13.3
Impact Factor
Theranostics 2014; 4(12):1176-1192. doi:10.7150/thno.8715 This issue Cite
Review
MicroRNAs Associated With HBV Infection And HBV-related HCC
1. The Department of Liver Surgery & Liver Transplantation Centre, West China Hospital, Sichuan University, Chengdu, 610041, China;
2. The Department of Burns and Plastic Surgery, West China Hospital, Sichuan University, Chengdu, 610041, China;
3. The Department of Pharmacy, West China Hospital, Sichuan University, Chengdu, 610041, China.
Received 2014-1-29; Accepted 2014-8-10; Published 2014-9-19
Abstract
Hepatitis B virus (HBV) infection is a global problem and a major risk factor for hepatocellular carcinoma (HCC). microRNAs (miRNAs) comprise a group of small noncoding RNAs regulating gene expression at the posttranslational level, thereby participating in fundamental biological processes, including cell proliferation, differentiation, and apoptosis. In this review, we summarize the roles of miRNAs in HBV infection, the recently identified mechanism underlying dysregulation of miRNAs in HBV-associated HCC, and their association with hepatocarcinogenesis. Moreover, we discuss the recent advances in the use of circulating miRNAs in the early diagnosis of HCC as well as therapies based on these aberrantly expressed miRNAs.
Keywords: MicroRNA, Hepatitis B virus, Hepatocellular carcinoma, Diagnosis, Therapy.
Introduction
miRNAs comprise a class of highly conserved noncoding RNAs of approximately 22 nucleotides in length. miRNAs regulate the expression of various genes and have important roles in various cellular processes, including cell proliferation, differentiation, and apoptosis [1]. The primary miRNA transcripts (pri-miRNAs) are synthesized by RNA polymerase II and cleaved by RNase III Drosha to release precursor miRNAs (pre-miRNAs) [2]. This processing is completed in the nucleus, after which pre-miRNAs are exported from the nucleus to the cytoplasm by the nuclear receptor exportin-5 [3, 4]. After the transport of pre-miRNAs to the cytoplasm, they are processed by the RNase III Dicer to release the approximately 22-nucleotide RNA duplexes that contain mature miRNAs and the passenger miRNA strand [5, 6]. Mature miRNAs can interact with the 3′-untranslated regions (UTRs) of target mRNAs to form RNA-induced silencing complexes (RISCs), resulting in the inhibition of translation or mRNA cleavage [5, 7-9]. Recent studies revealed that miRNAs can regulate gene expression at the posttranscriptional level and have a role in regulating epigenetic machinery [10-12]. By targeting proteins involved in DNA methylation and histone modification, miRNAs control whether chromatin is in its accessible form. Given the important roles of miRNAs in various pivotal biological processes, it is not surprising that dysregulated miRNA expression has been implicated in a variety of human diseases, including chronic hepatitis B virus (HBV) infection and hepatocarcinogenesis [13].
HBV is a small, enveloped virus with a partly double-stranded relaxed circular DNA (rcDNA) of 3.2 kb in length. HBV is the prototype member of the Hepadnaviridae family, which has a strong preference for liver cells. After infecting hepatocytes, HBV rcDNA is released into the nucleus and repaired to form a covalently closed circular DNA (cccDNA), which serves as the template for the transcription of all viral RNAs. The HBV genome contains four overlapping open reading frames (ORFs). The preS-S region encodes the envelope proteins S (HBsAg), M (pre-S2), and L (pre-S1), which contributes to HBV binding to host-cell receptors and its release from host cells. The preC-C region encodes the hepatitis B core antigen (HBcAg) and hepatitis B e antigen (HBeAg). HBcAg is the structural polypeptide of the viral capsid, while the function of HBeAg is unclear. The X region encodes the HBV x protein (HBx), which regulates viral and host gene expression to facilitate viral replication. Finally, the P region encodes viral reverse DNA polymerase, which participates in DNA synthesis and RNA encapsidation. There are two enhancers (enhancer I and II) and four promoters that control the HBV transcription.
Globally, approximately 240 million people are chronically infected with HBV [14], and approximately 25% of chronically HBV-infected individuals eventually develop HCC. Moreover, approximately 60% of cases of HCC are associated with HBV infection in Africa and Asia. Mounting evidence indicates that dysregulation of miRNA has important roles in HBV infection and HBV-associated HCC. In this review, we discuss the HBV-miRNA interaction in HBV infection and HBV-associated carcinogenesis. We focus on how miRNAs act as regulators of cellular defense systems against HBV infection, how HBV modulates cellular miRNAs to facilitate its replication, and the alteration of miRNA signatures and their roles in HBV-related tumorigenesis. Moreover, the recent advances in miRNA-based diagnostic techniques and therapies, primarily the nanoparticle (NP)-mediated delivery of miRNAs for HCC therapy, are also reviewed and discussed.
Cellular miRNAs involved in the HBV-host interaction
A large number of cellular miRNAs can modulate HBV gene expression and replication in a negative or positive manner depending on their own physiological functions (Table 1). This is achieved by directly targeting viral transcripts, indirectly regulating the critical transcription factors essential for HBV gene expression and replication (Figure 1), and modulating the host immune system. While most miRNAs exert an inhibitory effect on HBV replication, several miRNAs, including miR-1, were reported to enhance HBV replication. Taken together, considering all the positive and negative effects, the miRNA machinery appears to have a net negative impact on HBV infection [15].
Hepatitis B virus (HBV) genome and cellular microRNA (miRNA) target sites. The partially double-stranded genome is presented with transcription regulatory elements. The viral open reading frames are shown as arrows immediately surrounding the genome. Viral enhancer I and II, promoters, DR1, and DR2 are indicated. The binding positions of transcription factors and nuclear receptors on the HBV genome are shown (C/EBP: CAAT enhancer-binding protein; CREB: C-AMP-response element binding protein; FXRA: farnesoid X receptor alpha; PPARα: peroxisome proliferator-activated receptor alpha; NFIB: nuclear factor I/B). Cellular factors known for regulating hepatitis B virus (HBV) transcription and the positions of the binding sequences in HBV transcripts targeted by miRNAs are shown. Arrows indicate activation, while the bars indicate an inhibitory effect of miRNAs on HBV transcription.
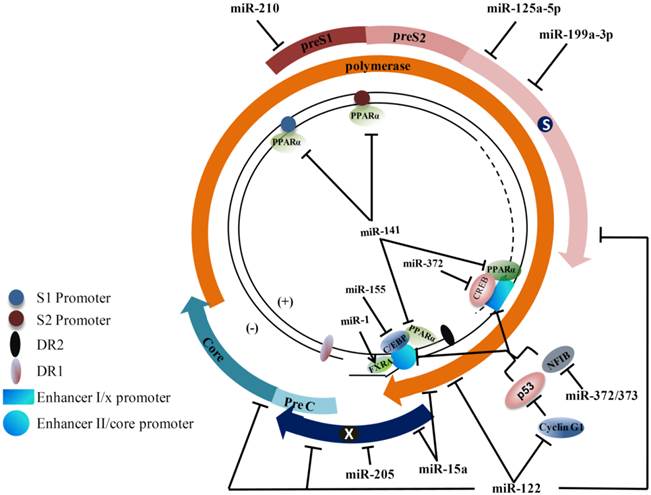
Dysregulated miRNAs involved in HBV infection.
| miRNA | Expression | Confirmed targets | Known Functions | Effects on HBV transcription and replication | Reference |
|---|---|---|---|---|---|
| miRNAs modulate host immune system | |||||
| miR-34a | Down | CCL22, Met | Metastasis (-) | Suppress HBV infection by inhibiting Tregs | [21] |
| miR-146a | Up | STAT1 | n/a | Promote HBV infection by suppressing T cell function | [20] |
| miR-155 | Up | SOCS1 | Proliferation (+) | Suppress HBV infection by augmenting IFN signaling pathway Suppress HBV transcription | [16] |
| miR-548 | Down | IFN-λ1 | n/a | Promote HBV infection by targeting the 3′UTR of IFN-λ1 | [17] |
| miRNAs target HBV transcripts | |||||
| miR-15a | Down | HBp and HBx mRNA | Proliferation (-) Apoptosis (+) | Suppress HBV infection by binding to HBx transcript | [15] |
| miR-17-92 | Up | HBV transcripts | Proliferation (+) | Suppress HBV infection by targeting HBV transcripts | [26] |
| miR-122 | Down | HBV mRNA | Proliferation (-), Invasion (-), Apoptosis (+) | Suppress HBV infection by targeting HBV mRNAs | [24, 66, 134] |
| miR125a-5p | Up | HBsAg mRNA | Proliferation (-) | Suppress HBV infection by interfere with the translation of HBsAg mRNAs | [27, 28, 135, 136] |
| miR199a-3p | Up | PAK4,mTOR MET, HBsAg S region | Proliferation (-) Liver fibrosis (+) Liver injury (+) | Suppress HBV infection by targeting sequences of HBsAg coding region | [23, 131, 137, 138] |
| miR-205 | Down | HBx mRNA | Proliferation (-) | Target HBx mRNA | [29] |
| miR-210 | Up | AIFM3, HBsAg pre-S1 region | Growth (+) | Suppress HBV infection by targeting sequences of HBsAg pre-S1 region | [23, 139] |
| miRNAs target regulators of HBV infection | |||||
| miR-1 | Down | HDAC4, MET | Proliferation (-) | Enhance HBV replication by augmenting FXRA Expression | [42, 43] |
| miR-22 | Up | HDAC4, ERα | Proliferation (-) | Inhibited HBsAg and HBeAg | [72, 86, 140] |
| miR-29c | Down | TNFAIP3 | Proliferation (-) Apoptosis (+) | Inhibit HBsAg/HBeAg secretion and HBV DNA replication | [141] |
| miR-122 | Down | Cyclin G1 HO-1 | Proliferation (-), Invasion (-), Apoptosis (+) | Promote HBV infection through inhibiting HO-1 expression | [38, 39] |
| miR-141 | n/a | PPARα | n/a | Represses HBV Replication | [36] |
| miR-155 | Up | C/EBP | Proliferation (+) | Suppress HBV infection by augmenting IFN signaling pathway Suppress HBV transcription | [32] |
| miR-152 | Down | DNMT-1 | Metastasis (-), Apoptosis (+) | [46] | |
| miR-372 | Up | NFIB | Liver cirrhosis (+) | Promote HBV expression | [35] |
| miR-501 | Up | HBXIP | n/a | Induced HBV replication | [40] |
(+): promotion; (-): inhibition; Up: upregulated; Down: downregulated; CCL22: chemokine (C-C motif) ligand 22; STAT1: signal transducer and activator of transcription 1; SOCS1: suppressor of cytokine signaling 1; C/EBP: CCAAT/enhancer binding protein; LKB1: liver kinase B1; PAK4: p21 protein (Cdc42/Rac)-activated kinase 4; mTOR: mechanistic target of rapamycin; AIFM3: apoptosis-inducing factor- mitochondrion-associated 3; PPARα: peroxisome proliferator-activated receptor alpha; NFIB: nuclear factor I/B; HO-1: heme oxygenase-1; HDAC: histone deacetylase; DNMT: DNA methyltransferase; ERα: estrogen receptor; TNFAIP3: tumor necrosis factor alpha-induced protein 3; HBXIP: hepatitis B virus x interacting protein; n/a: not available.
miRNAs modulate the host immune system during HBV infection
The host immune response is an important factor that partially determines whether HBV infection is cleared or maintained. In this setting, it is not surprising that HBV has developed strategies to suppress antiviral immune responses, thus facilitating its replication. miRNAs, whose roles in the immune system have been extensively studied since their discovery, were recently demonstrated to act as regulators of the immune response upon HBV infection [16, 17]. The results of several studies support the notion that mammalian organisms may, through the interferon (IFN) system, use miRNAs to defend against HBV infection. For instance, miR-155, by suppressing suppressor of cytokine signaling 1 expression and subsequently activating the JAK/signal transducer and activator of transcription (STAT) pathway, can enhance innate antiviral immunity and inhibit HBV gene expression. Accordingly, it is likely that the upregulation of miR-155 observed in HBV-infected patients with HCC may first serve to antagonize HBV infection; however, because of its oncogenic property, a persistent upregulation of miR-155 may finally contribute to hepatocarcinogenesis [16, 18]. Another miRNA, miR-548, unlike miR-155, can inhibit IFN-λ1 expression by targeting the 3′-UTR of IFN-λ1, thereby facilitating viral replication [17]. Interestingly, endogenous miR-548 levels are suppressed during HBV infection, and this could be a protective reaction of the host against viral infection. Although these miRNAs appear to play a role in antiviral defenses in humans, it is also argued that viruses may utilize these miRNAs to suppress their own replication to evade immune elimination and establish a persistent infection [19].
