13.3
Impact Factor
Theranostics 2014; 4(12):1153-1163. doi:10.7150/thno.9510 This issue Cite
Research Paper
Glycoproteomic Study Reveals Altered Plasma Proteins Associated with HIV Elite Suppressors
1. Department of Pathology, , Johns Hopkins University School of Medicine, Baltimore, Maryland, USA;
2. Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA;
3. Laboratory of Immunoregulation, Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH, Baltimore, Maryland, USA.
Received 2014-4-27; Accepted 2014-8-21; Published 2014-9-18
Abstract
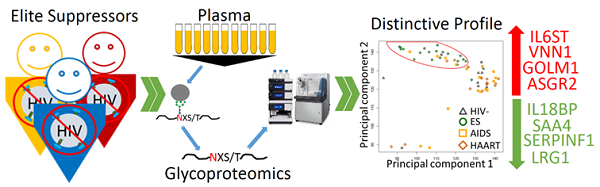
HIV elite suppressors (ES) or controllers are individuals achieving control of viremia by their natural immunological mechanisms without highly active antiretroviral therapy (HAART). Study of the mechanisms responsible for the immunological suppression of viremia in ES may lead to the detection of individuals with ES and the effective control of HIV infection. We hypothesize that plasma glycoproteins play essential roles in the immune system of ES since plasma proteins are critical and highly relevant in anti-viral immunity and most plasma proteins are glycoproteins. To examine glycoproteins associated with ES, plasma samples from ES individuals (n=20), and from individuals on HAART (n=20), with AIDS (n=20), and no HIV infection (n=10) were analyzed by quantitative glycoproteomics. We found that a number of glycoproteins changed between ES versus HAART, AIDS and HIV- individuals. In sharp contrast, the level of plasma glycoproteins in the HAART cohort showed fewer changes compared with AIDS and HIV- individuals. These results showed that although both ES and HAART effectively suppress viremia, ES appeared to profoundly affect immunologically relevant glycoproteins in plasma as consequence of or support for anti-viral immunity. Bioinformatic analysis revealed that altered proteins in ES plasma were mainly associated with inflammation. This analysis suggests that overlapping, while distinguishable, glycoprotein profiles for inflammation and immune activation appeared to be present between ES and non-ES (HAART+AIDS) cohorts, indicating different triggers for inflammation and immune activation between natural and treatment-related viral suppression.
Keywords: HIV, elite suppressor, HAART, AIDS, glycoprotein, glycoproteomics, inflammation, immune activation
Introduction
The global HIV epidemic remains a great threat to public health with approximately 1.7 million AIDS-related deaths in 2012 [1]. Elite suppression (ES) and highly active antiretroviral therapy (HAART) are two effective mechanisms to halt disease progression and represent the naturally immunological and treatment-related anti-viral mechanisms, respectively [2]. The rare ES cohort is able to naturally maintain normal CD4 cell count and undetectable viremia independent of HAART for more than 10 years. This natural immunological mechanism generates great interest in HIV research [3]. The proposed antiviral mechanism of ES varies among individuals, but specific HLA alleles and CD8+ T cell response are believed to be the primary factors [3]. Evidence, however, suggests that chronic inflammation and immune activation persist even after either efficient ES or HAART requiring therapeutics to normalize immune activation for long-term health management [4, 5].
Glycosylation is one of the most common protein modifications for secreted proteins in plasma including proteins in immune response [6]. Because of unique CD4+ T cell tropism of HIV and its overwhelming the impact on immune system, profile of glycoproteins in the infected plasma is prone to changes reflecting the host strategy to control the infection [7]. To profile glycoproteins, we applied hydrazide chemistry based solid phase capture of glycopeptides and releasing N-linked glycosite-containing peptides with the use of PNGase F for specific analysis of the N-linked glycosite-containing peptides [8]. The isolated glycosite-containing peptides were analyzed by LC separation followed by tandem mass spectrometry (MS/MS) to identify and quantify the changes of glycoproteins in a high throughput manner [9, 10].
