).
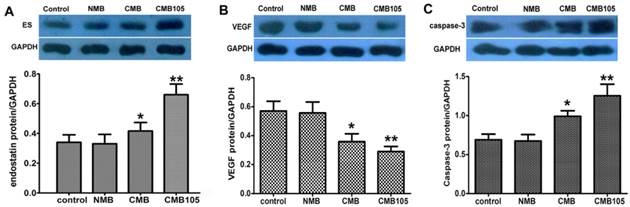
Western blot: expression of endostatin and effect on VEGF or caspase-3
To evaluate the delivery of the ES-GFP gene into HUVECs in vitro, the total amount of endostatin protein in the treated and control HUVECs was determined through a Western blot 48 h after UTMD-mediated gene transfer (Figure 11A). The endostatin protein level after being transferred with CMB105 was increased compared with CMB. The differences in endostatin protein levels between CMB105 and CMB were significant (P<0.05). However, the differences in the endostatin protein level between the control group and the NMB group were not significant (P>0.05). The results were consistent with the results for the transfection efficiency obtained by detecting the expression rate of GFP (Figure 8); both results suggest successful ES-GFP gene transfer by ultrasound with CMB105 and CMB.
To better evaluate the therapeutic effect after ES-GFP gene transfer, the protein levels of VEGF and caspase-3 were examined. With an increasing level of endostatin protein, the VEGF protein level decreased, whereas caspase-3 protein levels increased (Figure 11B, 11C), confirming the antiangiogenesis and apoptosis effects of endostatin.
Analysis of the effect of microbubbles on in vitro angiogenesis and tumor cell invasion
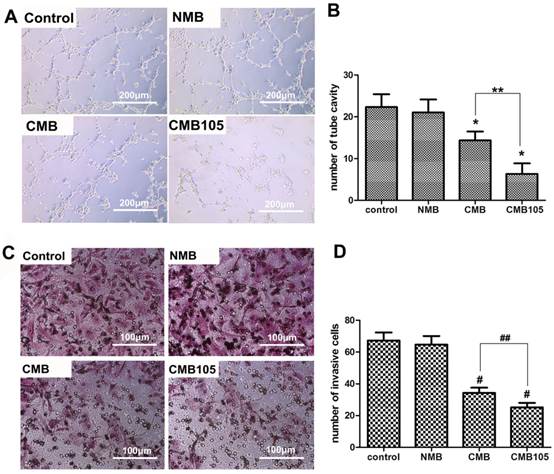
To evaluate the antiangiogenesis effect in vitro, a vascular formation experiment was performed after gene transfer by ultrasound with NMB, CMB or CMB105; the results are presented in Figure 12A. Vascular formation was clearly inhibited as observed with a microscope when cells were treated with CMB105 and CMB. In addition, the number of vascular tube cavities formed by HUVECs significantly decreased compared with the control and NMB groups (Figure 12B, P<0.01), indicating that endostatin protein levels were significantly increased with ultrasound-mediated ES-GFP gene transfer with CMB105 and CMB and that the increased endostatin protein levels could successfully inhibit angiogenesis in vitro. The antiangiogenesis effect was greater for CMB105 than for CMB (P<0.01). Therefore, ultrasound-mediated ES gene transfer with CMB105 is an efficient method to inhibit angiogenesis.
With the inhibition of angiogenesis, the number of invasive tumor cells decreased (Figures 12C and 12D). Many researchers have reported that endostatin can inhibit tumor growth and metastasis (31). In our report, our in vitro results are consistent with these data, and the following in vivo experiments confirmed the inhibitory therapy effect of the tumor growth.