In contrast, some miRNAs may be employed by the virus to impair the immune system. A recent study by Wang et al. revealed that upregulated miR-146a in patients with chronic HBV infection directly targets STAT1 mRNA, leading to impaired T-cell function, which may contribute to immune defects during chronic viral infection [20]. miR-34a, the direct regulator of chemokine (C-C motif) ligand 22 (CCL22), is suppressed in the presence of the HBV genome. Decreased expression of miR-34a results in the recruitment of regulatory T cells (Tregs) through CCL22, a chemo-attractant for Tregs, subsequently downregulating the antiviral activity of effector T cells [21].
miRNAs directly target HBV transcripts
miR-32 was the first mammalian miRNA demonstrated to exert antiviral activity [22]. This miRNA can target ORF-2 of the primate foamy virus type 1, thus inhibiting viral mRNA translation. Increasing numbers of studies documented that miRNAs could also repress HBV gene expression or replication by directly targeting its gene transcripts. In an attempt to screen the targets of cellular miRNAs in HBV transcripts that affect HBV replication, two different procedures have been used in previous studies. One procedure involves screening miRNAs that can modulate HBV gene expression and then pairing selected miRNAs with HBV sequences using computer software to determine whether binding sites exist for these miRNAs [23-26]. To identify host-coded miRNAs influencing HBV replication and gene expression, Zhang et al. employed a loss-of-function approach by transfecting HepG2 2.2.15 cells with antisense oligonucleotides (ASOs) of 328 identified human miRNAs and then measured the expression level of HBsAg. The results illustrated that transfection of ASOs of miR-184, miR-185, miR-196a, miR-199a-3p, miR-210, and miR-217 caused a 1.4-1.6-fold increase in HBsAg expression [23]. Further study using bioinformatics analysis identified putative binding sites for miR-199a-3p and miR-210 in the HBsAg coding region and HBV pre-S1 region, respectively. In another study, Chen et al. first demonstrated the inhibitory effect of miR-122 on HBV gene expression and replication, and then used computer programs (RNA22, miRanda, and TargetScan algorithms) to predict a target of miR-122 within the overlapping region of HBV pre-C mRNA and pregenomic RNA. To verify whether miR-122 binds the target HBV RNA sequence via base pairing interactions, they introduced mutations in the “seed region” of miR-122 and its corresponding positions in the target sequence of HBV, and then evaluated the effects of wild-type miR-122 and mutated miR-122 molecules on wild-type-HBV and mutated HBV gene expression. The results demonstrated that the HBsAg/HBeAg expression of wild-type HBV was not repressed by synthetic mutated miR-122 mimics, and wild-type miR-122 mimics did not significantly affect the protein expression of the mutated HBV. However, the HBsAg/HBeAg expression of mutated HBV was inhibited by mutated miR-122 mimics, which contained compensatory mutations [24]. In a similar manner, two recent studies found that miR-15a and the miR-17-92 polycistron could inhibit HBV replication by targeting the viral transcripts [25, 26].
By contrast, the second procedure firstly predicts the potential targets of human miRNAs in HBV sequences using computer software, followed by validation of these predicted targets for responsiveness to human miRNAs. In a study by Potenza et al. the computer program miRanda was used to scan genomes of HBV subtypes for the presence of target sites of human miRNAs listed in miRBase. The researchers found 7 promising targets site clustered in the viral polymerase ORF and the overlapping surface antigen ORF. Among these miRNAs, hsa-miR125a-5p was validated to silence the 3037-3065 segment of the HBV genome and interfere with the translation of viral surface antigen mRNAs, thus reducing the amount of secreted HBsAg [27]. Hsa-miR125a-5p, in turn, is upregulated by the HBx protein [28]. miR-205 has been predicted to target HBx by target prediction software. However, there was no experimental evidence to support this prediction until a recent study demonstrated that HBx was indeed a direct target of miR-205 [29].
miRNAs target regulators of HBV infection
HBV transcripts are under the control of four promoters and two enhancers (enhancer I and II). A number of transcription factors and nuclear receptors have been shown to interact with promoter/enhancer elements to regulate HBV transcription. Some miRNAs have the ability to target these transcription factors, thus inhibiting HBV gene expression and replication. The CAAT enhancer-binding protein (C/EBP) can bind and enhance HBV enhancer II and core promoter, thereby promoting HBV transcription [30, 31]. miR-155, apart from regulating innate immunity, has been demonstrated to target and negatively regulate C/EBP, resulting in the downregulation of HBV transcription [32]. Protein kinase cAMP-dependent catalytic β can induce CAMP response element-binding protein (CREB) phosphorylation and cause CREB to dissociate from the HBV promoter [33]. Because CREB binding to the HBV promoter is required for HBV replication and gene expression [34], indirect phosphorylation of CREB by its regulator miR-372 eventually results in the downregulation of HBV transcription. Furthermore, peroxisome proliferator-activated receptor alpha (PPARα), which binds and transactivates HBV promoters, has a critical role in HBV replication [35]. A recent study showed that transfection with miR-141 mimics could repress PPARα expression at both the transcriptional and translational levels, which subsequently inhibited HBV promoter transcription activities and thereby suppressed HBV replication [36]. p53 can bind to HBV enhancer elements and act as a suppressor of HBV replication [37]. The inhibitory effect of p53 on HBV replication is protected by miR-122, which prevents cyclin G1 from interacting with p53 and blocking its specific binding to HBV enhancer elements [38]. However, heme oxygenase-1 (HO-1), an anti-HBV enzyme, is simultaneously targeted by miR-122; thus, miR-122 may also promote HBV infection by repressing HO-1-mediated antiviral activity [39].
In constrast, another group of miRNAs may target proteins that negatively regulate HBV replication. HBXIP, a HBX-binding protein, can reduce HBV replication by interacting with a domain necessary for HBX transactivation. However, miR-501 may negatively regulate HBXIP expression and facilitate HBV replication [40]. Nuclear factor I/B (NFIB), a transcription factor, inhibits the enhancer I and core promoter of HBV (ENI-Cp) on the HBV genome, thus reducing viral HBsAg and HBeAg protein levels as well as viral core-associated DNA levels. However, the antiviral activity of NFIB was abolished by its regulator-miR-372/373. Knockdown of NFIB by small interfering RNA in HepG2 cells resulted in increased HBV gene expression, whereas enhanced expression of miR-372/373 stimulated the production of HBV proteins [41].
Epigenetic modifications usually include DNA methylation and histone modification. Many miRNAs are suggested to be involved in epigenetic modifications to their target genes during HBV infection. miR-1 may act on histone deacetylase 4 (HDAC4) and thereby enhance HBV replication in an indirect manner [42, 43]. Furthermore, transfection with miR-1 suppresses HDAC4 protein expression in HepG2.2.15 cells. Consequently, the downregulation of HDAC4 results in changes in the expression of many transcription factors, including an upregulation of farnesoid X receptor a, which can bind to the HBV core promoter and enhance HBV transcription and replication [44]. Besides their involvement in histone modification, miRNAs also influence viral cccDNA methylation, thereby regulating HBV replication [13]. A study by Zhang et al. showed decreased miR-152 expression in the livers of HBX transgenic mice [45]. Further research demonstrated that miR-152 could target DNA methyltransferase-1 (DNMT-1) and reduce its expression [46]. Alterations in the expression of DNMTs have been indicated to affect HBV gene expression and replication by promoting viral DNA methylation [47].
miRNAs and HBV-associated hepatocarcinogenesis
Chronic HBV infection is a risk factor for HCC. Increasing evidence indicates that HBV alters host gene expression by upregulating or downregulating selected miRNAs, which finally promote the development of HCC [48] (Table 2). Connolly et al. screened the miRNA profiles of HBV-associated cirrhotic livers, HCC tissue, and matched normal liver tissue; elevated expression of the miR-17-92 polycistron, and miR-21 was observed in cirrhotic livers and HCC tissue [49]. Gao et al. investigated the expression changes of 7 cancer-related miRNAs during the early stages of HBV-associated hepatocarcinogenesis. They found a persistent downregulation of miR-145 and miR-199b as well as an upregulation of miR-224 associated with pre-malignant dysplastic nodules and the development of HCC [50]. Further research demonstrated that these miRNAs might be involved in HCC cell proliferation, migration, and invasion, and changes in their expression were early events in the development of HCC [50-53].
Aberrantly expressed miRNAs in HBV-related hepatocellular carcinoma and their cellular targets.