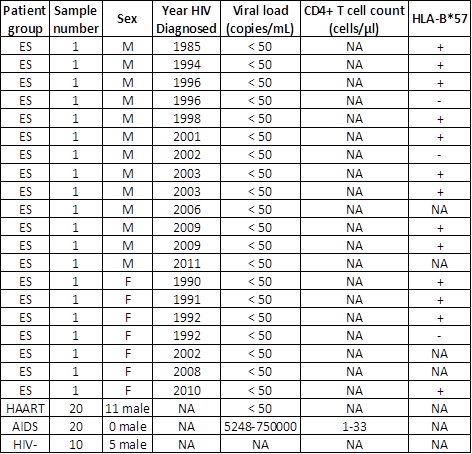
Clinical information of patients in normal and disease cohorts. (NA-not applicable)
The level and type of glycoproteins in plasma from ES (n=20), HAART (n=20), AIDS (n=20) and uninfected (HIV-, n=10) were studied using quantitative glycoproteomics. The role of the differentially expressed glycoproteins in ES plasma and their value in theranostics of HIV were analyzed by bioinformatics.
Materials and Methods
Chemicals: The bicinchoninic acid protein assay (BCA) assay kit was purchased from Pierce (Rockford, IL); Hydrazide resin and sodium periodate were purchased from Bio-Rad (Hercules, CA); trypsin with sequencing-grade was from Promega (Madison, WI); PNGase F was purchased from New England Biolabs (Ipswich, MA); Sep-Pak C18 1cc Vac Cartridge was from Waters (Milford, MA); Other chemicals such as Acetonitrile (ACN), trifluoroacetic acid (TFA), formic acid, urea, tris (2-carboxyethyl) phosphine (TCEP), iodoacetamide, 5M NaCl, sodium dodecyl sulfate (SDS), sodium sulfite, ammonia bicarbonate (AB), 1M Tris-HCl pH 8.0 buffer were purchased from Sigma-Aldrich (St. Louis, MO).
Patient plasmas: The 20 plasmas from ES cohort at Johns Hopkins University were infected with HIV-1 subtype B virus and the viral loads were below 50 copies/mL [11-13]. The 40 plasmas from patients on HAART (n=20, viral load < 50) or with AIDS (n=20, CD4+ T cell count < 50 cells/μl, viral load range 5,248-750,000) were collected from the Johns Hopkins HIV Clinical Practice Cohort at Johns Hopkins University [14]. The undetectable viral load in ES and HAART indicated suppressed viremia and CD4+ T cells counts were not available in these cohorts. The HLA class I types were described in previous studies [12, 13, 15-18]. These samples are described on Table 1. The plasma samples were stored in -80ºC until used. All participants provided written informed consent and Johns Hopkins Medicine Institutional Review Board (IRB) has approved the study.
Isolation of N-linked glycosite-containing peptides: Plasma (20 μl containing ~1.6 mg of proteins) was mixed with 90 μl of 8 M urea in 1 M AB buffer. Proteins were reduced in 10 mM TCEP at room temperature for 1 hour. Proteins were alkylated in 10 mM iodoacetamide and incubated at room temperature (RT) for 30 min in the dark. Samples were diluted into 6 mL using 0.2 M AB buffer to decrease the urea concentration to less than 2 M protein concentration. Trypsin (0.5 μg/μl, enzyme/protein ratio of 1/40 w/w) was added to digest proteins at 30ºC over night. After digestion, 5 μg of sample was resolved on a tricine gel and silver staining was used to determine the completion of tryptic digested peptides. The disappearance of high molecular bands and the appearance of a smear of bands under molecular weight of 10 kDa indicated completion of digestion. The peptides were desalted by a C18 cartridge according to the manufacturer's instructions and oxidized by incubating with 10 mM sodium periodate in 50% ACN (v/v) 0.1% TFA (v/v) at room temperature for 1 hour in the dark. The 60 mM sodium sulfite was used to quench the oxidation reaction. The samples were then mixed with 50 μl of (50% slurry) hydrazide support prewashed with 1 mL deionised water. The mixture was incubated with gentle shaking at RT over night for the coupling reaction. The glycopeptide conjugated hydrazide resin was sequentially washed with 800 μl of 50% ACN (v/v) 0.1% TFA (v/v), then 1.5 M NaCl and water, three times/solution to remove non-coupled peptides. The support was finally washed with 200 μl of 1X G7 buffer twice and incubated with 3 μl of PNGase F in 50 μl of 1X G7 buffer at 37ºC to specifically release formerly N-linked glycopeptides which were desalted by a C18 cartridge, dried in a speed-vac and re-suspended in 40 μl of 0.1% formic acid prior to MS analysis. For pooled glycopeptides, an equal volume (i.e. 1 μl) of the N-linked glycosite-containing peptides from each sample was pooled to become HIV-, HAART, AIDS and ES cohorts.