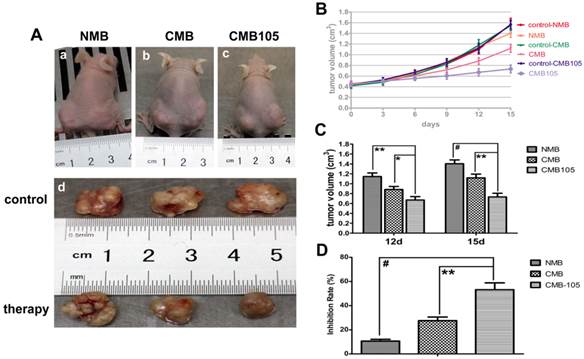
UTMD-mediated delivery of endostatin gene inhibited the growth of tumors in vivo
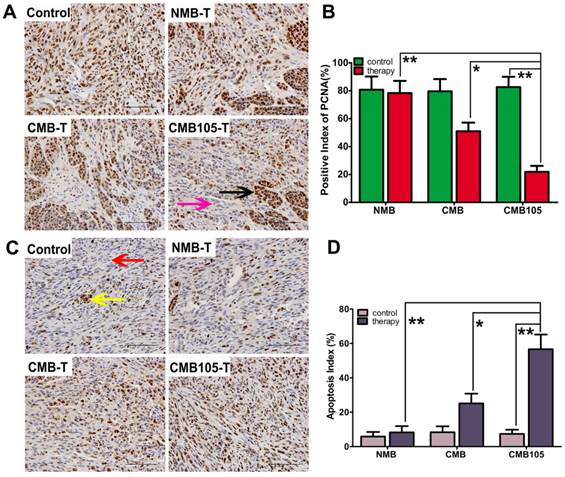
UTMD-mediated endostatin gene delivery with CMB105 significantly inhibited tumor growth 12 and 15 days after transfer (Figure 13). The inhibition rate of the CMB105 group was higher than the CMB group (Figure 13B, 13C and 13D). PCNA expression was decreased both in the CMB and CMB105 groups, and the positive index of PCNA in the CMB105 group was lower than other groups (Figure 14A and 14B); the expression of apoptosis-positive cells showed a significant tumor apoptosis in the CMB and CMB105 groups after gene therapy, and the apoptosis index in the CMB105 group was much higher than that of other groups (Figure 14C and 14D).
Effect of ES gene transfer on in vitro angiogenesis and tumor cell invasion. (A) Images of vascular tube formation in vitro after ultrasound-mediated ES gene transfer using NMB, CMB and CMB105 (10× objective). (B) Based on direct counting, the vascular formation was clearly inhibited in the CMB and CMB105 groups (*P<0.01 compared with control and NMB), and the antiangiogenesis effect was improved with CMB105 compared with CMB (**P<0.01 compared with CMB). (C) Images of invasive MDA-MB-231 cells after ultrasound-mediated ES gene transfer with NMB, CMB and CMB105 (20× objective). (D) The number of invasive tumor cells decreased significantly in the CMB and CMB105 groups (#P<0.01 compared with control and NMB, ##P<0.01 compared with CMB, n=3 per group).
Tumor volume after UTMD-mediated ES gene therapy. (A) a, b, c illustrate the pictures of mice at 15 days after transfer with NMB, CMB, and CMB105, respectively. Tumors in the right hind limbs served as control, and tumors in the left hind limbs were treated by ultrasound with NMB, CMB, CMB105, respectively; d shows the image of tumors at 15 days after gene therapy with different types of microbubbles. (B) Tumor growth curves. (C) Obvious inhibition of tumor growth was observed in the CMB105 and CMB groups at 12 and 15 days after therapy. Differences between the CMB and CMB105 groups were more significant at 15 days than 12 days (*P<0.05, **P<0.01, #P<0.001, n=6 per group). (D) The inhibition rates at 15 days after transfer. Tumors treated with UTMD by CMB105 had a significantly increased inhibition rate (**P<0.01, #P<0.001, n=6 per group).
Anti-tumor therapy effect with UTMD-mediated ES gene therapy. (A) The expression of PCNA in tumor tissue demonstrating the therapeutic effect (scale bar = 100 μm). Brown nuclei (black arrow) indicate PCNA-positive cells, and blue indicate negative cells (purple arrow). (B) A marked difference was observed between NMB, CMB and CMB105 after treatment with UTMD (*P<0.01, **P<0.001, n=6 per group). (C) Expression of TUNEL in tumor tissue (scale bar = 100 μm). Brown nuclei (yellow arrow) represent apoptosis-positive cells, and blue indicates negative (red arrow). (B) The apoptosis index was highest in the CMB105 group (*P<0.01, **P<0.001, n=6 per group).