| miRNA | Expression | Confirmed targets | Upstream regulators | Involvement in cellular process | Reference |
|---|---|---|---|---|---|
| Let-7 | Down | STAT3, collagen type I a2, c-myc, NGF, Bcl-xL, RAS, HMGA2 | Lin28 | Proliferation (-), migration (-), apoptosis (+) | [28, 76, 90, 108, 132, 142-144] |
| miR-1 | Down | HDAC4, MET | n/a | Proliferation (-) | [42, 43] |
| miR-15a | Down | BCL-2 | n/a | Proliferation (-), apoptosis (+) | [15, 25, 48] |
| miR-16 | Down | Cyclin D1, NCOR2 | c-Myc | Proliferation (-), clonogenicity (-), apoptosis (+) | [48, 90] |
| miR-17-92 | Up | E2F1 | E2F3,c-Myc | Proliferation (-), anchorage-independent growth (-) | [49, 145] |
| miR-18a | Up | ERα | n/a | Proliferation (+) | [73] |
| miR-21 | Up | PTEN, PDCD4 | BMP2, BMP4,BMP6 | Proliferation (-), anchorage-independent growth (-), apoptosis (+), | [49, 68, 69] |
| miR-22 | Down | CDKN1A, ERα HDAC4 | n/a | Proliferation (-) | [72, 86, 140] |
| miR-23b | Down | uPA, MET | n/a | Proliferation (-), migration (-). | [97] |
| miR-26a | Down | ERα, IL-6 Cyclin D2 Cyclin E2 | hAFP-TERT | Proliferation (-),metastasis (-) | [146, 147] |
| miR-29a | Up | PTEN | n/a | Migration (+) | [99] |
| miR-29c | Down | TNFAIP3 | n/a | Proliferation (-), apoptosis (+) | [141] |
| miR-34a | Down | CCL22, MET | TGF-β | Metastasis (-) | [21, 96] |
| miR-99a | Down | IGF-1R, mTOR | n/a | Proliferation (-) | [126] |
| miR-101 | Down | DNMT3A, FOS, MCL-1, EZH2- | c-Myc EZH2- | Metastasis (-), apoptosis (+), Induce aberrant DNA methylation | [11, 62, 148-150] |
| miR-122 | Down | PBF, ADAM10 Cyclin G1, Igf1R ADAM17, BCL-W NDGR3 | HBV mRNAs, HNF1A, HNF3A, HNF3B | Proliferation (-), invasion(-), apoptosis (+) | [38, 66, 79, 93, 109, 110, 134, 151] |
| miR-143 | Up | FNDC3B | NF-κB | Metastasis (+) | [45] |
| miR-145 | Down | HDAC2, ADAM17 | n/a | Proliferation (-), invasion (-) | [85, 152] |
| miR-148a | Down | HPIP, MET | P53,Myc | Proliferation (-), metastasis (-) | [56, 98, 102] |
| miR-152 | Down | DNMT1 | n/a | Metastasis (-), apoptosis(+) Induce aberrant DNA methylation | [46] |
| miR-155 | Up | SOX6 | n/a | Proliferation (+) | [18] |
| miR-199a-3p | Down | PAK4, mTOR | Histone modification | Proliferation (-),apoptosis (+) | [103, 131] |
| miR-222 | Up | PPP2R2A | n/a | Metastasis (+) | [101] |
| miR-224 | Up | API-5 Smad4 | NF-κB | Proliferation (+), metastasis (+) | [52, 55, 153] |
(+): promotion; (-): inhibition; Up: upregulated; Down: downregulated; HMGA2: high mobility group AT-hook 2; E2F1: E2F transcription factor 1; HPIP: pre-B cell leukemia transcription factor-interacting protein; NCOR2: nuclear co-repressor 2; STAT3: signal transducer and activator of transcription 3; NGF: nerve growth factor; PTEN: phosphatase and tensin homolog; HDAC: histone deacetylase; ADAM: disintegrin and metalloprotease; PBF: pituitary tumor-transforming gene 1 binding factor; IGF1R: insulin-like growth factor 1 receptor; NDGR3: N-myc downstream-regulated gene; CDKN1A: cyclin-dependent kinase inhibitor 1A; ERα: estrogen receptor; PDCD4: programmed cell death 4; PAK4: p21 protein (Cdc42/Rac)-activated kinase 4; mTOR: mechanistic target of rapamycin; DNMT: DNA methyltransferase; TNFAIP3: tumor necrosis factor alpha-induced protein 3; API-5: apoptosis inhibitor 5; CCL22: chemokine (C-C motif) ligand 22; uPA: urokinase-type plasminogen activator; FNDC3B: fibronectin type III domain containing 3B; PPP2R2A: protein phosphatase 2A subunit B; SOX6: sex-determining region Y box 6; n/a: not available.
HBV induces dysregulation of miRNAs in HBV-related hepatocarcinogenesis
What is the cause of the dysregulation of miRNAs in HBV-associated HCC? Although the exact mechanism is not fully understood, the present findings indicate a critical role of HBV, especially HBx, in regulating these miRNAs (Figure 2). HBx is a 17-kDa protein encoded by the X region of the HBV genome, and it is implicated in cell proliferation, cell cycle progress, apoptosis, metastasis, and genetic stability [54]. HBx affects miRNA expression by acting on multiple steps of miRNA processing.
At the transcriptional level, HBx modulates the upstream transcriptional factors essential for miRNA transcription. p53, a well-known tumor suppressor that is frequently mutated in human cancers, can bind to promoters of miRNAs, such as miR-23a, miR-34, miR-125b, miR-148a, miR-192, and miR-200, thus upregulating these miRNAs. However, HBx can interact with and inactivate p53, leading to decreased expression of these miRNAs. In addition to p53, HBx induces or inhibits the expression of several other transcription factors, such as nuclear factor kappa B (NF-κB) and c-Myc, subsequently regulating miRNA expression [21, 26, 45, 48, 55, 56].
Aberrant epigenetic modifications to miRNA gene promoters by HBx represents another important mechanism underlying dysregulated miRNAs. Shen et al. analyzed 254 cytosine-phosphate-guanosine (CpG) sites that covered 110 miRNAs from 62 patients with HCC, most of who were HBV-infected. The results indicated that a total of 54 CpG sites from 27 host genes displayed marked differences in DNA methylation levels between tumors and adjacent nontumor tissues, with 53 sites being significantly hypermethylated in tumor tissues. Further research demonstrated that the aberrant DNA methylation status was due to the dysregulated expression of miR-10a and miR-196b, which served as regulators of the epigenetic machinery [57]. The PPARγ and retinoid X receptor alpha (RXRα)-binding complex can bind to the miR-122 gene promoter and enhance miR-122 transcription. DNA methylation possibly has a negative role in miR-122 expression, because treating HCC cells with DNA methylation inhibitors can increase the association of PPARγ or RXRα with the miR-122 gene promoter and upregulate miR-122 [58]. HBx-induced promoter hypermethylation is also involved in the regulation of miR-205 and miR-132 expression [12, 29]. In addition to methylation of CpG islands on miRNA promoters, HBx-induced histone modification also contributes to dysregulation of miRNAs. Reportedly, cellular histone acetyltransferases, such as p300, and histone deacetylases, such as HDAC1, are regulated by HBx [59]. A subsequent study found that p300 overexpression might be partially responsible for the upregulation of miR-224 expression observed in patients with HCC [60]. Meanwhile, HBx can upregulate enhancer of zeste homolog 2 (EZH2), a histone lysine methyltransferase, which epigenetically silences several anti-metastasis miRNAs, including miR-139-5p, miR-125b, miR-101, and let-7c [61, 62]. Moreover, the step in which the pri-miRNA is cleaved into a pre-miRNA is also affected by HBx. As previous studies indicated, HBx has a role in regulating some key proteins of the miRNA processing machinery, such as Drosha, to induce dysregulation of miRNAs [63, 64].
Viral proteins other than HBx have also been found to influence cellular miRNA profiles. Overexpressed HBsAg in HepG2 cells could upregulate 133 miRNAs and downregulate 9 miRNAs [65]. Furthermore, the HBx viral RNA also mediates dysregulation of some miRNAs. As previously mentioned, two known tumor suppressors, miR-15a and miR-16, are directly downregulated by the HBx RNA [15, 25]. Additionally, a recent study showed that all HBV mRNAs harbored a miR-122 complementary site, allowing these mRNAs to act as sponges to bind and sequester endogenous miR-122 [66].
In a more direct mode of action, because miRNA-encoding genes are located in fragile sites of the genome, frequent integration of HBV DNA into host chromosomes may increase chromosomal instability, leading to dysregulation of miRNAs. For instance, genes encoding miR-15 and miR-16 are located in the region of chromosome 13q that is frequently deleted in HBV-induced HCC [67].
miRNAs deactivate tumor suppressors in HBV-related hepatocarcinogenesis
Tumor suppressors are often inactivated in the initiation and progression of HCC through genetic and epigenetic mechanisms. However, miRNA-mediated silencing may be another important mechanism underlying the loss of tumor suppressors (Figure 3). For instance, miR-21 has been identified as an oncogenic miRNA by targeting programmed cell death 4 (PDCD4), a tumor suppressor that induces cell apoptosis. Upregulation of miR-21 by HBx suppresses PDCD4 and eventually promotes hepatocarcinogenesis [68, 69].
Hepatitis B virus (HBV) infection alters microRNA (miRNA) profiles by acting on multiple steps of miRNA processing. At the transcriptional level, the HBV x protein (HBx) can interact with some transcription factors such as p53, nuclear factor-kappa B (NF-κB), and c-Myc, subsequently regulating miRNA transcription. HBx can also regulate the epigenetic machinery, which can determine whether the miRNA genes are accessible. Moreover, HBx may influence miRNA processing by inhibiting Drosha, which serves as a processor of the miRNA machinery. Finally, when mature miRNAs are released, they may be counteracted by HBV mRNAs, which act as sponges of endogenous miRNAs, forming the miRNA-induced silencing complexes (miRISC). Arrows indicate an activating effect of HBx on other transcriptional factors and proteins, whereas the bars indicate an inhibitory effect. The red crosses indicate an inhibitory effect on gene transcription.
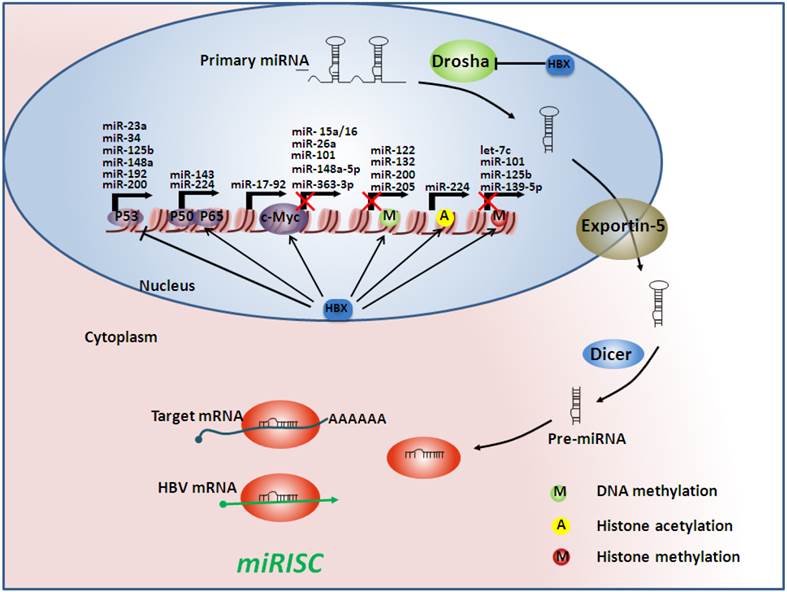
Aberrant microRNAs and their targets involved in HBV-related hepatocarcinogenesis. HMGA2: high mobility group AT-hook 2; E2F3: E2F transcription factor 3; STAT3: signal transducer and activator of transcription 3; PTEN: phosphatase and tensin homolog; HDAC: histone deacetylase; PBF: pituitary tumor-transforming gene 1 binding factor; CDKN2A: cyclin-dependent kinase inhibitor 2A; ERα: estrogen receptor; PDCD4: programmed cell death 4; DNMT: DNA methyltransferase; GSTP1: glutathione S-transferase pi; RASSF1: Ras association (RalGDS/AF-6) domain family member 1; CDH1: E-cadherin 1; TSLC1: tumor suppressor in lung cancer-1 gene; EZH2: enhancer of zeste homolog 2; FUT2: fucosyltransferase 2. Arrows indicate an activating effect of miRNAs on their targets, whereas the bars indicate an inhibitory effect. Green miRNA: upregulated; Red miRNA: downregulated.