LC-MS/MS analysis: The N-linked glycosite-containing peptides (~1 μg) were separated through a Dionex Ultimate 3000 RSLC nano system (Thermo Scientific) with a 75 µm x 15 cm Acclaim PepMap100 separating column (Thermo Scientific) protected by a 2 cm guarding column (Thermo Scientific). The mobile phase consisted of 0.1% formic acid in water (A) and 0.1% formic acid 95% acetonitrile (B). Flow rate was 300 nL/min. The gradient profile was set as following: 4-35% B for 70 min, 35-95% B for 5 min, 95% B for 10 min and equilibrated in 4% B for 15 min. MS analysis was performed using an Orbitrap Velos Pro mass spectrometer (Thermo Scientific). The spray voltage was set at 2.2 kV. Orbitrap spectra (AGC 1x106) were collected from 400-1800 m/z at a resolution of 60K followed by data-dependent HCD MS/MS (at a resolution of 7500, collision energy 35%, activation time 0.1 ms) of the ten most abundant ions using an isolation width of 2.0 Da. Charge state screening was enabled to reject unassigned and singly charged ions. A dynamic exclusion time of 35 sec was used to discriminate against previously selected ions.
Protein identification and quantification: Proteins were identified using Proteome Discoverer software (Thermo Fisher Scientific, version 1.3). A NCBI human database containing 36,430 protein entries was used [19]. The precursor mass tolerance was set at 20 ppm and the MS/MS tolerance at 0.06 Da. Parameters of the search were modified as follows: oxidized methionines (add Met with 15.995 Da), a (PNGase F-catalyzed) conversion of Asn to Asp (add Asn with 0.984 Da) and Cys modification (add cysteine with 57 Da). A maximum of two missed tryptic cleavage sites were allowed. The label-free, semi-quantitative differential expression analysis of the identified protein was conducted using SIEVE software (Thermo Scientific, version 2.0). The raw files were imported into SIEVE and the chromatograms were aligned. Two and five samples from AIDS and HAART respectively were excluded from the study due to low alignment score determined by the software. Frame parameters were set as follows: maximum frame 100,000, retention time from 0 to 60 mins, m/z from 350 to 1800, frame time retention time width 2.5 mins, frame m/z width 10 ppm, peak intensity threshold 5-E5. After framing, the Proteome Discoverer result files were imported with a 1% false-discovery rate (FDR) and only peptides unique to one protein were used for quantification. Proteins with at least 2 spectral counts were reported. The frames for the N-linked glycopeptides (i.e. peptides containing NXS/T, X≠P and deamidation at the N position) were targeted in SIEVE to reduce interference of non-glycopeptides for quantification. The relative intensity of peptides was normalized by total ion current (TIC) and used to calculate p-value. The average intensity of peptides was used to calculate fold change.
Statistics, pathway analysis and bioinformatics
Student t test was used to calculate the p-value of differentially expressed proteins. Hierarchical clustering was used to separate different plasma samples based on their relative abundance of glycopeptides using QCanvas version 1.2 [20]. Further, the principle component analysis (PCA) of the normalized intensity data matrix for peptides was used to obtain the first two orthogonal principle components, and the intensity data were then projected to the plane formed by these two principle components. The Database for Annotation, Visualization and Integrated Discovery (DAVID) was used for Gene Ontology (GO) and pathway analysis [21].
