13.3
Impact Factor
Theranostics 2015; 5(1):86-96. doi:10.7150/thno.10062 This issue Cite
Research Paper
Codon-optimized Human Sodium Iodide Symporter (opt-hNIS) as a Sensitive Reporter and Efficient Therapeutic Gene
1. Department of Nuclear Medicine, Seoul National University College of Medicine;
2. Biomedical Sciences, Seoul National University College of Medicine;
3. Cancer Research Institute, Seoul National University College of Medicine,
4. Tumor Microenvironment Global Core Research Center, Seoul National University;
5. Molecular Medicine and Biopharmaceutical Sciences, Seoul National University;
6. Cancer Imaging Center, Seoul National University Hospital, Seoul, South Korea;
7. Division of High-Risk Pathogen Research, Korea National Institute of Health, Osong, South Korea.
#These authors are equally contributed in this research.
Received 2014-7-8; Accepted 2014-9-17; Published 2015-1-1
Abstract
To generate a more efficient in vivo reporter and therapeutic gene, we optimized the coding sequence of the human sodium/iodide symporter (NIS) gene by replacing NIS DNA codons from wild type to new codons having the highest usage in human gene translation. The Codon Adaptation Index (CAI), representing the number of codons effective for human expression, was much improved (0.79 for hNIS, 0.97 for opt-hNIS). Both wild-type (hNIS) and optimized human NIS (opt-hNIS) were cloned into pcDNA3.1 and pMSCV vectors for transfection. Various cancer cell lines such as thyroid (TPC-1, FRO, B-CPAP), breast (MDA-MB-231), liver (Hep3B), cervical (HeLa), and glioma (U87MG) were transfected with pcDNA3.1/hNIS or pcDNA3.1/opt-hNIS. 125I uptake by opt-hNIS-expressing cells was 1.6 ~ 2.1 times higher than uptake by wild-type hNIS-expressing cells. Stable cell lines were also established by retroviral transduction using pMSCV/hNIS or pMSCV/opt-hNIS, revealing higher NIS protein levels and 125I uptake in opt-hNIS-expressing cells than in hNIS-expressing cells. Moreover, scintigraphic images from cell plates and mouse xenografts showed stronger signals from opt-hNIS-expressing cells than hNIS-expressing cells, and radioactivity uptake by opt-hNIS-expressing tumors was 2.3-fold greater than that by hNIS-expressing tumors. To test the efficacy of radioiodine therapy, mouse xenograft models were established with cancer cells expressing hNIS or opt-hNIS. 131I treatment reduced tumor sizes of hNIS- and opt-hNIS-expressing tumors to 0.57- and 0.27- fold, respectively, compared to their sizes before therapy, suggesting an improved therapeutic effect of opt-hNIS. In summary, this study shows that codon optimization strongly increases hNIS protein levels and radioiodine uptake, thus supporting opt-hNIS as a more sensitive reporter and efficient therapeutic gene.
Keywords: human sodium iodide symporter (hNIS), radioiodine therapy, codon optimization, reporter, gene therapy.
Introduction
The sodium iodide symporter (NIS) gene that facilitates iodide uptake driven by sodium ion gradients across the plasma membrane has been shown to be a simple and applicable reporter [1]. In thyroid cells, one iodide ion enters with two sodium ions through the NIS, and a sodium ion gradient is maintained by the sodium-potassium pump (Na+-K+ATPase) [2]. In addition to iodide, several anions such as ClO4-, SCN-, ClO3-, NO3- can be transported by the NIS [3]. Pertechnetate (TcO4-) and perrhenate (ReO4-) are also transported by the NIS, and radioactive forms (99mTc, 188Re, and 131I) of such anions are important for nuclear medicine imaging and radionuclide therapy [4-6]. In fact, radioiodine therapy was the first clinical application of targeted radionuclide therapy in NIS-expressing thyroid cancers [6].
For improved anti-cancer therapy, radioiodine gene therapy using an exogeneous NIS gene has been suggested [7-9]. Cloning and characterization of the human sodium iodide symporter gene [10,11] has facilitated diagnostic and therapeutic applications of NIS [12-15], and radioiodine accumulation was highly improved in various cancer cells following ectopic NIS gene transfection [16-18].
Because radioiodine uptake is proportional to total NIS protein levels [19], enhancing NIS protein expression improves reporter sensitivity and the therapeutic efficacy of radioiodine therapy. To achieve this goal, several strategies have been suggested. Thyroid-stimulating hormone [20] and various compounds such as retinoic acid [21], valporic acid [22], the PPAR gamma agonist Troglitazone [23], the PI3K inhibitor LY294002 [24], and the heat-shock protein inhibitor Tanespimycin [25] were reported to elevate NIS protein levels by up-regulating NIS gene expression [26]. This type of approach is closely related to modulation of promoter activity for the intrinsic NIS gene.
On the other hand, applications of an exogenous NIS gene as an extrinsic reporter and therapeutic gene require different strategies. Because both higher numbers of NIS mRNA transcripts and higher translation efficiency are required to increase NIS protein synthesis, the promoters of ectopic expression vectors should be constitutive, with strong transcriptional activity in their target cells [27-29]. However, ectopic expression by constitutive promoters is restricted to the regulation of transcription (and not translation) for increasing mRNA transcripts.
Recent advances have improved eukaryotic translational efficiency by optimization of balanced codon usage [30]. Codon optimization is a method that alters gene sequences to contain the highest frequency of codons for tRNAs in the host organism to maximize the amount of protein synthesis [31-34]. In this study, we developed a highly efficient human NIS gene (opt-hNIS) as a sensitive reporter and a therapeutic gene by codon optimization, and evaluated its expression and activity in vitro and in vivo.