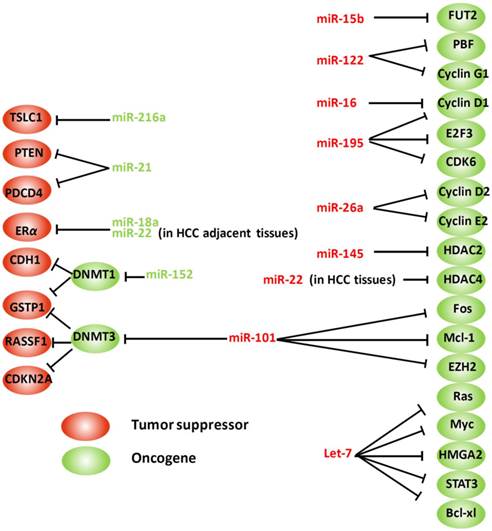
Estrogen receptor α (ERα) protects females from hepatocarcinogenesis through the estrogen signaling pathway, which partially explains the sex disparity characteristic in HCC [70, 71]. ERα is significantly downregulated in HCCs, potentially because of overexpression of some miRNAs, such as miR-22 and miR-18a, which directly target ERα, thereby attenuating the protective effects of estrogens [72, 73]. By contrast, overactivation of the androgen pathway functions as a tumor-promoting factor in hepatocarcinogenesis in males. Chen et al. documented that androgen receptor (AR) expression was significantly elevated in HBV-related male HCC liver tissues, suggesting a high activity of androgen pathway in these patients. AR, as a transcription factor, activates its target genes through binding with the corresponding androgen response element (ARE) residues within the promoter regions. An ARE has also been identified in the promoter of miR-216a. Thus, activation of the androgen pathway leads to the upregulation of miR-216a, which acts as oncogenic miRNA by targeting the mRNA of the tumor suppressor in lung cancer-1 gene (TSLC1) [74].
miRNAs permit oncogene expression in HBV-related hepatocarcinogenesis
The upregulated expression of oncogenes is another important factor that contributes to hepatocarcinogenesis. In previous studies, we identified a cluster of miRNAs that upon downregulation are directly responsible for the upregulation of some oncogenes (Figure 3). For instance, let-7, the second miRNA discovered, has been shown to regulate the expression of a cluster of oncogenes, including c-Myc, RAS, high mobility group AT-hook 2, and STAT3 [28, 75-77]. However, in the presence of HBx, let-7 expression is suppressed, resulting in overexpression of its target oncogenes and the promotion of hepatocarcinogenesis [28].
The pituitary tumor-transforming gene 1 (PTTG1) binding factor (PBF) promotes the transcriptional activities of PTTG1 by facilitating its nuclear translocation. This in turn enhances the expression of the target genes of PTTG1, such as VEGF, c-Myc, and MMP-2, leading to HCC cell growth and invasion. Normally, the expression of PBF is suppressed by miR-122, the dominant miRNA in the liver. However, with the downregulation of miR-122 in HCC, PBF may be ectopically expressed because of the relief of repression [66]. The expression of miR-15b is suppressed by HBx, which permits the expression of the targeted gene of miR-15b, fucosyltransferase 2 (FUT2). Upregulated FUT2 then activates its downstream protein Globo H, resulting in HCC cell proliferation [78]. These results suggest that HBx promotes tumorigenesis by employing miRNAs as its agents.
miRNAs can also interact with key cell cycle regulators and regulate cell cycle progression. Cyclin G1 is regulated by miR-122, and downregulation of miR-122 has been shown to accelerate the cell cycle [79]. The miR-16 family can target cyclin D1 and thereby arrest the cell cycle in the G1 phase [48]. miR-26a and miR-195 can block G1/S transition by repressing multiple molecules, such as cyclin D1, cyclin D2, cyclin E2, CDK6, and E2F transcription factor 3 [80, 81]. Therefore, the downregulation of these miRNAs causes defects in cell cycle control, resulting in the development and progression of HCC.
Abnormal epigenetic control is a common early event in HCC progression. In addition to the epigenetic regulation of miRNA expression by HBx, many miRNAs themselves can regulate the epigenetic machinery and epigenetically silence some tumor suppressors, subsequently promoting hepatocarcinogenesis [82, 83]. miR-152 has been shown to be involved in epigenetic modification by regulating DNMT1 (maintenance DNMT). Compared with its expression in adjacent noncancerous liver tissues, miR-152 is frequently downregulated in HBV-related HCC tissues. Reduced expression of miR-152 results in elevated expression of its target gene DNMT1, which then increases methylation levels of two tumor suppressor genes, glutathione S-transferase pi 1 (GSTP1) and E-cadherin 1 [46]. DNMT3A, a DNA methyltransferase, is regulated by miR-101, an miRNA, which when suppressed may cause abnormal DNA methylation of several tumor-suppressor genes, including GSTP1, Ras association (RalGDS/AF-6) domain family member 1, and cyclin-dependent kinase inhibitor 2A, in HCC cells [11]. Moreover, EZH2, a histone lysine methyltransferase, is also a direct target of the tumor suppressor miR-101 [84].
Meanwhile, HDACs (enzymes that remove acetyl groups from histone proteins) are proposed to act as oncogenes by inhibiting tumor suppressor gene transcription. HDAC2 is regulated by miR-145 [85], and HDAC4 is a target of miR-22 [86]. Loss or suppressed expression of these miRNAs results in overexpression of histone deacetylases and the promotion of HCC tumorigenesis.
Additionally, miRNAs can also modulate the expression of genes participating in the regulation of protein phosphorylation, which usually results in the activation of a signaling pathway involved in tumor onset and progression. NF-κB-inducing kinase (NIK) is a member of the mitogen-activated protein kinase family that may either directly or indirectly phosphorylate or activate the IκB kinase α/β, leading to the phosphorylation and degradation of IκBα, followed by NF-κB activation. Zhang et al. demonstrated that NIK was a direct target of miR-520e; thus, the reduced expression of miR-520e may be partially responsible for the activation of the NF-κB pathway in HCC [87]. Another example is that miR-637 may suppress autocrine leukemia inhibitory factor (LIF) expression and exogenous LIF-triggered STAT3 phosphorylation, subsequently promoting the proliferation and metastasis of HCC [88].
miRNAs and the metastasis of HBV-related HCC
Tumor metastasis is one of the most significant processes that affect the clinical management and long-term survival of patients with HCC. Investigators have identified miRNAs that may be involved in HCC metastasis using miRNA expression screening (Table 2). Budhu et al. examined the miRNA expression profiles of 482 cancerous and noncancerous specimens of 241 patients with HCC, and they built a unique 20-miRNA metastasis signature that could significantly predict primary HCC tissues with venous metastasis from metastasis-free solitary tumors. Furthermore, these miRNAs showed a significant correlation with patient survival and disease relapse [89]. By comparing the sequence information and parallel miRNA and mRNA expression data between patients with HBV-related HCC with or without venous metastasis, Zeng et al. identified miR-16, miR-30a, let-7e, and miR-204 as key miRNA regulators that contribute to HBV-related HCC metastasis [90].
miRNAs and epithelial-mesenchymal transition (EMT) in HBV-related HCC
The EMT process, in which polarized immotile epithelial cells are converted into motile mesenchymal cells, is frequently activated and implicated in the promotion of tumor invasion and metastasis [91]. EMT is characterized by the loss of cell adhesion, downregulation of epithelial markers (E-cadherin, occludins, and claudins), and upregulation of mesenchymal markers (vimentin, fibronectin, N-cadherin). Several oncogenic pathways, including the transforming growth factor-β (TGF-β), Wnt, and receptor tyrosine kinase (RTK)-mediated pathways, have been shown to induce EMT. Meanwhile, numerous miRNAs are involved in the regulation of these pathways to regulate EMT-related HCC metastasis (Figure 4).
TGF-β signaling
TGF-β is a multifunctional factor that has critical roles in HBV-related liver pathogenesis, including HCC metastasis [92]. Growing evidence indicates that TGF-β promotes tumor metastasis by activating its downstream mediators, which usually leads to EMT. Ras homolog family member A (RHOA) is a crucial downstream mediator for TGF-β-mediated EMT. It has been reported that miR-122 directly targets RHOA, and that the downregulation of miR-122 will relieve RHOA, which greatly promotes the migration and invasion of cancer cells through cytoskeletal rearrangements [93]. Moreover, miR-140-5p can suppress tumor metastasis by directly targeting the TGF-β receptor 1 [94]. Zinc finger E-box 1 (ZEB1) and ZEB2 are two downstream transcription factors of TGF-β signaling. They are involved in TGF-β-mediated EMT by inhibiting E-cadherin transcription and promoting vimentin transcription. Kim et al. documented that p53 regulated EMT by repressing the expression of ZEB1 and ZEB2 in HBV-related HCC, and this regulation was mediated by miR-200 and miR-192 family members, which directly targeted ZEB1 and ZEB2, respectively [95].
Involvement of microRNAs (miRNAs) in epithelial-mesenchymal transition (EMT)-related metastasis. miRNAs can regulate EMT by acting on several EMT-related pathways, including transforming growth factor-β (TGF-β) signaling, receptor tyrosine kinase (RTK)-mediated pathways, and Wnt/β-catenin signaling. miR-140-5p can directly target TGF-β receptor 1, miR-122 can target Ras homolog family member A (RHOA), and miR-192 and miR-200 can negatively regulate zinc finger E-box 1 (ZEB1) and ZEB2, respectively, thus inhibiting TGF-β-mediated metastasis. The RTK-related pathways, including the Ras-Raf-MEK-ERK cascades and phosphatidylinositol 3-kinase (PI3K)-Akt pathway, are also regulated by miRNAs. miR-1, miR-23b, miR-34a, and miR-148a can target c-Met, a RTK for hepatocyte growth factor (HGF). MiR-199a/b-3P can target p21 protein (Cdc42/Rac)-activated kinase 4 (PAK4), subsequently inhibiting Raf-MEK-ERK cascades. On the contrary, phosphatase and tensin homolog (PTEN) can be negatively regulated by miR-21 and miR-29a, and miR-222 and miR-222 simultaneously target PP2A. miR-148a can also negatively regulate pre-B cell leukemia transcription factor-interacting protein (HPIP), an activator of both ERK and AKT. Moreover, miR-148a can target Wnt1 mRNA, thus blocking EMT mediated by Wnt/β-catenin-signaling in hepatocellular carcinoma. EGF: epidermal growth factor; FGF: fibroblast growth factor. Arrows indicate an activating effect of miRNAs on their targets, whereas the bars indicate an inhibitory effect. Green miRNA: upregulated; Red miRNA: downregulated.
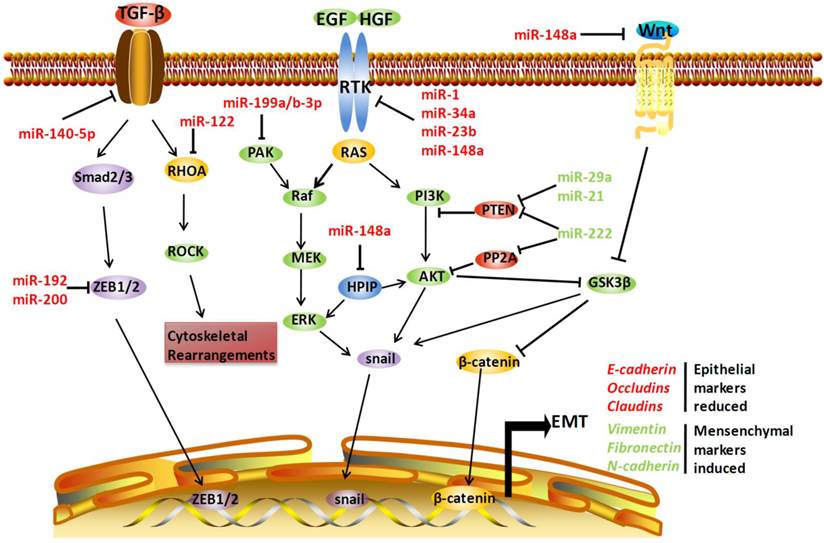
RTK-mediated pathways
c-Met is a RTK for the hepatocyte growth factor (HGF). Activation of HGF by c-Met will induce downstream signaling, including MAPK pathway activation and inhibitory phosphorylation of glycogen synthase kinase 3β (GSK-3β), which consequently increases the nuclear accumulation of Snail, a transcription factor that promotes EMT. c-Met is frequently upregulated in many cancers including HCC, which has recently been demonstrated to be caused by dysregulation of several miRNAs, including miR-1, miR-34a, miR-23b, and miR-148a. These miRNAs are downregulated in HBV-related HCC tissue, and they can directly target c-Met and inhibit EMT in hepatoma cells [43, 96-98].