Discussion
In our previous studies on ultrasound-mediated gene delivery (UMGD) with NMB (28, 29), the gene transfection efficiency was modest, similar to the reports of other researchers (32, 33). However, because of its advantages of noninvasive, non-viral and targeted transfection, UMGD has been continuously investigated since its development. The advantages of non-viral transfection could make clinical therapy a reality, except for its modest transfection efficiency. Therefore, improving the transfection efficiency is becoming the key point for future studies. According to previous experiments concerning cationic liposome-mediated gene transfection, CMB were developed to enhance the loading capacity of DNA. Most CMB were prepared by incorporating different types of cationic lipids, such as DSTAP, DOTAP, DOTMA, DMTMP, DPTAP, DDAB or Stearic-PEI600 into the lipid microbubble shell. Then, the loading capacity was increased through charge-coupling, and the transfection efficiency was also enhanced compared with NMB (5, 7, 8, 34, 35, 36). This approach appears to be effective in increasing the gene transfection efficiency due to the increase of the local DNA concentration. To further increase the local DNA concentration and transfection efficiency, targeted CMB were developed and used here. Because microbubbles cannot cross the gap between endothelial cells, endothelial cells were selected as targets for in vitro and in vivo gene delivery.
In contrast to other reports, we selected DC-Chol to make the surface of the microbubbles positively charged. This lipid is one of the most efficient and commonly used cationic lipids and has been widely used in gene transfection and drug delivery (37). In our experiment, 0.5 mg of DC-Chol was added to construct the CMB, and the mean zeta-potential was 26.44±2.13 mV. The microbubbles were stable in suspension for one month at 4℃. Upon increasing the amount of DC-Chol, the zeta-potential of the CMB increased, whereas the stability decreased. The zeta-potentials of CMB reported by other researches were different (5, 21, 38, 39); no previous study has reported that the stability of microbubbles was affected by the potential, even at potentials greater than 60 mV. We speculated that this result might be because DC-Chol is more rigid than DSTAP, which previous researchers used to construct CMB. Thus, determining which lipid to use in making CMB requires further research. Here, the plasmid loading capacity of CMB was slightly higher than the value reported by Panje et al (21). However, we also reported that the combination of plasmid reached saturation. In contrast to our results, Sun et al (8) reported no saturation for the plasmid loading capacity, even when 80 μg plasmid was added, and they did not report the number of microbubbles used in their experiments. The zeta-potentials reported by each researcher were different, as well as the plasmid loading capacities and transfection efficiencies. Thus, the relationships between the zeta-potential, plasmid loading capacity and transfection efficiency of CMB also require further investigation.
One notable feature of our CMB is that it was made of DSPE-PEG2000-Biotin rather than DSPE-PEG2000 and had a biotinylated cationic shell. We selected DSPE-PEG2000-Biotin instead of DSPE-PEG2000 to construct CMB not only because incorporating the biotin tag permitted molecular targeting strategies but also because the concentration of CMB was higher and stability was better. We hypothesize that increased concentration and improved stability could be attributed to the biotin component. Other researchers have also observed this phenomenon and reported that the potential of CMB decreased from 64±5 mV to 56±2 mV with the addition of DSPE-PEG-biotin to the shell (38). This finding further confirmed that the stability of CMB was related to the potential. In our preliminary experiment, most HUVECs were dead after treatment with CMB at a zeta-potential higher than 60 mV through ultrasound destruction, indicating that a high potential may be damaging to HUVECs and have a negative effect on cell viability. Avidin is an exogenous protein and has been regarded as a potential antigen to induce immunological reactions when used in vivo. The formation of immune complexes in the basement membrane of kidney is easy and limits the use of avidin in human body. Avidin was still chosen in our experiment due to the high specificity and stability of the reaction between avidin and biotin to ensure the connection between microbubbles and antibody; additionally, we have extensive experience in constructing targeted microbubbles in this manner. If CMB105 could be used in human body, the method to connect CD105 antibody to CMB should be improved.