Results
Plasma from ES Has Distinctive Profile of Glycoproteins
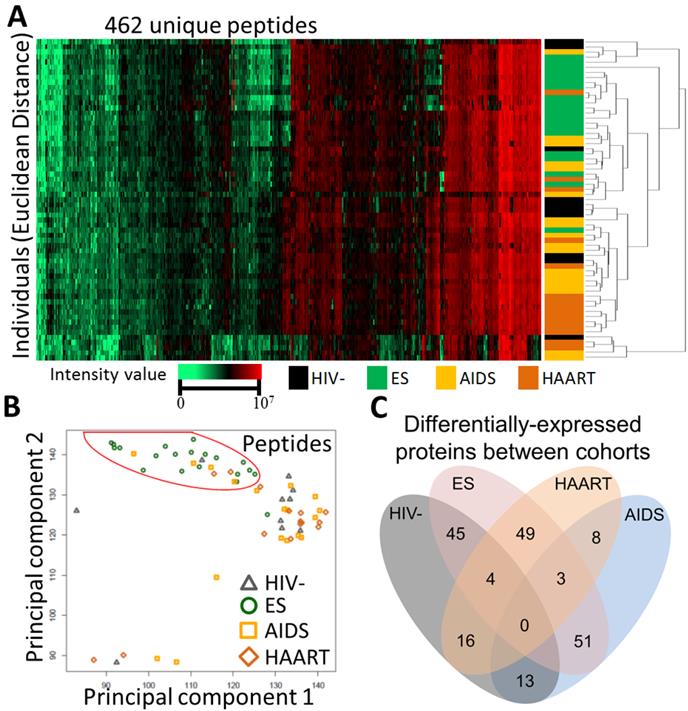
To profile glycoproteins, plasmas collected from individuals with elite suppression (n=20), HAART (n=20), AIDS (n=20) and uninfected individuals (HIV-, n=10) were subjected to glycoproteomics described in materials and methods. Proteome Discoverer identified 563 unique glycopeptides from 247 glycoproteins with 1% FDR (Additional file 1: Supplementary Table 1). Of these proteins, SIEVE was able to quantify 462 unique glycopeptides from 185 proteins with the same FDR and at least two spectral counts per protein (Additional file 2: Supplementary Table 2). Unsupervised hierarchical clustering was performed using relative intensity of the 462 unique peptides (Fig. 1A). Strikingly, most of the individuals with ES were separated from others (Fig. 1A). This result was visualized using PCA where most individuals with ES were again separated (Fig. 1B). Changes of glycosite-containing peptides detected by our method can be caused, separately or in combination, by different changes in glycoproteins including changes in glycoprotein expression, glycosylation occupancy or glycoprotein isoforms. Using a cut off ratio of 1.5-fold and p-value ≤ 0.05 in the comparison, 45, 16 and 13 proteins were changed in ES, HAART and AIDS, respectively versus HIV- (Fig. 1C and Table 2). The striking number of changed proteins in ES group suggested that the plasma glycoprotein profile of ES appeared to differ significantly from HIV- than that of HAART and AIDS. Furthermore, pair-wise comparison was performed amongst ES, HAART and AIDS (Fig. 1C and Table 2). Again, a clear difference between ES and non-ES (HAART and AIDS) was observed, and 49, 51, 35 proteins changed in ES/HAART, ES/AIDS and ES/HAART+AIDS respectively (Fig. 1C and Table 2). Relatively fewer number of proteins (i.e. 8 proteins) were changed between HAART and AIDS suggesting that HAART had a relatively limited effect on the level of plasma glycoproteins (Fig. 1C and Table 2). This result indicated that the profile of plasma glycoproteins of ES significantly differed from HIV- and other HIV+ cohorts. The differentially expressed glycoproteins might participate in or simply reflect a consequence of the anti-viral immunity in ES.
Separation of ES from other cohorts. (A) Unsupervised hierarchical clustering using relative intensity of peptides for individuals. Euclidean Distance and Spearman Rand were used for individuals and peptides respectively. (B) Principal component analysis using relative intensity of peptides from individuals. Red circle highlighted ES in segregation from other groups. (C) Venn diagram showing the number of significantly changed proteins with a cut off ratio of 1.5-fold and p-value ≤ 0.05 among cohorts.
Heat map and differentially expressed glycoproteins among cohorts.