The phosphatidylinositol 3-kinase (PI3K)/AKT signaling pathway is another component of RTK-mediated pathways that promotes EMT-related metastasis. Normally, PI3K/AKT signaling is negatively regulated by phosphatase and tensin homolog (PTEN) and protein phosphatase 2A (PP2A). Silencing of PTEN and PP2A will activate the PI3K/AKT pathway and promote metastasis. In HBV-related HCC, upregulated miR-29a, miR-21, and miR-222 may target PTEN [68, 99, 100], and miR-222 can simultaneously target PP2A [101], leading to tumor cell metastasis. Moreover, miR-148a may regulate PI3K/AKT signaling by targeting hematopoietic pre-B cell leukemia transcription factor-interacting protein, which activates AKT and ERK and thus promotes EMT [102]. The inhibitory effect of miR-148a on EMT was validated by another study, which verified that Met/Snail signaling is a downstream signaling pathway regulated by miR-148a [98].
In addition, RTK transduction also activates downstream Raf/MEK/ERK signaling cascades. However, a study by Hou et al. showed that miR-199a/b-3p could regulate the Raf/MEK/ERK pathway by targeting p21 protein (Cdc42/Rac)-activated kinase 4, a direct regulator of Raf, thus suppressing HCC progression [103].
Wnt/β-catenin signaling
Another pathway that mediates EMT in HCC is Wnt/β-catenin signaling. Activation of Wnt/β-catenin signaling inhibits GSK-3β, which phosphorylates β-catenin, targeting the protein for degradation. A recent study demonstrated that miR-148a could target Wnt1 mRNA, thus blocking EMT of HCCs [104].
Some of the EMT markers are directly or indirectly regulated by miRNAs in other ways. miR-373 acts as a positive regulator of the expression of E-cadherin, an epithelial marker discussed previously. Downregulation of miR-373 by HBx has been found to be correlated with repressed expression of E-cadherin in both HBx-expressing cells and tissue sections from HBV-infected patients, which subsequently leads to EMT [105]. Moreover, miR-9 can downregulate KLF17 protein expression by directly targeting the 3′-UTR region of the KLF17 gene. As a transcription factor, KLF17 may interact with the promoters of EMT-related genes (vimentin and fibronectin) in HCC cell lines and enhance EMT-related metastasis [106]. Another study reported that overexpression of miR-194 suppressed the invasion and migration of mesenchymal-like cancer cells by suppressing the expression of N-cadherin [107].
miRNAs regulate metastasis-related genes
Microarray analysis has identified sets of genes that are associated with HCC invasion and metastasis. miRNAs may act as upstream regulators of these genes and play important roles in HCC metastasis. let-7g acts as a suppressor of HCC metastasis by targeting collagen type I a2, a soluble collagen that contributes to HCC cell invasion and metastasis [108]. Zhang et al. identified the upregulation of miR-143 by NF-κB in HBV-related HCC. Upon upregulation, miR-143 suppressed the expression of fibronectin type III domain containing 3B, a regulator of cell motility, and thereby promoted HCC invasion and metastasis [45]. Loss of miR-122 expression in liver cancer has also been found to be correlated with the metastatic properties of HCC [93]. This may be partially caused by the upregulation of a disintegrin and metalloprotease 17 (ADAM17) and ADAM10, two oncogenes that are targeted by miR-122 and involved in regulating cell adhesion, migration, and metastasis [109, 110].
miRNAs can also participate in immune-suppressive Foxp3(+) Treg recruitment, thereby creating a repressed immune surveillance microenvironment that facilitates the intrahepatic metastasis of HCC. In a mouse model of acute HBV infection, initiation of HBV replication is accompanied by increased numbers of Tregs in the liver. These Tregs can downregulate the antiviral activity of effector T cells and suppress the recruitment of innate immune cells, thereby limiting immune-mediated liver damage in the phase of acute infection and simultaneously promoting persistent HBV infection [111]. When HBV-infected patients develop HCC, the recruitment of Tregs may facilitate the immune escape of HCC cells, leading to the formation of intrahepatic metastasis. Yang et al. found that this pathologic process was mediated by a miRNA, miR-34a. The presence of the HBV genome could cause suppression of miR-34a expression, subsequently relieving the expression of its target,CCL22, a chemo-attractant for Tregs, and promoted intrahepatic metastasis [21].
Diagnostic and therapeutic potential of miRNAs
miRNAs in the diagnosis of HBV infection and HBV-related HCC
It is proposed that cellular miRNAs may leak from cells followingcell injury and death, reach the blood circulation and stably exist in serum. It is hypothesized that the circulating miRNAs, in addition to functioning within cells, may be secreted by specific tissues or cells to act as “messengers” and mediate cell-cell communication. This notion is supported by a recent study that showed that circulating miRNAs were taken up and then bound by Toll-like receptors of recipient cells to induce a prometastatic inflammatory response [112]. In this setting, the serum miRNA profile may reflect some pathological conditions, including chronic HBV infection and HBV-related HCC. Ji et al. compared miRNA expression profiles in HBV-infected serum with that in control serum, and identified 9 differentially expressed miRNAs in HBV-infected serum. Among these miRNAs, the expression of liver-specific miR-122 was significantly upregulated. Further research indicated that miRNA-122 serum concentrations could help in differentiating individuals with or without HBV infection as well as inactive carriers with high (>3500 IU/mL) or low (<3500 IU/mL) HBsAg levels [113]. Additionally, miR-122 serum levels have been found to be correlated with alanine aminotransferase levels, HBV DNA expression, and liver injuries [114]. These results suggest that serum miRNAs may serve as biomarkers for diagnosis of HBV infection, particularly in patients with occult hepatitis B virus infection (OBI). OBI is characterized by the presence of HBV infection without detectable HBsAg in the serum, and >20% of OBI carriers are negative for all serum markers of HBV infection [115]. Current blood screening practices largely rely on HBsAg detection, and the detection of HBV DNA does not always indicate infectivity. Thus, the presence of OBI may increase the risk of HBV infection during blood transfusion processes and organ transplantation [116]. Nevertheless, aberrant serum miRNA profiles provide clues for the identification of these types of patients. Chen et al. identified 4 miRNAs, let-7c, miR-23b, miR-122, and miR-150, which were differentially expressed in serum from patients with OBI compared with that from controls. These miRNAs had a high accuracy (area under the curve, AUC = 0.999) in discriminating patients with OBI from those without OBI [117].
The early diagnosis of HCC is critically important for achieving a good prognosis. Currently, the diagnosis of HCC mainly relies on α-fetoprotein (AFP) and imaging technology, including abdominal ultrasonography, computed tomography, and magnetic resonance imaging [118]. These imaging techniques can detect HCC lesions as small as 1 cm in diameter. However, their diagnostic accuracy depends on the tumor size and vascularity, and the associated high cost makes them unsuitable as routine screening tools. AFP has long been used in the diagnosis of HCC and prediction of recurrence. However, its sensitivity is low (approximately 30%), and its use for early HCC detection is limited. There is an urgent need to develop a high-validity, accurate, and cost-effective method for the early diagnosis and recurrence surveillance of HCC. Recent studies indicated that miRNA profiles might represent a new modality for this purpose [118]. To identify miRNAs of clinical value for HBV-related HCC diagnosis, the first step is to establish a miRNA signature that is specific for HBV-related HCC. Li et al. compared the miRNA expression profiles of HBV-infected serum with those of control serum, and they identified 13 differentially expressed miRNAs in HBV-infected serum. This serum miRNA-based biomarker profile could separate patients with HBV-positive HCC from HBV-infected, HCV-infected, and control patients, with only 3 of 65 patients with HBV-positive HCC being incorrectly classified. Furthermore, they employed an age-, gender-, and etiology-matched control group and identified 6 miRNAs that were significantly upregulated in HCC samples. Three of these 6 serum miRNAs could help to clearly separate the HBV-related HCC group from the control group, displaying an AUC of 99.6% with a sensitivity and specificity of 97.9% and 99.1%, respectively [119]. In another study, Tomimaru et al. investigated miR-21 levels in patients with HCC and chronic hepatitis and compared miR-21 levels in patients with HCC before and after curative resection. The results showed a significant reduction in miR-21 levels in postoperative compared with preoperative plasma samples. Meanwhile, miR-21 levels in patients with HCC were significantly higher than those in patients with chronic hepatitis, exhibiting an AUC of 0.773 with a sensitivity and specificity of 61.1% and 83.3%, respectively. Moreover, the authors suggested that the use of plasma miR-21 combined with AFP as a biochemical marker might be useful for HCC detection [120]. These two studies determined that some miRNAs could help in differentiating patients with HCC from those with viral hepatitis. However, both studies lacked validation of the diagnostic accuracy of these miRNA signatures in independent patient cohorts. External validation of each signature is an absolutely essential step for identifying miRNAs that are valuable for clinical use. Zhou et al. used microarray analysis to screen 723 miRNAs that were differently expressed in plasma samples obtained from patients with HBV-related HCC compared with those obtained from healthy individuals, patients with chronic hepatitis virus infection, and patients with cirrhosis. Fifteen candidate miRNAs were identified in this phase and further tested in an independent cohort of plasma samples from 407 patients. From these data, 7 miRNAs were selected and further used to predict the probability of an HCC diagnosis in an independent validation cohort comprising plasma samples from 390 patients. The results showed that the identified miRNA panel (miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a, and miR-801) could provide a high diagnostic accuracy (AUCs for Barcelona Clinic Liver Cancer stages 0, A, B, and C were 0.888, 0.888, 0.901, and 0.881, respectively) for HCC regardless of disease stage [121]. This study is more informative than those of circulating miRNAs in diagnosing HCC, because it additionally included a cirrhosis group and compared the diagnosis accuracy for different tumor staging groups.
Collectively, these data illustrate the potential application of circulating miRNAs as biomarkers for HCC diagnosis. However, some critical points must be considered before their use can be translated into clinical practice. First, a strategy for obtaining reproducible results in different study groups must be established. Apart from a comparable and appropriate sample, the experimental strategies should also be improved. Another issue is that more data is required from early-stage patients to facilitate early diagnoses, requiring collection and analysis of more early-stagesamples. Finally, for patients who undergo curative resection for HCC, there is still a high risk of recurrence. Thus, identifying a serum miRNA profile that could help predict in advance the recurrence of HCC in these patients would also be useful.
miRNAs in the treatment of HBV-related HCC
In addition to the diagnostic implications, the specific signatures of aberrantly expressed miRNAs also have promising potential in HCC therapy. Novel therapeutic strategies based on modulating miRNA function are being devised to downregulate the expression of miRNAs that act as oncogenes and upregulate the expression of miRNAs that exert tumor suppression. However, the delivery of active miRNAs to the specific target site in the body requires controllable, sustained, and tissue-specific processes, and still faces many obstacles. To improve the efficacy of miRNA therapy, several strategies have been explored focusing on the delivery system. Notably, NPs have been widely used in vector delivery methods because they help to establish a nonimmunogenic, cell- and tissue-specific delivery system for cancer therapy. Both synthetic miRNA mimics and anti-miRNAs can be conjugated or complexed with NPs, which makes them more resistant to nuclease degradation and helps to reduce the immune response associated with naked miRNA delivery. Another point is that when miRNAs are delivered using nanocarriers, a lower concentration of miRNAs is required to achieve target suppression compared with naked miRNA delivery. Moreover, because the targeting ligand can be attached to the surface of NPs, the delivery of therapeutic miRNA using NPs is more specific compared with naked miRNA delivery [122].