The combination of CMB with the CD105 antibody had no effect on the plasmid binding ability of CMB (Figure 6A, 6B). Thus, we concluded that the existence of the biotin tag and combination with the CD105 antibody had little or no effect on plasmid binding, which has also been confirmed by other researchers (5, 38). Further, the targeting ability of CMB105 to HUVECs was not affected by the combination of plasmids (Figure 6C). The mechanism of targeting to HUVECs was different between CMB and CMB105. We observed only a slight non-specific interaction between NMB and HUVECs, whereas a substantial amount of targeted CMB attached to HUVECs; the difference between the NMB and CMB groups was significant. This finding confirmed that CMB could target to HUVECs through electrostatic interaction and biotinylation, as has been observed by others (5). The primary mechanism of CMB105 targeting to HUVECs was the reaction between the antigen and antibody; the number of attached CMB105 was 1.35-fold higher than for CMB. The mechanism of CMB105 targeting to HUVECs may still include electrostatic interaction; however, we believe that the reaction between the antigen and antibody is the main mechanism and real target reaction, which was further confirmed in our next experiment. By analyzing whether DNA binding would affect CMB interaction with HUVECs, we observed that the number of CMB targeted to HUVECs decreased slightly (P<0.05); however, the difference was not as large as other researchers have reported (5), potentially because the potential of our CMB was higher compared with previous studies. Thus, even if some change in potential occurred after plasmid binding, the effect of targeting to cells was smaller than previous studies. One important result was that the number of CMB targeted to HUVECs appeared to remain unchanged (P>0.05), further confirming our previous conclusion that the primary mechanism of CMB105 targeting to HUVECs was the reaction between the antigen and antibody.
In the cell viability test, we observed that the HUVECs viability decreased after adding CMB and CMB105; cell viability after treatment with CMB105 was lower than for CMB, confirming that increasing the number of target-attached microbubbles can decrease the cell viability, as has been reported previously (5, 34, 40). Cell membrane permeability and transfection efficiency increased with decreasing cell viability. The cell membrane permeability and transfection efficiency reported here were different from the results reported previously because the transfection method was different. To better investigate the advantages of targeted and CMB in UMGD and better use their targeting ability and plasmid loading capacity, the inverted and upright cell experimental upset (Figure 2) was used for the transfer experiment. This method ensures that the microbubbles used in the transfer process were attached to HUVECs and that the plasmid was loaded by those microbubbles. We regarded this method as the best to utilize the targeting ability and plasmid loading capacity of our microbubbles. A limitation of this experiment was that a parallel flow chamber was not used because such a device was not available. Most have used the inverted method for the upward floating characteristic of microbubbles and did not wash out any microbubbles that did not target and attach to HUVECs. Thus, the advantage of CMB or targeted microbubbles could not be accurately assessed. Therefore, in our report, the transfection efficiency with NMB was lower than the value previously reported by us and other researchers (5, 8, 21).
Through analyzing the therapy effect in vitro, these data confirmed that the treatment effect was remarkable with both CMB and CMB105. Although the therapy effect with CMB105 was better than with CMB, the biological effect was not ideal. Therefore, because using CMB induced an obvious therapeutic effect, the reason for using CMB105 was unclear. Although the therapeutic effect with CMB105 was better than the effect using CMB in vitro, the advantage of CMB105 when used in vivo is more important. Although CMB could target HUVECs in vitro through electrostatic interaction, this advantage disappeared in vivo, and because of the increased phagocytosis by reticuloendothelial system, the CMB would be quickly metabolized, as confirmed in the ultrasound imaging (Supplementary Material: Figure S2). The above two limitations of CMB went against UMGD in vivo. Because CMB was unable to target aggregate in the therapy areas, the advantage of using CMB105 in vivo was much better than in vitro. The inhibition rate with CMB105 was nearly double that with CMB, and an obvious inhibition of tumor growth was observed (Figure 14). The differences in the therapy effect between CMB and CMB105 in vivo were more significant than that in vitro. Except for the enhanced transfection efficiency, the outstanding antiangiogenesis effect of endostatin was also attributed to the excellent treatment effect. After treatment with CMB and CMB105 loaded with ES-GFP plasmid, the expression of VEGF decreased with increasing endostatin expression, and the formation of blood vessels in vitro also decreased. Endostatin has been shown to inhibit the proliferation and angiogenesis of endothelial cells through multiple pathways, not only through the VEGF pathway but also by down regulation of the expression of anti-apoptotic protein (41, 42, 43). The effect of endostatin appears to be better than that observed with VEGF antibodies. Therefore, endostatin gene therapy using UMGD may offer an efficient method for antiangiogenesis therapy because it could overcome the defect of endostatin instability. Many drugs are used to treat tumors, and angiogenesis inhibitors are commonly used as adjuvant chemotherapy. If this method is inefficient, other drugs should be considered, such as multispecific tyrosine-kinase inhibitors, etc.