Methods and Materials
Cell lines
Human papillary thyroid cancer TPC-1, B-CPAP and human anaplastic thyroid cancer FRO cell lines were cultured in RPMI-1640 medium (WelGENE, Daegu, South Korea) containing 5% heat-inactivated fetal bovine serum (Gibco, Grand Island, NY, USA). Thyroid cancer cell lines were kindly provided by Dr. Shunichi Yamashita and Dr. Norisato Mitsutake (Department of Molecular Medicine, Atomic Bomb Disease Institute, Nagasaki University Graduate School of Biomedical Sciences, Nagasaki, Japan). Human breast cancer MDA-MB-231, human cervical cancer HeLa, and human hepatoma cancer Hep3B cell lines were obtained from the Korean Cell Line Bank (Seoul, South Korea) and cultured in Dulbecco's Modified Eagle Medium (DMEM) (WelGENE). The glioma U87MG cell line was obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in Minimum Essential Medium (MEM). Cells were incubated in culture media containing 10% heat-inactivated fetal bovine serum and 1% penicillin/streptomycin (Gibco) at 37oC in a 5% CO2 chamber.
Codon-optimization of human sodium iodide symporter gene
The nucleotide sequence of the human NIS (GenBank Accession Number NM_000453) was obtained from the NCBI database. Codon optimization was performed using GeneOptimizer® expert software (GENEART®, Regensburg, Germany) [24]. The Codon Adaptation Index (CAI) was first advocated by Sharp et al. [34] as a directional measure of codon usage, relative to reference optimal codons, for a specific species. The CAI of the wild type human NIS gene (hNIS) and sequence-optimized NIS gene (opt-hNIS) were analyzed by GenScript Rare Codon Analysis Tool software (GenScript Corporation, Piscataway, NJ, USA). Sequence-optimized hNIS (opt-hNIS) genes were synthesized and cloned into pMK-RQ plasmids flanked by Nco I and Sfi I restriction sites.
Construction of recombinant plasmids
The wild type hNIS gene was kindly provided by Dr. Sissy M. Jhiang of The Ohio State University College of Medicine (Columbus, OH, USA). The wild type hNIS gene and purified synthetic opt-hNIS were inserted into pGEM-T vectors and then transformed into E. coli DH5α cells, according to the manufacturer's protocol (Promega, Madison, WI, USA). The recombinant plasmids were characterized by digestion using the restriction enzymes XhoI and Hpa I. One recombinant pGEM-T transformant that carried no mutation was selected as the source for further subcloning into pcDNA3.1(+) (Invitrogen, Grand Island, NY, USA) and pMSCVpuro (Clontech, Mountain View, CA, USA) plasmids. The recombinant plasmids were designated as pcDNA3.1/hNIS or pcDNA3.1/opt-hNIS and pMSCV/hNIS or pMSCV/opt-hNIS.
Transient transfection and retroviral transduction
Tumor cells such as TPC-1, FRO, B-CPAP, MDA-MB-231, Hep3B, HeLa, and U87MG cell lines were transiently transfected with pcDNA3.1/hNIS or pcDNA3.1/opt-hNIS using Lipofectamine® 2000 (Invitrogen), according to the manufacturer's instructions. For normalizing transfection efficiency, pcDNA3.1/Luc was co-transfected with hNIS or opt-hNIS expressing vectors. Luciferase activities were measured using the ONE-Glo luciferase assay system (Promega, Madison, WI, USA). Retroviruses were generated using a modification of the Retro-X universal packaging system (BD Biosciences Clontech, Palo Alto, CA, USA), with 293FT packaging cells cotransfected with 5 µg pMSCV/hNIS or pMSCV/opt-hNIS vector, 5 µg pcDNA3-gag-pol vector, and 5 µg of envelope vector, using Lipofectamine 2000 (Invitrogen). Supernatants were collected at 72 hr post-transfection and concentrated using Amicon Ultra 100K Filters (Millipore, Bedford, MA, USA), followed by filtering through 0.45-µm polyvinylidene difluoride (PVDF) filters (Millipore). Viral supernatants were titered and stored at -80°C until required.
Firefly luciferase-expressing MDA-MB-231 and FRO cell lines were stably transduced with pMSCV/hNIS or pMSCV/opt-hNIS using retroviruses, and were thus designated MDA-MB-231/hNIS, MDA-MB-231/opt-hNIS, FRO/hNIS, or FRO/opt-hNIS. Tumor cells expressing hNIS or opt-hNIS were pooled after puromycin selection, and monoclonal cells with similar number of hNIS or opt-hNIS transcripts were isolated for further analysis.
RT-PCR
Total RNA was isolated from cancer cells using TRIZOL reagent (Invitrogen), according to the manufacturer's protocol. The sequences of the forward and reverse hNIS primers were 5'-TGGGCGGCATGAAGGCTGTG and 5'-CTCTGTAGGCACAGGCCAGG, respectively. The sequences of the forward and reverse primers of GAPDH were 5'-GAGAAGGCTGGGGCTCATTT and 5'-CCTTCCGTGTCCCCACTG, respectively. The PCR reaction for hNIS (94°C for 30 sec, 56.5°C for 60 sec, 72°C for 30 sec) was run for 30 cycles, after an initial single step of 94°C for 5 min to activate the Taq polymerase. For GAPDH, all PCR conditions were the same except the anealing temperature, 60°C. PCR products were separated by 1% agarose gel electrophoresis and visualized by Loading Star (DyneBioInc, Seoul, Korea) stain.
Real-time PCR
Quantitative real-time PCR in 96-well optical plates was conducted and analyzed in an ABI Prism 7000 Sequence Detection System (Applied Biosystems, Lincoln, CA, USA). The PCR conditions used were as follows. After initial activation of uracil-N-glycosylase at 50oC for 2 min, AmpliTaq Gold was activated for 10 min at 95oC, and the subsequent PCR reaction consisted of 40 cycles of denaturation at 95oC for 15 sec and annealing/extension at 60oC for 1 min per cycle. Since we used pMSCVpuro vectors for stable transduction of pMSCV/hNIS or pMSCV/opt-hNIS, PCR primers and probe sets for puromycin (Assay ID: AI70L25, Applied Biosystems) were used for copy number measurements. The expression levels of the amplified genes were normalized to human 18S ribosomal rRNA, and were represented as relative expression levels. All experiments were performed in triplicate.