Restoring miRNA levels using oligonucleotides, including miRNA mimics or miRNAs encoded in expression vectors, is a commonly employed approach for boosting levels of particular miRNAs. miRNA mimics are synthetic oligonucleotides that may regulate target mRNAs in an miRNA-like manner. miRNAs that act as tumor suppressors and that are downregulated can be exogenously delivered to return the cells to a normal state. Hsu et al. used a cationic lipid NP formulation as a vehicle to restore miR-122 expression in HCC cells and a mouse model of HCC. This NP-mediated miR-122 delivery resulted in >95% downregulation of miR-122 target genes in vitro and approximately 50% growth suppression of HCC xenografts within 30 days without causing systemic toxicity [123]. Meanwhile, Wang et al. found that the inclusion of oleic acid in lipid-based NPs significantly enhanced the delivery efficacy for miR-122 and decreased the expression of Bcl-w, a target of miR-122 [124]. Liang et al. employed a new type of poly(d, l-lactide-co-glycolide)-based NPs as vectors for miR-26a transfection and induced cell cycle arrest in HepG2 cells. These nanocomplexes exhibited a higher transfection efficiency with lower cytotoxicity and accumulated in tumor cells after transfection, indicating that these complexes contribute to the cellular uptake of pDNA and enhance the treatment effect of miR-26a by inducing cell cycle arrest [125]. In another study, Li et al. reported that the intratumoral injection of cholesterol-conjugated miR-99a mimics significantly inhibited tumor growth as well as reduced AFP levels in HCC-bearing nude mice [126]. Meanwhile, viral delivery of miRNAs has also been investigated. Using an adeno-associated virus as a vector, Kota et al. demonstrated that systemic miR-26a administration resulted in the inhibition of cancer cell proliferation, induction of tumor-specific apoptosis, and dramatic protection from disease progression with minimal toxicity in a mouse model of HCC [81]. This study demonstrated that restoring the expression of a dysregulated miRNA could specifically affect the tumor cells in which its expression was aberrant, whereas the normal tissue was not affected. Among these anticancer miRNA agents, the one that is furthest in development is MRX34, a liposome-formulated miR-34 mimic, which is in clinical trials for the treatment of liver cancer [127].
While restoring tumor-suppressive miRNAs is one approach for HCC therapy, suppressing or blocking oncogenic miRNAs with antisense oligonucleotides (also known as anti-miRs) may be considered another treatment strategy. As cellular uptake and incorporation into the cytoplasm are necessary for the function of anti-miRs, the transfection efficiency of the carrier is a major concern. NPs that are specifically engineered for delivery to specific cells will help with this goal. miR-155 is an oncogenic miRNA that is frequently overexpressed in HCC. Zhang et al. demonstrated that the delivery of anti-miR-155 using lactosylated gramicidin-containing lipid NPs could markedly upregulate the expression of miR-155 target genes in HCC cells, suggesting the potential of anti-miR therapy using a liver-specific delivery vehicle [128]. Additionally, anti-miR-155 delivery using polymer-NPs has also been used to study its tumor-suppressing effects in lymphoma-bearing mice. These studies suggested that when therapeutic molecules are delivered using NPs, the dosage needed for a positive therapeutic response is less than that needed with uncoated molecules [129].
In addition to direct effects in HCC treatment, miRNA therapy may be combined with chemotherapy, thus promoting the sensitization of tumors to these anti-cancer agents. miR-122 can sensitize HCC cells to doxorubicin treatment by regulating the expression of multidrug resistance genes [130]. Similarly, miR-199a-3p enhances the effect of doxorubicin treatment on HCC by influencing mTOR and c-Met expression [131]. The let-7 family can potentiate sorafenib-induced apoptosis via the inhibition of Bcl-xL expression in HCC cells [132]. Moreover, miR-101 synergized doxorubicin and fluorouracil treatment by inducing apoptosis and inhibiting autophagy in tumor cells [84]. In contrast, miRNA therapy combined with antiviral drugs could be a promising area of exploration for HBV treatment. This concept has been put into practice in a phase II clinical trial that combined miRavisen (an anti-miR) with the antiviral drugs telaprevir and ribavirin in treating HCV infection [133].
Conclusions
In this review, we have focused on the recent advances of miRNAs involved in HBV infection and HBV-associated tumorigenesis. It is clear that the host-virus interaction in HBV infection is mediated not only by immune responses but also by miRNAs. The host and virus may up- or downregulate a group of miRNAs for their own benefit. Consequently, sustained dysregulation of these miRNAs, some of which act as oncogenes or oncosuppressors, may contribute to the onset and progression of cancer. In the past 3 years, some progress has been made in understanding the mechanisms responsible for aberrant miRNA expression, such as HBx-induced dysregulation of miRNAs and epigenetically mediated dysregulation of miRNA. Meanwhile, more targets and upstream regulators of these dysregulated miRNAs have been unveiled. However, much work is still requirede to obtain more detailed and comprehensive knowledge about these processes.
From the clinical perspective, the diagnosis and treatment of HCC based on aberrant miRNAs appears to have great potential. Given that aberrant miRNA expression is an early event during HBV-associated hepatocarcinogenesis, an accurate diagnosis before the tumor moves to metaphase is possible to achieve. With the development of targeted miRNA delivery technology, HBV-related disease therapies based on restoring miRNA levels or blocking miRNA functions are expected to successfully reach the clinic in the very near future.
Acknowledgements
This work was supported by grants from the Science and Technology Support Program of Sichuan Province (2014SZ0002-4), the National 973 Basic Research Program of China (2013CB911300, 2012CB518900), the National Science and Technology Major Project (2012ZX09501001-003), and the Natural Science Foundation of China (81225015, 81072022, and 81172173).
Author contributions
Kun-Lin Xie and Yan-Ge Zhang contributed equally to this work; Kun-Lin Xie, Yan-Ge Zhang, and Jun Liu wrote the manuscript; Yong Zeng and Hong Wu gave suggestions and revised the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Huang S, He X. The role of micrornas in liver cancer progression. Br J Cancer. 2011;104:235-240
2. Lee Y, Ahn C, Han J. et al. The nuclear rnase iii drosha initiates microrna processing. Nature. 2003;425:415-419
3. Lund E, Guttinger S, Calado A. et al. Nuclear export of microrna precursors. Science. 2004;303:95-98
4. Kiss T. Small nucleolar rnas: An abundant group of noncoding rnas with diverse cellular functions. Cell. 2002;109:145-148
5. Kim VN. Microrna biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376-385
6. Bernstein E, Caudy AA, Hammond SM. et al. Role for a bidentate ribonuclease in the initiation step of rna interference. Nature. 2001;409:363-366
7. Yekta S, Shih IH, Bartel DP. Microrna-directed cleavage of hoxb8 mrna. Science. 2004;304:594-596
8. Zeng Y. Principles of micro-rna production and maturation. Oncogene. 2006;25:6156-6162
9. Kim VN, Han J, Siomi MC. Biogenesis of small rnas in animals. Nat Rev Mol Cell Biol. 2009;10:126-139
10. Braconi C, Huang N, Patel T. Microrna-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology. 2010;51:881-890
11. Wei X, Xiang T, Ren G. et al. Mir-101 is down-regulated by the hepatitis b virus x protein and induces aberrant DNA methylation by targeting DNA methyltransferase 3a. Cell Signal. 2013;25:439-446
12. Wei X, Tan C, Tang C. et al. Epigenetic repression of mir-132 expression by the hepatitis b virus x protein in hepatitis b virus-related hepatocellular carcinoma. Cell Signal. 2013;25:1037-1043
13. Liu WH, Yeh SH, Chen PJ. Role of micrornas in hepatitis b virus replication and pathogenesis. Biochim Biophys Acta. 2011;1809:678-685
14. Ott JJ, Stevens GA, Groeger J. et al. Global epidemiology of hepatitis b virus infection: New estimates of age-specific hbsag seroprevalence and endemicity. Vaccine. 2012;30:2212-2219
15. Wang Y, Jiang L, Ji X. et al. Hepatitis b viral rna directly mediates down-regulation of the tumor suppressor microrna mir-15a/mir-16-1 in hepatocytes. J Biol Chem. 2013;288:18484-18493
16. Su C, Hou Z, Zhang C. et al. Ectopic expression of microrna-155 enhances innate antiviral immunity against hbv infection in human hepatoma cells. Virol J. 2011;8:354
17. Li Y, Xie J, Xu X. et al. Microrna-548 down-regulates host antiviral response via direct targeting of ifn-lambda1. Protein Cell. 2013;4:130-141
18. Xie Q, Chen X, Lu F. et al. Aberrant expression of microrna 155 may accelerate cell proliferation by targeting sex-determining region y box 6 in hepatocellular carcinoma. Cancer. 2012;118:2431-2442
19. Mahajan VS, Drake A, Chen J. Virus-specific host mirnas: Antiviral defenses or promoters of persistent infection? Trends Immunol. 2009;30:1-7
20. Wang S, Zhang X, Ju Y. et al. Microrna-146a feedback suppresses t cell immune function by targeting stat1 in patients with chronic hepatitis b. J Immunol. 2013;191:293-301
21. Yang P, Li QJ, Feng Y. et al. Tgf-beta-mir-34a-ccl22 signaling-induced treg cell recruitment promotes venous metastases of hbv-positive hepatocellular carcinoma. Cancer Cell. 2012;22:291-303
22. Lecellier CH, Dunoyer P, Arar K. et al. A cellular microrna mediates antiviral defense in human cells. Science. 2005;308:557-560
23. Zhang GL, Li YX, Zheng SQ. et al. Suppression of hepatitis b virus replication by microrna-199a-3p and microrna-210. Antiviral Res. 2010;88:169-175
24. Chen Y, Shen A, Rider PJ. et al. A liver-specific microrna binds to a highly conserved rna sequence of hepatitis b virus and negatively regulates viral gene expression and replication. FASEB J. 2011;25:4511-4521
25. Liu N, Zhang J, Jiao T. et al. Hepatitis b virus inhibits apoptosis of hepatoma cells by sponging mir-15a/16 cluster. J Virol. 2013
26. Jung YJ, Kim JW, Park SJ. et al. C-myc-mediated overexpression of mir-17-92 suppresses replication of hepatitis b virus in human hepatoma cells. J Med Virol. 2013;85:969-978
27. Potenza N, Papa U, Mosca N. et al. Human microrna hsa-mir-125a-5p interferes with expression of hepatitis b virus surface antigen. Nucleic Acids Res. 2011;39:5157-5163
28. Wang Y, Lu Y, Toh ST. et al. Lethal-7 is down-regulated by the hepatitis b virus x protein and targets signal transducer and activator of transcription 3. J Hepatol. 2010;53:57-66