Targeted transfection was possible through ultrasound molecular imaging using CMB105, and evaluating CD105 expression levels, determining whether the tumor was benign or malignant, and assessing the therapeutic effect or prognosis was possible because the expression of CD105 has been reported to be correlated with poor prognosis and elevated levels of soluble CD105 in the plasma have been linked to metastasis (44, 45). This agent appears to be a multifunctional contrast agent, and we next plan to confirm its multifunctional abilities.
Conclusions
In conclusion, a targeted cationic microbubble (CMB105) was successfully constructed. Changes in the plasmid loading capacity and targeting to endothelial cells and ultrasound-mediated gene transfer with NMB, CMB, and CMB105 were carefully compared both in vitro and in vivo. The plasmid loading capacity increased significantly with both CMB and CMB105; the addition of CD105 antibody to the CMB did not significantly affect plasmid loading capacity. CMB105 exhibited excellent ability for targeting endothelial cells both in vitro and in vivo and were not affected after loading the plasmid because the main mechanism was the reaction between the antigen and antibody. In contrast, for CMB, the mechanism involved electrostatic interaction in vitro. Thus, CMB may not target endothelial cells in vivo. Cell membrane permeability and transfection efficiency increased with increasing quantities of microbubbles targeting HUVECs, whereas cell viability decreased. With increasing transfection efficiency, the expression of endostatin and caspase-3 increased after treatment with CMB and CMB105. In contrast, VEGF was decreased, and the cell cycle was inhibited in the G1 phase. The therapeutic effect of CMB105 was better than CMB; angiogenesis and tumor cell invasion were inhibited successfully in vitro, and apoptosis and tumor growth inhibition were observed in vivo. Overall, these data demonstrated the excellent advantages associated with using targeted CMB to enhance ultrasound-mediated gene transfer in vitro and in vivo. The transfer method selected for our in vitro experiment could better evaluate this advantage. When targeted cationic microbubbles (CMB105) were used in vivo, better therapeutic effect was achieved compared with CMB. We suggest that CMB105 is a very useful multifunctional ultrasound contrast agent.
Supplementary Material
Fig.S1 - Fig.S3.
Acknowledgements
The authors are grateful to Dr. Zipeng He and Dr. Jinshun Xu for assistance with ultrasound imaging. This work was supported in part by the following National Natural Science Foundation of China Grants: 81161120548, 81130025, 81371578, 81371579, 30770565, 81227801, 81202089, 81170858, 81401503 and 61205203; the National Key Basic Research (973) Program of China: 2014CB744503; the International S&T Cooperation Program of China: 2014DFG32800; and the fund of Chongqing university scientific innovation team 2013(KJTD201303).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Taniyama Y, Azuma J, Kunugiza Y. et al. Therapeutic option of plasmid-DNA based gene transfer. Curr Top Med Chem. 2012;12(15):1630-7
2. Porter TR, Iversen PL, Li S. et al. Interaction of diagnostic ultrasound with synthetic oligonucleotide-labeled perfluorocarbon exposed sonicated dextrose albumin microbubbles. J. Ultrasound Med. 1996;15(8):577-84
3. Greenleaf WJ, Bolander ME, Sarkar G. et al. Artificial cavitation nuclei significantly enhance acoustically induced cell transfection. Ultrasound Med Biol. 1998;24(4):587-95
4. Miller DL, Pislaru SV, Greenleaf JE. Sonoporation: mechanical DNA delivery by ultrasonic cavitation. Somat Cell Mol Genet. 2002;27(1-6):115-34
5. Nomikou N, Tiwari P, Trehan T. et al. McHale. Studies on neutral, cationic and biotinylated cationic microbubbles in enhancing ultrasound-mediated gene delivery in vitro and in vivo. Acta Biomaterialia. 2012;8(3):1273-80