Western blot analysis
Cells were lysed in radio-immunoprecipitation assay (RIPA) buffer (Sigma, St. Louis, MO, USA) containing protease inhibitor cocktail (Roche Diagnostics, Basel, Switzerland), and subsequently cleared via centrifugation (14,000 × g for 20 min at 4oC). Protein concentrations were determined using BCA protein assay kits (Thermo Scientific, Palm Springs, CA, USA). Total protein (30 μg) mixed with 4X polyacrylamide gel electrophoresis sample buffer (Invitrogen) was separated via NuPAGE Novex 4%-12% gradient Bis-Tris mini gels (Invitrogen) and transferred to nitrocellulose membranes. Membranes were blocked with 3% skim milk for 1 hr at room temperature, and then incubated with following primary antibody overnight at 4oC: anti-hNIS (Koma Biotech, Seoul, South Korea; diluted 1:1000) or beta-actin (Sigma; diluted 1:5000). Antigen-antibody complexes were visualized with HRP-conjugated secondary antibody (Sigma; diluted 1:2000) and enhanced chemiluminescence detection reagent (Thermo Fisher Scientific, Rockford, IL, USA). Signal intensities were then measured using an LAS-3000 imaging system (Fujifilm, Tokyo, Japan).
125I uptake assays
Cells were seeded in 24-well plates, washed with warmed Hank's balanced salt solution (HBSS), and incubated for 30 min at 37oC with 500 μL warmed HBSS containing 0.1 µCi of 125I and 10 μM non-radioactive NaI. The cells were then washed twice with cold HBSS and lysed for 5 min in 1% SDS. Cell lysates were collected and radioactivity measured using a gamma counter (Canberra-Packard, Meriden, USA). Radioactivity was normalized to the amount of total protein at the time of the assay. All experiments were performed in triplicate.
Animal experiments
Six-week-old male BALB/c nude mice were obtained from Orient Bio, Inc. (Seoul, Korea). All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Seoul National University Hospital. For establishment of tumor xenograft mouse models, tumor cells (1×106) expressing hNIS and opt-hNIS were transplanted subcutaneously into the left and right thighs, respectively, to allow comparison of tumors expressing the wild-type and optimized transporter genes.
Bioluminescence Imaging
For bioluminescence imaging acquisition, an IVIS100 imaging system (Caliper Life Sciences, Hopkinton, MA, USA), which includes an optical CCD camera mounted on a light-tight specimen chamber, was used. D-luciferin potassium salt was diluted to 0.3 mg/mL in PBS before use, and 100 μL of the D-luciferin solution was intraperitoneally injected into mice. Bioluminescence images were serially acquired every 5 min until maximum signals were reached. To quantify emitted light, regions of interest (ROI) were drawn over tumor regions. Total photon flux was expressed as photons per cm2 per second per steradian (p/cm2/sec/sr).
Scintigraphic Imaging
For scintigraphic imaging acquisition, cell culture plates containing cell monolayers (105 cells/well) were imaged after addition of a radiotracer. After 30 min incubation of 6 different doses of 99mTc-pertechnetate, ranging from 0.625 to 50 μCi, cell culture plates were washed with HBSS buffer and imaged using a γ-ray camera (ON-410, Ohio Nuclear, Solon, OH, USA). Planar imaging techniques were applied, with at least 50000 total counts per image collected. Images were processed with a modified version of NIH Image software (NucMed†Image, Mark D Wittry, St Louis University, St Louis, MO, USA). Scintigraphic images of tumor-xenografted mice were also acquired by a γ-ray camera after intraperitoneal 99mTc-pertechnetate (0.5 mCi/100 μL in saline) injection. All mouse experiments were performed on at least three mice per group.
SPECT/CT imaging
Single-photon emission-computed tomography (SPECT) was performed with a four-headed multiplexing multipinhole NanoSPECT/CT (Bioscan, Poway, CA, USA) using a mouse aperture. For SPECT/CT, cells expressing hNIS or opt-hNIS gene were exposed to five different doses of 99mTc-pertechnetate, ranging from 3.12 to 50 μCi, for one hour, and 106 cells were subcutaneously injected into the nude mice to test imaging sensitivity. An acquisition time of 30 s per view was chosen for SPECT, resulting in acquisition times ranging from 30 to 40 min per animal. The 9-min CT imaging was performed immediately following whole-body SPECT imaging at 50-μm resolution. Reconstruction of the images was performed using HiSPECT software (Bioscan) without attenuation correction. Reconstructed data from SPECT and CT were co-registered using InVivoScope (Bioscan) for further analysis and interpretation.
Radioiodine therapy using mouse tumor xenograft models
Tumor sizes were serially measured every week with calipers. At day 14, tumor-bearing mice were intraperitoneally administered 1.5 mCi 131I radioiodine therapy. Bioluminescence imaging, as well as tumor size measurements, was performed at 0, 14, 21, and 28 days after tumor challenge. Tumor sizes were measured from at least three mice per group.
Statistical analysis
All data were expressed as means ± standard deviation (SD) and are representative of at least two separate biological experiments performed in triplicate. Statistical significance was determined by unpaired Student's t test. A value of p < 0.05 was considered statistically significant.
Results
Comparative sequence analysis of wild-type and codon-optimized NIS genes
Codon substitutions from wild type human NIS (hNIS) for sequence-optimized NIS (opt-hNIS) genes were performed based on highest usage frequencies in humans, and regions which might negatively influence human NIS protein expression were adjusted. RNA instability motifs, very high (>80%) or very low (<30%) GC content regions and cis-acting sequence motifs were also removed. GC content was adjusted to prolong mRNA half-life and the average GC content of opt-hNIS sequences was adjusted to 64%.