29. Zhang T, Zhang J, Cui M. et al. Hepatitis b virus x protein inhibits tumor suppressor mir-205 through inducing hypermethylation of mir-205 promoter to enhance carcinogenesis. Neoplasia. 2013;15:1282-1291
30. Lopez-Cabrera M, Letovsky J, Hu KQ. et al. Transcriptional factor c/ebp binds to and transactivates the enhancer element ii of the hepatitis b virus. Virology. 1991;183:825-829
31. Lopez-Cabrera M, Letovsky J, Hu KQ. et al. Multiple liver-specific factors bind to the hepatitis b virus core/pregenomic promoter: Trans-activation and repression by ccaat/enhancer binding protein. Proc Natl Acad Sci U S A. 1990;87:5069-5073
32. Wang B, Majumder S, Nuovo G. et al. Role of microrna-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in c57bl/6 mice. Hepatology. 2009;50:1152-1161
33. Wang J, Liu X, Wu H. et al. Creb up-regulates long non-coding rna, hulc expression through interaction with microrna-372 in liver cancer. Nucleic Acids Res. 2010;38:5366-5383
34. Kim BK, Lim SO, Park YG. Requirement of the cyclic adenosine monophosphate response element-binding protein for hepatitis b virus replication. Hepatology. 2008;48:361-373
35. Raney AK, Johnson JL, Palmer CN. et al. Members of the nuclear receptor superfamily regulate transcription from the hepatitis b virus nucleocapsid promoter. J Virol. 1997;71:1058-1071
36. Hu W, Wang X, Ding X. et al. Microrna-141 represses hbv replication by targeting ppara. PloS one. 2012;7:e34165
37. Ori A, Zauberman A, Doitsh G. et al. P53 binds and represses the hbv enhancer: An adjacent enhancer element can reverse the transcription effect of p53. EMBO J. 1998;17:544-553
38. Wang S, Qiu L, Yan X. et al. Loss of microrna 122 expression in patients with hepatitis b enhances hepatitis b virus replication through cyclin g(1) -modulated p53 activity. Hepatology. 2012;55:730-741
39. Qiu L, Fan H, Jin W. et al. Mir-122-induced down-regulation of ho-1 negatively affects mir-122-mediated suppression of hbv. Biochem Biophys Res Commun. 2010;398:771-777
40. Jin J, Tang S, Xia L. et al. Microrna-501 promotes hbv replication by targeting hbxip. Biochem Biophys Res Commun. 2013;430:1228-1233
41. Guo H, Liu H, Mitchelson K. et al. Micrornas-372/373 promote the expression of hepatitis b virus through the targeting of nuclear factor i/b. Hepatology. 2011;54:808-819
42. Zhang X, Zhang E, Ma Z. et al. Modulation of hepatitis b virus replication and hepatocyte differentiation by microrna-1. Hepatology. 2011;53:1476-1485
43. Datta J, Kutay H, Nasser MW. et al. Methylation mediated silencing of microrna-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049-5058
44. Reese V, Ondracek C, Rushing C. et al. Multiple nuclear receptors may regulate hepatitis b virus biosynthesis during development. Int J Biochem Cell Biol. 2011;43:230-237
45. Zhang X, Liu S, Hu T. et al. Up-regulated microrna-143 transcribed by nuclear factor kappa b enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50:490-499
46. Huang J, Wang Y, Guo Y. et al. Down-regulated microrna-152 induces aberrant DNA methylation in hepatitis b virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52:60-70
47. Kim JW, Lee SH, Park YS. et al. Replicative activity of hepatitis b virus is negatively associated with methylation of covalently closed circular DNA in advanced hepatitis b virus infection. Intervirology. 2011;54:316-325
48. Wu G, Yu F, Xiao Z. et al. Hepatitis b virus x protein downregulates expression of the mir-16 family in malignant hepatocytes in vitro. Br J Cancer. 2011;105:146-153
49. Connolly E, Melegari M, Landgraf P. et al. Elevated expression of the mir-17-92 polycistron and mir-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am J Pathol. 2008;173:856-864
50. Gao P, Wong CC, Tung EK. et al. Deregulation of microrna expression occurs early and accumulates in early stages of hbv-associated multistep hepatocarcinogenesis. J Hepatol. 2011;54:1177-1184
51. Li H, Li Y, Liu D. et al. Mir-224 is critical for celastrol-induced inhibition of migration and invasion of hepatocellular carcinoma cells. Cell Physiol Biochem. 2013;32:448-458
52. Lan SH, Wu SY, Zuchini R. et al. Autophagy suppresses tumorigenesis of hepatitis b virus-associated hepatocellular carcinoma through degradation of mir-224. Hepatology. 2013
53. Wang C, Song B, Song W. et al. Underexpressed microrna-199b-5p targets hypoxia-inducible factor-1alpha in hepatocellular carcinoma and predicts prognosis of hepatocellular carcinoma patients. J Gastroenterol Hepatol. 2011;26:1630-1637
54. Zhang WY, Cai N, Ye LH. et al. Transformation of human liver l-o2 cells mediated by stable hbx transfection. Acta Pharmacol Sin. 2009;30:1153-1161
55. Scisciani C, Vossio S, Guerrieri F. et al. Transcriptional regulation of mir-224 upregulated in human hccs by nfkappab inflammatory pathways. J Hepatol. 2012;56:855-861
56. Han H, Sun D, Li W. et al. A c-myc-microrna functional feedback loop affects hepatocarcinogenesis. Hepatology. 2013;57:2378-2389
57. Shen J, Wang S, Zhang YJ. et al. Genome-wide aberrant DNA methylation of microrna host genes in hepatocellular carcinoma. Epigenetics. 2012;7:1230-1237
58. Song K, Han C, Zhang J. et al. Epigenetic regulation of microrna-122 by peroxisome proliferator activated receptor-gamma and hepatitis b virus x protein in hepatocellular carcinoma cells. Hepatology. 2013
59. Belloni L, Pollicino T, De Nicola F. et al. Nuclear hbx binds the hbv minichromosome and modifies the epigenetic regulation of cccdna function. Proc Natl Acad Sci U S A. 2009;106:19975-19979
60. Wang Y, Toh HC, Chow P. et al. Microrna-224 is up-regulated in hepatocellular carcinoma through epigenetic mechanisms. FASEB J. 2012;26:3032-3041
61. Shi XY, Zhang YY, Zhou XW. et al. Hepatitis b virus x protein regulates the mezh2 promoter via the e2f1-binding site in aml12 cells. Chin J Cancer. 2011;30:273-279
62. Au SL, Wong CC, Lee JM. et al. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor micrornas to promote liver cancer metastasis. Hepatology. 2012;56:622-631
63. Ren M, Qin D, Li K. et al. Correlation between hepatitis b virus protein and microrna processor drosha in cells expressing hbv. Antiviral Res. 2012;94:225-231
64. Liu AM, Zhang C, Burchard J. et al. Global regulation on microrna in hepatitis b virus-associated hepatocellular carcinoma. OMICS. 2011;15:187-191
65. Wu J, Zhang XJ, Shi KQ. et al. Hepatitis b surface antigen inhibits mica and micb expression via induction of cellular mirnas in hepatocellular carcinoma cells. Carcinogenesis. 2014;35:155-163
66. Li C, Wang Y, Wang S. et al. Hepatitis b virus mrna-mediated mir-122 inhibition upregulates pttg1-binding protein, which promotes hepatocellular carcinoma tumor growth and cell invasion. J Virol. 2013;87:2193-2205
67. Longerich T, Mueller MM, Breuhahn K. et al. Oncogenetic tree modeling of human hepatocarcinogenesis. Int J Cancer. 2012;130:575-583
68. Liu C, Yu J, Yu S. et al. Microrna-21 acts as an oncomir through multiple targets in human hepatocellular carcinoma. J Hepatol. 2010;53:98-107
69. Qiu X, Dong S, Qiao F. et al. Hbx-mediated mir-21 upregulation represses tumor-suppressor function of pdcd4 in hepatocellular carcinoma. Oncogene. 2013;32:3296-3305
70. Nakatani T, Roy G, Fujimoto N. et al. Sex hormone dependency of diethylnitrosamine-induced liver tumors in mice and chemoprevention by leuprorelin. Jpn J Cancer Res. 2001;92:249-256
71. Yu MW, Chang HC, Chang SC. et al. Role of reproductive factors in hepatocellular carcinoma: Impact on hepatitis b- and c-related risk. Hepatology. 2003;38:1393-1400
72. Jiang R, Deng L, Zhao L. et al. Mir-22 promotes hbv-related hepatocellular carcinoma development in males. Clin Cancer Res. 2011;17:5593-5603
73. Liu WH, Yeh SH, Lu CC. et al. Microrna-18a prevents estrogen receptor-alpha expression, promoting proliferation of hepatocellular carcinoma cells. Gastroenterology. 2009;136:683-693
74. Chen PJ, Yeh SH, Liu WH. et al. Androgen pathway stimulates microrna-216a transcription to suppress the tumor suppressor in lung cancer-1 gene in early hepatocarcinogenesis. Hepatology. 2012;56:632-643
75. Sampson VB, Rong NH, Han J. et al. Microrna let-7a down-regulates myc and reverts myc-induced growth in burkitt lymphoma cells. Cancer Res. 2007;67:9762-9770
76. Johnson SM, Grosshans H, Shingara J. et al. Ras is regulated by the let-7 microrna family. Cell. 2005;120:635-647
77. Lee YS, Dutta A. The tumor suppressor microrna let-7 represses the hmga2 oncogene. Genes Dev. 2007;21:1025-1030
78. Wu CS, Yen CJ, Chou RH. et al. Downregulation of microrna-15b by hepatitis b virus x enhances hepatocellular carcinoma proliferation via fucosyltransferase 2-induced globo h expression. Int J Cancer. 2013
79. Gramantieri L, Ferracin M, Fornari F. et al. Cyclin g1 is a target of mir-122a, a microrna frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092-6099
80. Xu T, Zhu Y, Xiong Y. et al. Microrna-195 suppresses tumorigenicity and regulates g1/s transition of human hepatocellular carcinoma cells. Hepatology. 2009;50:113-121
81. Kota J, Chivukula RR, O'Donnell KA. et al. Therapeutic microrna delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005-1017
82. Park IY, Sohn BH, Yu E. et al. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis b virus x protein. Gastroenterology. 2007;132:1476-1494
83. Tong A, Gou L, Lau QC. et al. Proteomic profiling identifies aberrant epigenetic modifications induced by hepatitis b virus x protein. J Proteome Res. 2009;8:1037-1046
84. Xu L, Beckebaum S, Iacob S. et al. Microrna-101 inhibits human hepatocellular carcinoma progression through ezh2 downregulation and increased cytostatic drug sensitivity. J Hepatol. 2013
85. Noh JH, Chang YG, Kim MG. et al. Mir-145 functions as a tumor suppressor by directly targeting histone deacetylase 2 in liver cancer. Cancer Lett. 2013;335:455-462
86. Zhang J, Yang Y, Yang T. et al. Microrna-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br J Cancer. 2010;103:1215-1220
87. Zhang S, Shan C, Kong G. et al. Microrna-520e suppresses growth of hepatoma cells by targeting the nf-kappab-inducing kinase (nik). Oncogene. 2012;31:3607-3620
88. Zhang JF, He ML, Fu WM. et al. Primate-specific microrna-637 inhibits tumorigenesis in hepatocellular carcinoma by disrupting signal transducer and activator of transcription 3 signaling. Hepatology. 2011;54:2137-2148
89. Budhu A, Jia HL, Forgues M. et al. Identification of metastasis-related micrornas in hepatocellular carcinoma. Hepatology. 2008;47:897-907
90. Zeng L, Yu J, Huang T. et al. Differential combinatorial regulatory network analysis related to venous metastasis of hepatocellular carcinoma. BMC Genomics. 2012;13(Suppl 8):S14
91. Firrincieli D, Boissan M, Chignard N. Epithelial-mesenchymal transition in the liver. Gastroenterol Clin Biol. 2010;34:523-528
92. Reichl P, Haider C, Grubinger M. et al. Tgf-beta in epithelial to mesenchymal transition and metastasis of liver carcinoma. Curr Pharm Des. 2012;18:4135-4147