6. Nomikou N, McHale AP. Exploiting ultrasound-mediated effects in delivering targeted, site-specific cancer therapy. Cancer Lett. 2010;296(2):133-43
7. Wang DS, Panje C, Pysz MA. et al. Cationic versus neutral microbubbles forultrasound-mediated gene delivery in cancer. Radiology. 2012;264(3):721-32
8. Sun L, Huang CW, Wu J. et al. The use of cationic microbubbles to improve ultrasound-targeted gene delivery to the ischemic myocardium. Biomaterials. 2013;34(8):2107-16
9. Kiessling F, Huppert J, Palmowski M. Functional and molecular ultrasound imaging: concepts and contrast agents. Curr Med Chem. 2009;16(5):627-42
10. Klibanov AL. Ligand-carrying gas-filled microbubbles: ultrasound contrast agents for targeted molecular imaging. Bioconjug Chem. 2005;16(1):9-17
11. Dayton PA, Rychak JJ. Molecular ultrasound imaging using microbubble contrast agents. Front Biosci. 2007;12:5124-42
12. Korpanty G, Carbon JG, Grayburn PA. et al. Monitoring response to anticancer therapy by targeting microbubbles to tumor vasculature. Clin Cancer Res. 2007;13(1):323-30
13. Bzyl J, Lederle W, Rix A. et al. Molecular and functional ultrasound imaging in differently aggressive breast cancer xenografts using two novel ultrasound contrast agents (BR55 and BR38). Eur Radiol. 2011;21(9):1988-95
14. Sorace AG, Saini R, Mahoney M. et al. Molecular ultrasound imaging using a targeted contrast agent for assessing early tumor response to antiangiogenic therapy. J Ultrasound Med. 2012;31(10):1543-50
15. Minhajat R, Mori D, Yamasaki F. et al. Organ-specific endoglin (CD105) expression in the angiogenesis of human cancers. Pathol Int. 2006;56(12):717-23
16. Duff SE, Li C, Garland JM. et al. CD105 is important for angiogenesis: evidence and potential applications. FASEB J. 2003;17(9):984-92
17. Krämer I, Lipp HP. Bevacizumab, a humanized anti-angiogenic monoclonal antibody for the treatment of colorectal cancer. J Clin Pharm Ther. 2007;32(1):1-14
18. Force T, Krause DS, Van Etten RA. Molecular mechanisms of cardiotoxicity of tyrosine kinase inhibition. Nat Rev Cancer. 2007;7(5):332-44
19. Feldman AL, Libutti SK, Chen Q-R. et al. Liposomes complexed to plasmids encoding angiostatin and endostatin inhibit breast cancer in nude mice. Cancer Res. 2000;60(5):1463-4
20. Calvo A, Yokoyama Y, Smith LE. et al. Inhibiton of the mammary carcinoma angiogenic switch C3(1)/SV40 transgenic mice by a mutated form of human endostatin. Int J Cancer. 2002;101(3):224-34
21. Panje CM, Wang DS, Pysz MA. et al. Ultrasound-mediated gene delivery with cationic versus neutral microbubbles: effect of DNA and microbubble dose on in vivo transfection efficiency. Theranostics. 2012;2(11):1078-91
22. Pochon S, Tardy I, Bussat P. et al. BR55: A Lipopeptide-based VEGFR2-targeted ultrasound contrast agent for molecular imaging of angiogenesis. Invest Radiol. 2010;45(2):89-95
23. Anderson CR, Hu X, Zhang H. et al. Ultrasound molecular imaging of tumor angiogenesis with an integrin targeted microbubble contrast agent. Invest Radiol. 2011;46(4):215-24
24. Niu C, Wang Z, Zuo G. et al. Poly(Lactide-Co-Glycolide) Ultrasonographic Microbubbles Carrying Sudan Black for Preoperative and Intraoperative Localization of Lymph Nodes. Clin Breast Cancer. 2012;12(3):199-206
25. Sun Y, Zheng Y, Ran H. et al. Superparamagnetic PLGA-iron oxide microcapsules for dual-modality US/MR imaging and high intensity focused US breast cancer ablation. Biomaterials. 2012;33(24):5854-64
26. Zhou Y, Zhou XY, Wang ZG. et al. Elevation of plasma membrane permeability upon laser irradiation of extracellular microbubbles. Lasers Med Sci. 2010;25(4):587-94
27. Umebayashi Y, Miyamoto Y, Wakita M. et al. Elevation of plasma membrane permeability on laser irradiation of extracellular latex particles. J Biochem. 2003;134(2):219-24