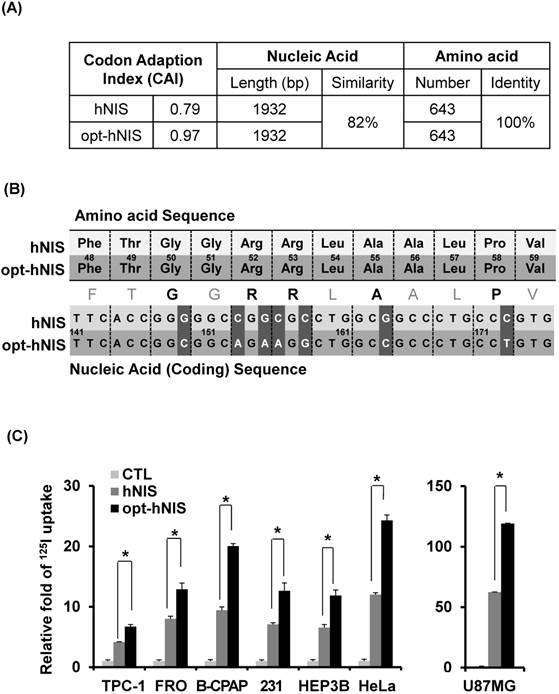
To investigate the general validity of codon optimization, the sequences of hNIS and opt-hNIS were analyzed (Figure 1A). The predictive level of gene expression by tRNA abundance in humans is described by the Codon Adaptation Index (CAI) [34]. The maximum possible CAI is set to 1, corresponding to the quality value of the most frequently used codon for a given amino acid in the desired expression system. The CAI for wild-type hNIS was verified as 0.79, but increased to 0.97 in opt-hNIS after codon optimization. The coding sequences of hNIS and opt-hNIS consisted of 1932 base pairs (bp) to produce 643 amino acids. Though both the hNIS and opt-hNIS genes encode the same protein with 100% identical amino acid sequence homology, they have only 82% similarity in terms of nucleic acid sequence. Figure 1B demonstrates sequence comparison of one specific portion (141 to 176 bp) of the nucleic acid sequence (FTGGRRLAALPV) of hNIS vs. opt-hNIS. For example, the 'GGG' codon in hNIS, for the 50th amino acid Gly of the NIS protein, is changed to 'GGC' in opt-hNIS to improve the expression of hNIS protein by introducing a more abundant codon for the human glycine tRNA. Despite these differences between the hNIS and opt-hNIS coding sequences, the amino acids produced are identical.
Comparison between the wild-type human NIS (hNIS) and codon-optimized (opt-hNIS) gene. (A) The predictive level of gene expression by tRNA abundance in humans is described by the Codon Adaptation Index (CAI). The maximum possible CAI is set to 1.0, corresponding to the quality value of the most frequently used codon for a given amino acid at the desired expression. Despite differences in coding sequence, the amino acids produced are identical. (B) Sequence comparison of a specific portion of the hNIS and opt-hNIS genes, i.e., amino acid sequence 48 to 59 (FTGGRRLAALPV) corresponding to nucleic acid sequence 141 to 176 bp. Bold characters in the amino acid sequence indicate amino acids with their codons changed after optimization. Dark gray blocks in the nucleic acid sequences indicate the location of the changed codons having high frequency in humans by optimization. (C) Increased radioiodine uptake in cancer cells by hNIS and opt-hNIS transfection. Various cancer cells such as thyroid (FRO, TPC-1, B-CPAP), breast (MDA-MB-231), liver (Hep3B), cervical (HeLa), and glioma (U87MG) were transiently transfected with pcDNA3.1/hNIS or opt-hNIS to evaluate functional changes of hNIS expression (P < 0.01, N=3).
Increased radioiodine uptake in various cancer cells by transient transfection of a codon-optimized hNIS gene
To evaluate the effect of codon optimization on the function of the NIS protein as an iodine transporter, we synthesized opt-hNIS and established a mammalian expression vector for transfection. Various cancer cells, such as TPC-1, FRO, B-CPAP (thyroid), MDA-MB-231 (breast), Hep3B (liver), HeLa (cervical), and U87MG (glioma), were transiently transfected with pcDNA3.1/hNIS or pcDNA3.1/opt-hNIS, and radioiodine uptake by those transfected cell lines was evaluated. Though each cell line had different transfection efficiencies, 125I uptake of opt-hNIS-expressing cells was significantly increased by 1.6- to 2.1-fold over uptake by wild-type hNIS-expressing cells (Figure 1C).
Increased levels of NIS protein and radioiodine uptake in codon-optimized hNIS-expressing cells
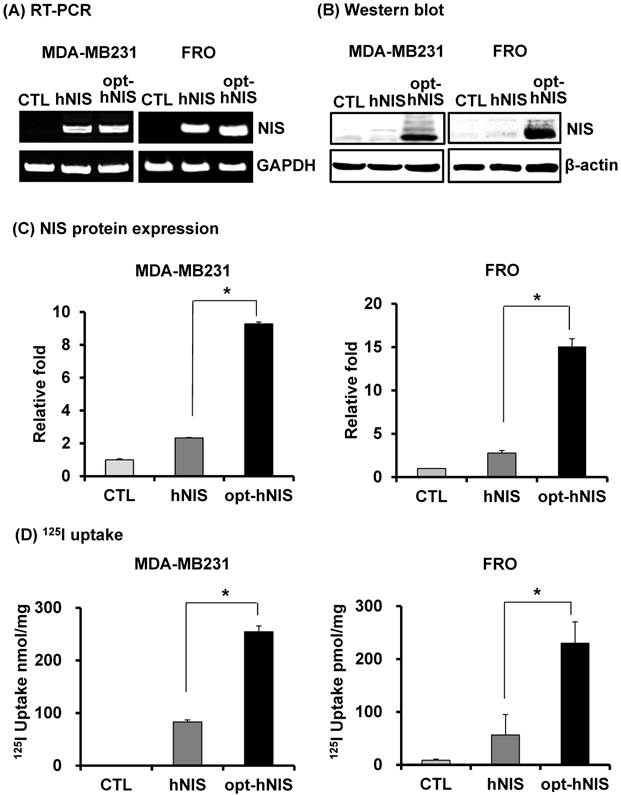
To remove the effect of variable factors from transient transfection such as transfection efficiency and copy number variation, stable FRO and MDA-MB-231 cell lines were established by retroviral transduction using pMSCV/hNIS or pMSCV/opt-hNIS. Since the natural NIS protein is located in the basolateral membrane of normal thyroid and lactating mammary glands, FRO and MDA-MB-231 cells were selected as representatives for thyroid and breast cancer. By real-time quantitative PCR, monoclonal FRO or MDA-MB-231 cells expressing similar number of hNIS mRNA transcripts were selected to evaluate functional hNIS production. NIS mRNAs were clearly detected at a similar level in both hNIS- and opt-hNIS-expressing cells (Figure 2A). The NIS protein appeared as multiple bands consisting of its major unglycosylated (72 kDa) and glycosylated (97 kDa or higher) forms in both hNIS- and opt-hNIS-transduced cells (Figure 2B). However, more NIS protein was produced in opt-hNIS-expressing cells than in hNIS-expressing cells. Quantitative analysis of western blot data verified that opt-hNIS protein expression increased 4- to 5.4-fold over that of wild-type hNIS (Figure 2C). 125I uptake by opt-hNIS-expressing cells was also 2.4 to 3.1 times higher than uptake by hNIS-expressing cells (Figure 2D).