93. Coulouarn C, Factor VM, Andersen JB. et al. Loss of mir-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526-3536
94. Yang H, Fang F, Chang R. et al. Microrna-140-5p suppresses tumor growth and metastasis by targeting transforming growth factor beta receptor 1 and fibroblast growth factor 9 in hepatocellular carcinoma. Hepatology. 2013;58:205-217
95. Kim T, Veronese A, Pichiorri F. et al. P53 regulates epithelial-mesenchymal transition through micrornas targeting zeb1 and zeb2. J Exp Med. 2011;208:875-883
96. Li N, Fu H, Tie Y. et al. Mir-34a inhibits migration and invasion by down-regulation of c-met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275:44-53
97. Salvi A, Sabelli C, Moncini S. et al. Microrna-23b mediates urokinase and c-met downmodulation and a decreased migration of human hepatocellular carcinoma cells. FEBS J. 2009;276:2966-2982
98. Zhang JP, Zeng C, Xu L. et al. Microrna-148a suppresses the epithelial-mesenchymal transition and metastasis of hepatoma cells by targeting met/snail signaling. Oncogene. 2013
99. Kong G, Zhang J, Zhang S. et al. Upregulated microrna-29a by hepatitis b virus x protein enhances hepatoma cell migration by targeting pten in cell culture model. PloS one. 2011;6:e19518
100. Garofalo M, Di Leva G, Romano G. et al. Mir-221&222 regulate trail resistance and enhance tumorigenicity through pten and timp3 downregulation. Cancer Cell. 2009;16:498-509
101. Wong QW, Ching AK, Chan AW. et al. Mir-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing akt signaling. Clin Cancer Res. 2010;16:867-875
102. Xu X, Fan Z, Kang L. et al. Hepatitis b virus x protein represses mirna-148a to enhance tumorigenesis. J Clin Invest. 2013;123:630-645
103. Hou J, Lin L, Zhou W. et al. Identification of mirnomes in human liver and hepatocellular carcinoma reveals mir-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232-243
104. Yan H, Dong X, Zhong X. et al. Inhibitions of epithelial to mesenchymal transition and cancer stem cells-like properties are involved in mir-148a-mediated anti-metastasis of hepatocellular carcinoma. Mol Carcinog. 2013
105. Arzumanyan A, Friedman T, Kotei E. et al. Epigenetic repression of e-cadherin expression by hepatitis b virus x antigen in liver cancer. Oncogene. 2012;31:563-572
106. Sun Z, Han Q, Zhou N. et al. Microrna-9 enhances migration and invasion through klf17 in hepatocellular carcinoma. Mol Oncol. 2013;7:884-894
107. Meng Z, Fu X, Chen X. et al. Mir-194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology. 2010;52:2148-2157
108. Ji J, Zhao L, Budhu A. et al. Let-7g targets collagen type i alpha2 and inhibits cell migration in hepatocellular carcinoma. J Hepatol. 2010;52:690-697
109. Tsai WC, Hsu PW, Lai TC. et al. Microrna-122, a tumor suppressor microrna that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571-1582
110. Bai S, Nasser MW, Wang B. et al. Microrna-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284:32015-32027
111. Stross L, Gunther J, Gasteiger G. et al. Foxp3+ regulatory t cells protect the liver from immune damage and compromise virus control during acute experimental hepatitis b virus infection in mice. Hepatology. 2012;56:873-883
112. Fabbri M, Paone A, Calore F. et al. Micrornas bind to toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110-2116
113. Waidmann O, Bihrer V, Pleli T. et al. Serum microrna-122 levels in different groups of patients with chronic hepatitis b virus infection. J Viral Hepat. 2012;19:e58-65
114. Arataki K, Hayes CN, Akamatsu S. et al. Circulating microrna-22 correlates with microrna-122 and represents viral replication and liver injury in patients with chronic hepatitis b. J Med Virol. 2013;85:789-798
115. Raimondo G, Pollicino T, Romano L. et al. A 2010 update on occult hepatitis b infection. Pathol Biol (Paris). 2010;58:254-257
116. Raimondo G, Pollicino T, Cacciola I. et al. Occult hepatitis b virus infection. J Hepatol. 2007;46:160-170
117. Chen Y, Li L, Zhou Z. et al. A pilot study of serum microrna signatures as a novel biomarker for occult hepatitis b virus infection. Med Microbiol Immunol. 2012;201:389-395
118. Chen CJ, Lee MH. Early diagnosis of hepatocellular carcinoma by multiple micrornas: Validity, efficacy, and cost-effectiveness. J Clin Oncol. 2011;29:4745-4747
119. Li LM, Hu ZB, Zhou ZX. et al. Serum microrna profiles serve as novel biomarkers for hbv infection and diagnosis of hbv-positive hepatocarcinoma. Cancer Res. 2010;70:9798-9807
120. Tomimaru Y, Eguchi H, Nagano H. et al. Circulating microrna-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol. 2012;56:167-175
121. Zhou J, Yu L, Gao X. et al. Plasma microrna panel to diagnose hepatitis b virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29:4781-4788
122. Muthiah M, Park IK, Cho CS. Nanoparticle-mediated delivery of therapeutic genes: Focus on mirna therapeutics. Expert Opin Drug Deliv. 2013;10:1259-1273
123. Hsu SH, Yu B, Wang X. et al. Cationic lipid nanoparticles for therapeutic delivery of sirna and mirna to murine liver tumor. Nanomedicine. 2013;9:1169-1180
124. Wang X, Yu B, Ren W. et al. Enhanced hepatic delivery of sirna and microrna using oleic acid based lipid nanoparticle formulations. J Control Release. 2013;172:690-698
125. Liang GF, Zhu YL, Sun B. et al. Plga-based gene delivering nanoparticle enhance suppression effect of mirna in hepg2 cells. Nanoscale Res Lett. 2011;6:447
126. Li D, Liu X, Lin L. et al. Microrna-99a inhibits hepatocellular carcinoma growth and correlates with prognosis of patients with hepatocellular carcinoma. J Biol Chem. 2011;286:36677-36685
127. Bader AG. Mir-34 - a microrna replacement therapy is headed to the clinic. Front Genet. 2012;3:120
128. Zhang M, Zhou X, Wang B. et al. Lactosylated gramicidin-based lipid nanoparticles (lac-gln) for targeted delivery of anti-mir-155 to hepatocellular carcinoma. J Control Release. 2013;168:251-261
129. Babar IA, Cheng CJ, Booth CJ. et al. Nanoparticle-based therapy in an in vivo microrna-155 (mir-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci U S A. 2012;109:E1695-1704
130. Xu Y, Xia F, Ma L. et al. Microrna-122 sensitizes hcc cancer cells to adriamycin and vincristine through modulating expression of mdr and inducing cell cycle arrest. Cancer Lett. 2011;310:160-169
131. Fornari F, Milazzo M, Chieco P. et al. Mir-199a-3p regulates mtor and c-met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70:5184-5193
132. Shimizu S, Takehara T, Hikita H. et al. The let-7 family of micrornas inhibits bcl-xl expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol. 2010;52:698-704
133. Ling H, Fabbri M, Calin GA. Micrornas and other non-coding rnas as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847-865
134. Fan CG, Wang CM, Tian C. et al. Mir-122 inhibits viral replication and cell proliferation in hepatitis b virus-related hepatocellular carcinoma and targets ndrg3. Oncol Rep. 2011;26:1281-1286
135. Coppola N, Potenza N, Pisaturo M. et al. Liver microrna hsa-mir-125a-5p in hbv chronic infection: Correlation with hbv replication and disease progression. PloS one. 2013;8:e65336
136. Park SO, Kumar M, Gupta S. Tgf-beta and iron differently alter hbv replication in human hepatocytes through tgf-beta/bmp signaling and cellular microrna expression. PloS one. 2012;7:e39276
137. Lee CG, Kim YW, Kim EH. et al. Farnesoid x receptor protects hepatocytes from injury by repressing mir-199a-3p, which increases levels of lkb1. Gastroenterology. 2012;142:1206-1217
138. Henry JC, Park JK, Jiang J. et al. Mir-199a-3p targets cd44 and reduces proliferation of cd44 positive hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2010;403:120-125
139. Yang W, Sun T, Cao J. et al. Downregulation of mir-210 expression inhibits proliferation, induces apoptosis and enhances radiosensitivity in hypoxic human hepatoma cells in vitro. Exp Cell Res. 2012;318:944-954
140. Shi C, Xu X. Microrna-22 is down-regulated in hepatitis b virus-related hepatocellular carcinoma. Biomed Pharmacother. 2013;67:375-380
141. Wang CM, Wang Y, Fan CG. et al. Mir-29c targets tnfaip3, inhibits cell proliferation and induces apoptosis in hepatitis b virus-related hepatocellular carcinoma. Biochem Biophys Res Commun. 2011;411:586-592
142. Piskounova E, Polytarchou C, Thornton JE. et al. Lin28a and lin28b inhibit let-7 microrna biogenesis by distinct mechanisms. Cell. 2011;147:1066-1079
143. Nam Y, Chen C, Gregory RI. et al. Molecular basis for interaction of let-7 micrornas with lin28. Cell. 2011;147:1080-1091
144. Park SM, Shell S, Radjabi AR. et al. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene hmga2. Cell Cycle. 2007;6:2585-2590
145. Aguda BD, Kim Y, Piper-Hunter MG. et al. Microrna regulation of a cancer network: Consequences of the feedback loops involving mir-17-92, e2f, and myc. Proc Natl Acad Sci U S A. 2008;105:19678-19683
146. Chen L, Zheng J, Zhang Y. et al. Tumor-specific expression of microrna-26a suppresses human hepatocellular carcinoma growth via cyclin-dependent and -independent pathways. Mol Ther. 2011;19:1521-1528
147. Yang X, Liang L, Zhang XF. et al. Microrna-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-stat3 pathway. Hepatology. 2013;58:158-170
148. Wang L, Zhang X, Jia LT. et al. C-myc-mediated epigenetic silencing of microrna-101 contributes to dysregulation of multiple pathways in hepatocellular carcinoma. Hepatology. 2013
149. Su H, Yang JR, Xu T. et al. Microrna-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135-1142
150. Li S, Fu H, Wang Y. et al. Microrna-101 regulates expression of the v-fos fbj murine osteosarcoma viral oncogene homolog (fos) oncogene in human hepatocellular carcinoma. Hepatology. 2009;49:1194-1202
151. Lin CJ, Gong HY, Tseng HC. et al. Mir-122 targets an anti-apoptotic gene, bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2008;375:315-320
152. Yang XW, Zhang LJ, Huang XH. et al. Mir-145 suppresses cell invasion in hepatocellular carcinoma cells: Mir-145 targets adam17. Hepatol Res. 2013
153. Wang Y, Lee AT, Ma JZ. et al. Profiling microrna expression in hepatocellular carcinoma reveals microrna-224 up-regulation and apoptosis inhibitor-5 as a microrna-224-specific target. J Biol Chem. 2008;283:13205-13215
Author contact
![]() Corresponding author: Department of Liver Surgery & Liver Transplantation Centre, West China Hospital, Sichuan University, No. 37, Guo Xue Xiang, Chengdu 610041, Sichuan Province, China. Telephone: 86-28-85422323; Fax: 86-28-85422323 E-mail: wuhongedu.cn.
Corresponding author: Department of Liver Surgery & Liver Transplantation Centre, West China Hospital, Sichuan University, No. 37, Guo Xue Xiang, Chengdu 610041, Sichuan Province, China. Telephone: 86-28-85422323; Fax: 86-28-85422323 E-mail: wuhongedu.cn.
 Global reach, higher impact
Global reach, higher impact