28. Zheng MM, Zhou XY, Wang LP. et al. Experimental research of RB94 gene transfection into retinoblastoma cells using ultrasound-targeted microbubble destruction. Ultrasound Med Biol. 2012;38(6):1058-66
29. Gao R, Zhou X, Yang Y. et al. Transfection of wtp53 and Rb94 genes into retinoblastomas of nude mice by ultrasound-targeted microbubble destruction. Ultrasound Med Biol. 2014;40(11):2662-70
30. Jia J, Chengbo L, Yuping G. et al. India Ink Incorporated Multifunctional Phase-transition Nanodroplets for Photoacoustic/Ultrasound Dual-modality Imaging and Photoacoustic Effect Based Tumor Therapy. Theranostics. 2014;4(10):1026-1038
31. Hanai J, Dhanabal M, Karumanchi SA. et al. Endostatin causes G1 arrest of endothelial cells through inhibition of cyclin D1. J Biol Chem. 2002;277(19):16464-9
32. Bekeredjian R, Grayburn PA, Shohet RV. Use of ultrasound contrast agents for gene or drug delivery in cardiovascular medicine. J Am Coll Cardiol. 2005;45(3):329-35
33. Chen S, Shohet RV, Bekeredjian R. et al. Optimization of ultrasound parameters for cardiac gene delivery of adenoviral or plasmid deoxyribonucleic acid by ultrasound-targeted microbubble destruction. J Am Coll Cardiol. 2003;42(2):301-8
34. Tlaxca JL, Anderson CR, Klibanov AL. et al. Analysis of in vitro transfection by sonoporation using cationic and neutral microbubbles. Ultrasound Med Biol. 2010;36(11):1907-18
35. Christiansen JP, French BA, Klibanov AL. et al. Targeted tissue transfection with ultrasound destruction of plasmid-bearing cationic microbubbles. Ultrasound Med Biol. 2003;29(12):1759-67
36. Jin Q, Wang Z, Yan F. et al. A novel cationic microbubble coated with stearic acid-modified polyethylenimine to enhance DNA loading and gene delivery by ultrasound. PLoS One. 2013;8(9):e76544
37. Caracciolo G, Callipo L, De Sanctis SC. et al. Surface adsorption of protein corona controls the cell internalization mechanism of DC-Chol-DOPE/DNA lipoplexes in serum. Biochim Biophys Acta. 2010;1798(3):536-43
38. Xie A, Belcik T, Qi Y. et al. Ultrasound-mediated vascular gene transfection by cavitation of endothelial-targeted cationic microbubbles. JACC Cardiovasc Imaging. 2012;5(12):1253-62
39. Christiansen JP, French BA, Klibanov AL. et al. Targeted tissue transfection with ultrasound destruction of plasmid-bearing cationic microbubbles. Ultrasound Med Biol. 2003;29(12):1759-67
40. Li YS, Davidson E, Reid CN. et al. Optimising ultrasound-mediated gene transfer (sonoporation) in vitro and prolonged expression of a transgene in vivo: potential applications for gene therapy of cancer. Cancer Lett. 2009;273(1):62-9
41. Yamaguchi N, Anand-Apte B, Lee M. et al. Endostatin inhibits VEGF-induced endothelial cell migration and tumor growth independently of zinc binding. EMBO J. 1999;18(16):4414-23
42. Lee SJ, Jang JW, Kim YM. et al. Endostatin binds to the catalytic domain of matrix metalloproteinase-2. FEBS Lett. 2002;519(1-3):147-52
43. Sauter BV, Martinet O, Zhang WJ. et al. Adenovirus-mediated gene transfer of endostatin in vivo results in high level of transgene expression and inhibition of tumor growth and metastases. Proc Natl Acad Sci USA. 2000;97(9):4802-7
44. Minhajat R, Mori D, Yamasaki F. et al. Organ-specific endoglin (CD105) expression in the angiogenesis of human cancers. Pathol Int. 2006;56(12):717-23
45. Li C, Guo B, Bernabeu C. et al. Angiogenesis in breast cancer: the role of transforming growth factor beta and CD105. Microsc Res Tech. 2001;52(4):437-49
Author contact
![]() Corresponding author: Tel.: +86 23 63693602; Fax: +86 23 63822696; E-mail: wangdongedu.cn
Corresponding author: Tel.: +86 23 63693602; Fax: +86 23 63822696; E-mail: wangdongedu.cn
 Global reach, higher impact
Global reach, higher impact