Enhanced sensitivity of opt-hNIS as a reporter in vitro and in vivo
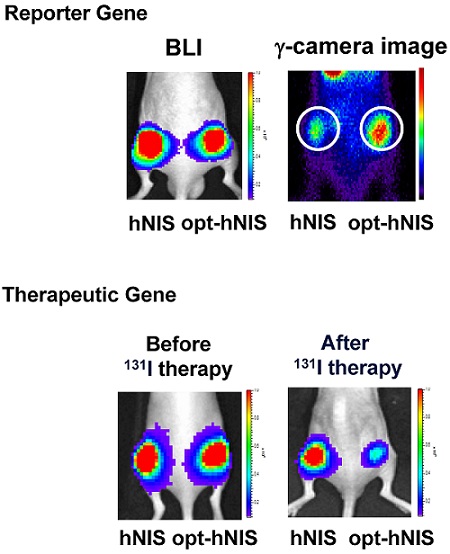
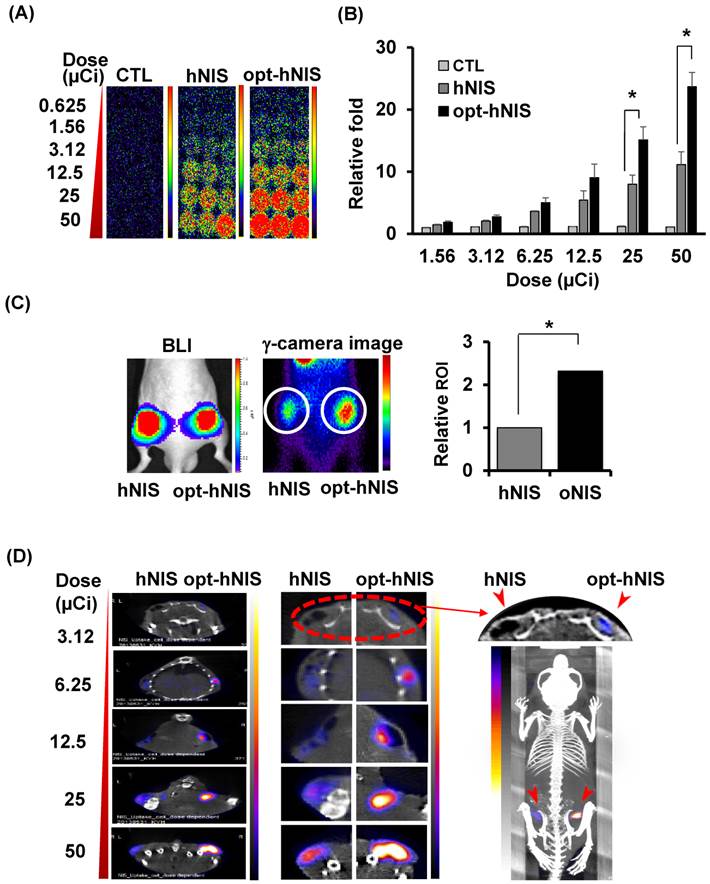
To evaluate the enhanced sensitivity of opt-hNIS as a reporter construct, 99mTc-pertechnetate uptake by either hNIS- or opt-hNIS-expressing cells was measured in vitro and in vivo. γ-camera imaging showed that 99mTc-pertechnetate uptake by both hNIS- and opt-hNIS-expressing cells increased in a dose-dependent manner. Particularly, the image signals from opt-hNIS-expressing cells were more sensitive than those from hNIS-expressing cells under the same dose of radiation exposure, requiring only 3.12 μCi of 99mTc-pertechnetate to clearly visualize opt-hNIS cells by γ-camera imaging (Figure 3A). Moreover, the amount of 99mTc-pertechnetate uptake by opt-hNIS-expressing cells was significantly higher than uptake by hNIS-expressing cells when they were exposed to 25 or 50 μCi of 99mTc-pertechnetate (Figure 3B).
For monitoring enhanced in vivo tumor uptake of 99mTc-pertechnetate, MDA-MB-231 cells expressing luciferase (MDA-MB-231/luc) with hNIS or opt-hNIS were implanted into the left and right thigh flanks, respectively, of nude mice. Two weeks later, tumor growth was measured by caliper and visualized by bioluminescence imaging, showing the sizes of the tumors expressing hNIS or opt-hNIS to be fairly similar. However, higher 99mTc-pertechnetate uptake by opt-hNIS xenografts (right flanks) was observed by γ-camera scintigraphic imaging. Quantitative analysis of specific regions of interest verified that the opt-hNIS xenografts showed significantly higher uptake (2.3-fold) than hNIS xenografts (Figure 3C). To visualize the enhanced sensitivity of opt-NIS for in vivo imaging, 106 cells of MDA-MB-231 expressing hNIS or opt-hNIS were exposed to different doses of 99mTc-pertechnetate and subcutaneously implanted into nude mice. In SPECT/CT imaging, opt-hNIS-expressing cells showed stronger 99mTc-pertechnetate signals than hNIS-expressing cells. Moreover, opt-hNIS-expressing cells (but not hNIS-expressing cells) treated with 3.12 µCi 99mTc-pertechnetate were clearly observed at the tumor injection site (Figure 3D).
Improved therapeutic efficacy of opt-hNIS in mouse xenograft models
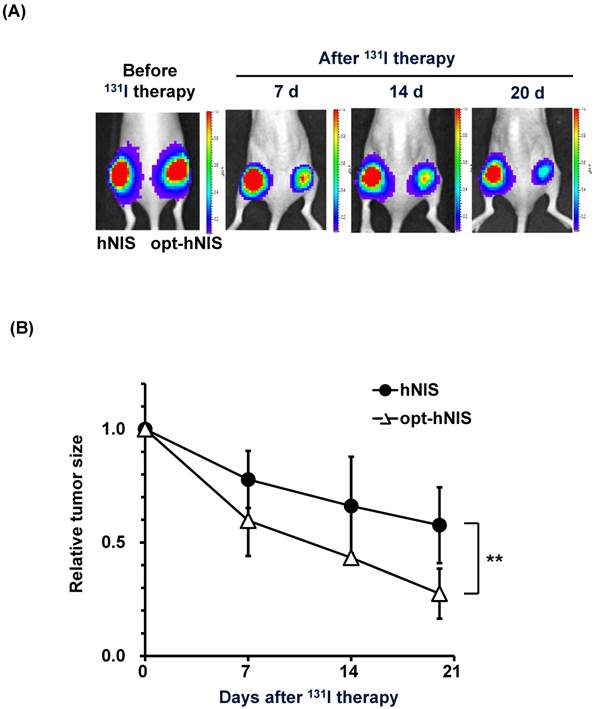
To monitor the therapeutic effects of codon optimization, tumors expressing hNIS and opt-hNIS genes were grown in nude mice. After challenge with 131I, the sizes of both hNIS- and opt-hNIS-expressing tumors decreased. However, the size of the opt-hNIS-expressing tumor (right flank) was more prominently reduced, compared to the hNIS-expressing tumor (left flank). The sizes of the hNIS and opt-hNIS tumors at 20 d after therapy were relatively decreased by 0.57- and 0.27-fold, respectively, compared to the sizes of the tumors before therapy (Figure 4).
Comparative analysis of NIS expression between cells expressing hNIS or opt-hNIS genes. (A) RT-PCR analysis of NIS transcripts in hNIS-/or opt-hNIS-expressing cells (MDA-MB-231 and FRO cells) established from monoclonal selections of retrovirus-mediated stably transduced cells. GAPDH was used as an internal control. Marked overexpression of hNIS mRNA was observed in both hNIS- and opt-hNIS-expressing cells. No expression was observed in the parental cells (CTL). (B) Western blot analysis of NIS protein in hNIS- or opt-hNIS-expressing cells. Multiple bands of glycosylated NIS were observed in both hNIS- and opt-hNIS-expressing cells, but more NIS protein was produced in opt-hNIS-expressing cells. (C) Quantitative analysis of protein expression in hNIS- or opt-hNIS-expressing cells. (D) 125I uptake assay in hNIS- or opt-hNIS-expressing cells. All experiments were performed in triplicate, and bars represent means ±SD (*, P<0.001).
Monitoring the sensitivity of hNIS- or opt-hNIS gene-expressing tumor cells in vitro and in vivo. (A) γ-camera images of cell culture plates. 99mTc-pertechnetate uptake increased in a dose-dependent manner in both hNIS- and opt-hNIS-expressing cells, but signals from opt-NIS-expressing cells were stronger than those from hNIS-expressing cells under the same dose of radiation. (B) The amount of 99mTc-pertechnetate uptake by opt-hNIS-expressing cells was significantly higher than uptake by hNIS-expressing cells. All experiments were performed in triplicate, and bars represent means ±SD (*, P<0.001). (C) In vivo tumor xenograft models were established using MDA-MB-231/luc cells expressing hNIS or opt-hNIS. Increased uptake of 99mTc-pertechnetate was observed in opt-hNIS xenografts, compared to hNIS xenografts (n=5; *, P<0.001). Regions of interest (ROIs) were drawn over tumor regions to quantify emitted luminescence. (D) SPECT/CT imaging of hNIS- or opt-hNIS-expressing tumors exposed to different doses of 99mTc-pertechnetate for one hour and subcutaneously implanted in a nude mouse. Only 3.12 μCi of 99mTc-pertechnetate was necessary to visualize cells expressing opt-hNIS in subcutaneous grafts.
Monitoring of therapeutic effects of xenografted tumors expressing hNIS or opt-hNIS. (A) Therapeutic effects of codon optimization were monitored by bioluminescence imaging. Tumors expressing hNIS and opt-hNIS were grown for 2 weeks in nude mice. After 131I therapy, both hNIS- and opt-hNIS-expressing tumor sizes were reduced, but the size reduction of the opt-hNIS-expressing tumor (right flank) was more prominent than that of the hNIS-expressing tumor (left flank). (B) Sizes of hNIS-expressing and opt-hNIS-expressing tumors after 131I challenge were measured with calipers. At 20 d after radioiodine therapy, tumor sizes relatively decreased by 0.57- and 0.27-fold for hNIS- and opt-hNIS-expressing tumors, respectively, compared to the tumor sizes before therapy (n=3, P<0.05).
Discussion
The use of an exogenous NIS gene as a reporter and a therapeutic gene, for thyroid cancer, has several advantages. First, various radiolabeled probes, such as radioiodines and Tc-99m, are available, and their metabolism is well understood. NIS seems to less perturb cells because iodide is not metabolized by most tissues, and no adverse effects have been observed. Because most nuclear medicine departments have easy access to a γ-camera, reporter gene imaging by NIS may be quite convenient. Unlike other nuclear reporter systems using radiolabeled ligands, such as Herpes Simplex Virus-thymidine kinase (HSV-tk), NIS has higher labeling stability because the NIS symporter directly uses radioiodine or Tc-99m. Another advantage of hNIS is that it is a less immunoactive human protein naturally expressed at high levels in the thyroid, stomach, and urinary tract. Previously, we established a transgenic mouse with myocardial cell-specific NIS expression, and did not find any significant injury or adverse effects on cardiac function, suggesting that exogenous NIS is safe in living subjects [36]. Since NIS is localized on cell surfaces, reporter probes can bind to cells easily. In addition, radioiodines and Tc-99m pertechnetate are highly selective for NIS-expressing cells and are rapidly cleared from background tissues not expressing NIS [3].
Expression of functional NIS protein, located on the surface of cellular membranes, enables cells to concentrate radioiodine from plasma, and the continuously active transport of radiotracers amplifies signals in NIS-expressing cells [6-8]. However, lower NIS protein levels lead only to partial radioiodine accumulation in cells for a short period of time. Indeed, the amount of NIS symporter expression is an important variable for using exogenous NIS as a reporter and therapeutic gene.
Several pharmaceutical drugs have been studied for enhancing radioiodine uptake by improving transcription efficiency [20-26]. However, for use of NIS as an exogenous reporter and therapeutic gene by ectopic expression, improving transcription is highly effective because most delivery vectors for exogenous gene expression have constitutive promoters with strong transcriptional activity. To additionally increase protein levels, codon optimization can improve translation efficiency by using codons most frequently used by host tRNAs [30, 31]. For this reason, we performed codon optimization of hNIS to increase NIS expression in humans for developing a more sensitive and efficient reporter gene.
A particular codon is used to specify a certain amino acid based on best codon usage for human expression, which was calculated by Wada et al from an analysis of 1490 human genes (601,683 codons) [35]. For example, we changed the codon for alanine residues from GCG to GCC because while 10.3% of alanine residues are specified by GCG, 41.6% of alanine residues are specified by GCC. The parameter Codon Adaption Index (CAI) [34] describes how well codons match the codon usage preference within the target organism. Codon usage of our opt-hNIS gene adapted to the bias of Homo sapiens resulted in a CAI value of 0.97, indicating that opt-hNIS should be highly expressed in human cells and tissues. Since the CAI of perfect codon usage is 1.0, a CAI of > 0.9 is generally considered very good [35]. Besides codon usage, GC content and regulatory sequences related to RNA stability can also optimize a DNA sequence to generate high levels of protein.
When exogenous NIS was expressed in a prokaryotic system, such as Escherichia coli, polypeptides having 50-kDa are produced [37]. In a eukaryotic system by contrast, the most abundantly observed NIS isoform was 72-kDa, with more glycosylated forms of NIS between 72-kDa to 130-kda also observed (Fig. 2D). Native forms of human NIS protein in eukaryotic systems are highly modified after protein synthesis. Glycosylation and phosphorylation especially are very important to localize NIS to the cellular membrane, a mandatory requirement for iodine uptake.
Synthesized opt-hNIS showed a greater level of NIS protein synthesis and enhanced radioiodine uptake, resulting in increased sensitivity and therapeutic response. Moreover, opt-hNIS similarly enhanced levels of signal intensity for imaging, using lower amounts of radioiodine, thus reducing radioiodine exposure. Effective tumor reduction by radioiodine therapy was also observed in opt-hNIS-expressing tumors, leading to prolonged survival in tumor-bearing mice.
While this study demonstrated an effective way to increase hNIS expression, there are still several issues to be resolved. Most importantly, a safe and reliable NIS gene delivery method must be developed. Though adenoviruses have many obstacles, they still represent a promising method of gene delivery. Since oncolytic viruses are able to selectively target and kill cancer cells rather than healthy cells, the combination of NIS gene-expressing oncolytic viruses and radioiodine therapies could have a synergistic effect on cancer treatment. However, potential toxicity and immune activation are other obstacles to be solved.
Conclusion
In this study, we established a codon-optimized human NIS (opt-hNIS) gene, and found that levels of NIS protein and radioiodine uptake were successfully enhanced. These findings demonstrate that opt-hNIS is a more sensitive reporter than wild-type hNIS, and thus opt-hNIS may be a more efficient therapeutic gene, and higher radioiodine uptake by opt-hNIS will improve the efficacy of iodine therapy.
Acknowledgements
This work was supported by the National Research Foundation (NRF) for the Global Core Research Center (GCRC), funded by Ministry of Science and ICT & Future Planning (MSIP) (No. 2011-0030680), and partly supported by grants (A101446, HI13C0826, HI14C1072) of the Korea Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea. This research also supported by grant no. 03-2010-0230 from the SNUH Research Fund.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chung JK. Sodium iodide symporter: its role in nuclear medicine. J Nucl Med. 2002;43:1188-1200
2. Filetti S, Bidart JM, Arturi F. et al. Sodium/iodide symporter: a key transport system in thyroid cancer cell metabolism. Eur J Endocrinol. 1999;141:443-457
3. Van Sande J, Massart C, Beauwens R. et al. Anion selectivity by the sodium iodide symporter. Endocrinology. 2003;144:247-252
4. Verburg FA, de Keizer B, Lips CJ. et al. Prognostic significance of successful ablation with radioiodine of differentiated thyroid cancer patients. Eur J Endocrinol. 2005;152:33-37
5. Hackshaw A, Harmer C, Mallick U. et al. 131-I activity for remnant ablation in patients with differentiated thyroid cancer: A systematic review. J Clin Endocrinol Metab. 2007;92:28-38
6. Chung J-K, Youn HW, Kang JH. et al. Sodium Iodide Symporter and the Radioiodine Treatment of Thyroid Carcinoma. Nuclear Medicine and Molecular Imaging. 2010;44:4-14
7. Klutz K, Willhauck MJ, Wunderlich N. et al. Sodium iodide symporter (NIS)-mediated radionuclide (131I, 188Re) therapy of liver cancer after transcriptionally targeted intratumoral in vivo NIS gene delivery. Hum Gene Ther. 2011;22:1403-1412
8. Kogai T, Brent GA. The sodium iodide symporter (NIS): regulation and approaches to targeting for cancer therapeutics. Pharmacol Ther. 2012;135:355-370
9. Kogai T, Taki K, Brent GA. Enhancement of sodium/iodide symporter expression in thyroid and breast cancer. Endocr Relat Cancer. 2006;13:797-826
10. Dai G, Levy O, Carrasco N. Cloning and characterization of the thyroid iodide transporter. Nature. 1996;379:458-460
11. Smanik PA, Liu Q, Furminger TL. et al. Cloning of the human sodium lodide symporter. Biochem Biophys Res Commun. 1996;226:339-34
12. Spitzweg C, Dietz AB, O'Connor MK. et al. In vivo sodium iodide symporter gene therapy of prostate cancer. Gene Ther. 2001;8:1524-1531
13. Dingli D, Russell SJ, Morris JC. In vivo imaging and tumor therapy with the sodium iodide symporter. J Cell Biochem. 2003;90:1079-1086
14. Chung JK, Kang JH. Translational research using the sodium/iodide symporter in imaging and therapy. Eur J Nucl Med Mol Imaging. 2004;31:799-802
15. Penheiter AR, Russell SJ, Carlson SK. The sodium iodide symporter (NIS) as an imaging reporter for gene, viral, and cell-based therapies. Curr Gene Ther. 2012;12:33-47
16. Grunwald GK, Klutz K, Willhauck MJ. et al. Sodium iodide symporter (NIS)-mediated radiovirotherapy of hepatocellular cancer using a conditionally replicating adenovirus. Gene Ther. 2013;20:625-633
17. Trujillo MA, Oneal MJ, McDonough S. et al. A steep radioiodine dose response scalable to humans in sodium-iodide symporter (NIS)-mediated radiovirotherapy for prostate cancer. Cancer Gene Ther. 2012;19:839-844
18. Penheiter AR, Wegman TR, Classic KL. et al. Sodium iodide symporter (NIS)-mediated radiovirotherapy for pancreatic cancer. AJR Am J Roentgenol. 2010;195:341-349
19. Vadysirisack DD, Shen DH, Jhiang SM. Correlation of Na+/I- symporter expression and activity: implications of Na+/I- symporter as an imaging reporter gene. J Nucl Med. 2006;47:182-190
20. Kogai T, Endo T, Saito T. et al. Regulation by thyroid-stimulating hormone of sodium/iodide symporter gene expression and protein levels in FRTL-5 cells. Endocrinology. 1997;138:2227-2232
21. Schmutzler C, Winzer R, Meissner-Weigl J. et al. Retinoic acid increases sodium/iodide symporter mRNA levels in human thyroid cancer cell lines and suppresses expression of functional symporter in nontransformed FRTL-5 rat thyroid cells. Biochem Biophys Res Commun. 1997;240:832-838
22. Furuya F, Shimura H, Suzuki H. et al. Histone deacetylase inhibitors restore radioiodide uptake and retention in poorly differentiated and anaplastic thyroid cancer cells by expression of the sodium/iodide symporter thyroperoxidase and thyroglobulin. Endocrinology. 2004;145:2865-2875
23. Park JW, Zarnegar R, Kanauchi H. et al. Troglitazone, the peroxisome proliferator-activated receptor-gamma agonist, induces antiproliferation and redifferentiation in human thyroid cancer cell lines. Thyroid. 2005;15:222-231
24. Hou P, Bojdani E, Xing M. Induction of thyroid gene expression and radioiodine uptake in thyroid cancer cells by targeting major signaling pathways. J Clin Endocrinol Metab. 2010;95:820-828
25. Yu KH, Youn H, Song MG. et al. The Effect of Tanespimycin (17-AAG) on Radioiodine Accumulation in Sodium-Iodide Symporter Expressing Cells. Nuclear Medicine and Molecular Imaging. 2012;46:239-246
26. Kogai T, Taki K, Brent GA. Enhancement of sodium/iodide symporter expression in thyroid and breast cancer. Endocr Relat Cancer. 2006;13:797-826
27. Norrman K, Fischer Y, Bonnamy B. et al. Quantitative comparison of constitutive promoters in human ES cells. PLoS One. 2010;5:e12413
28. Qin JY, Zhang L, Clift KL. et al. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS One. 2010;5:e10611
29. Schlabach MR, Hu JK, Li M. et al. Synthetic design of strong promoters. Proc Natl Acad Sci U S A. 2010;107:2538-43
30. Qian W, Yang JR, Pearson NM. et al. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012;8:e1002603
31. Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346-353
32. Angov E. Codon usage: nature's roadmap to expression and folding of proteins. Biotechnol J. 2011;6:650-659
33. Raab D, Graf M, Notka F. et al. The GeneOptimizer Algorithm: using a sliding window approach to cope with the vast sequence space in multiparameter DNA sequence optimization. Syst Synth Biol. 2010;4:215-225
34. Sharp PM, Li WH. The codon Adaptation Index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281-1295
35. Wada K, Wada Y, Ishibashi F. et al. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1992;20(Suppl):2111-8
36. Kang JH, Lee DS, Paeng JC. et al. Development of a sodium/iodide symporter (NIS)-transgenic mouse for imaging of cardiomyocyte-specific reporter gene expression. J Nucl Med. 2005;46:479-83
37. Dohan O, De la Vieja A, Paroder V. et al. The sodium/iodide symporter (NIS): characterization, regulation, and medical significance. Endocr Rev. 2003;24:46-47
Author contact
![]() Corresponding author: Hyewon Youn, Ph.D. Cancer Imaging Center, Seoul National University Hospital, #207-4, Samsung Cancer Research Bldg., 101 Daehak-ro, Jongno-gu, Seoul, Korea, 110-799 E-mail: hwyounac.kr; Tel: +82-2-3668-7026; Fax: +82-2-745-7690.
Corresponding author: Hyewon Youn, Ph.D. Cancer Imaging Center, Seoul National University Hospital, #207-4, Samsung Cancer Research Bldg., 101 Daehak-ro, Jongno-gu, Seoul, Korea, 110-799 E-mail: hwyounac.kr; Tel: +82-2-3668-7026; Fax: +82-2-745-7690.
 Global reach, higher impact
Global reach, higher impact