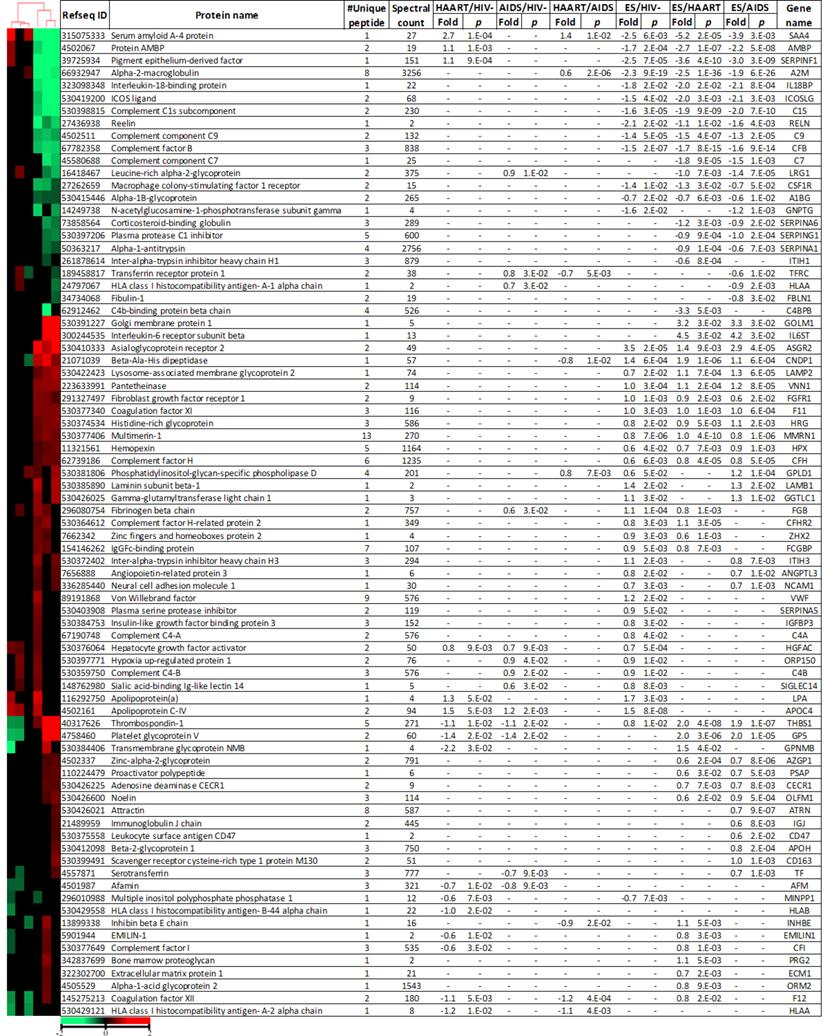
Log2 fold change and p-value were calculated using relative intensity of peptides described in Materials and Methods. Proteins with p-value ≤ 0.05, at least 1.5-fold change and two spectral counts were shown. Euclidian distance and city-block distance were used to cluster cohorts and proteins respectively. Minus (-) indicated not significant.
Differentially expressed Glycoproteins in ES
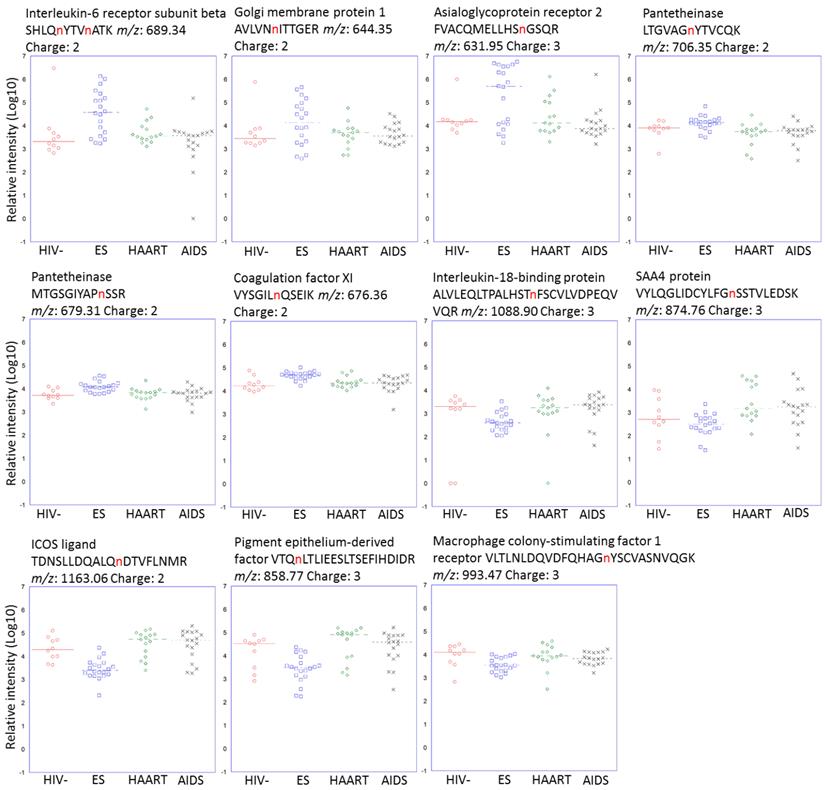
Glycoproteins discriminating ES from HAART and AIDS suggested a potential involvement in or a consequence of HIV immunological suppression. There were 18 glycoproteins increased in ES versus both HAART and AIDS. Of these proteins, interleukin-6 receptor subunit beta (IL6ST) with peptide SHLQnYTVnATK (n represents the N-linked glycosite) had the highest upregulation of 23 and 18-fold in ES versus HAART and AIDS respectively, (Fig. 2). Golgi membrane protein 1 (GOLM1) had fold changes of 9 and 10 for ES/HAART and ES/AIDS respectively (Fig. 2). Sixteen proteins had less significant fold changes including asialoglycoprotein receptor 2 (ASGR2), platelet glycoprotein V (GP5), thrombospondin-1 (THBS1), lysosome-associated membrane glycoprotein 2 (LAMP2), pantetheinase (VNN1), beta-Ala-His dipeptidase (CNDP1), histidine-rich glycoprotein (HRG), coagulation factor XI (F11), noelin (OLFM1), hemopexin (HPX), multimerin-1 (MMRN1), complement factor H (CFH), proactivator polypeptide (PSAP), zinc-alpha-2-glycoprotein (AZGP1), adenosine deaminase CECR1 (CECR1) and fibroblast growth factor receptor 1 (FGFR1). Seventeen proteins exhibited downregulation in ES versus both HAART and AIDS. Serum amyloid A-4 protein (SAA4) with peptides VYLQGLIDCYLFGnSSTVLEDSK showed fold changes of 38 and 15 in ES versus HAART and AIDS, respectively (Fig. 2).
Altered glycosite-containing peptides for each cohort. Glycosite-containing peptides from six up-regulated and five down-regulated proteins specific for ES cohort were plotted. The sites for N-linked glycosylation were highlighted in red.
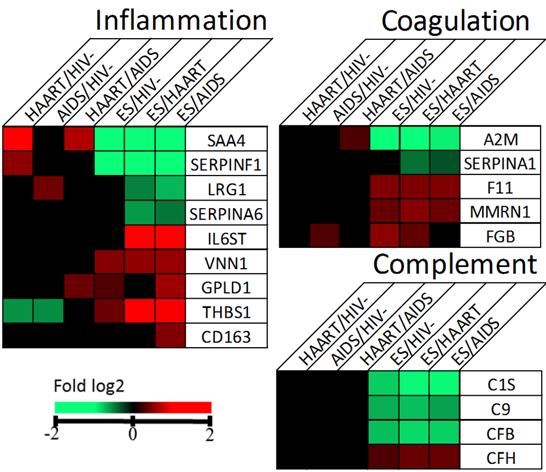
Changed proteins in ES associated with inflammation, coagulation and complement. Proteins with p-value ≤ 0.05 and fold change ≥ 1.5 were considered significantly different and coloured in red or green.
The pigment epithelium-derived factor (SERPINF1) had reductions of 12 and 8-fold in ES versus HAART and AIDS, respectively (Fig. 2). Other downregulated proteins included complement C1s subcomponent (C1S), alpha-2-macroglobulin (A2M), complement factor B (CFB), reelin (RELN), complement component C7 (C7), leucine-rich alpha-2-glycoprotein (LRG1), complement component C9 (C9), plasma protease C1 inhibitor (SERPING1), corticosteroid-binding globulin (SERPINA6), macrophage colony-stimulating factor 1 receptor (CSF1R), alpha-1B-glycoprotein (A1BG), alpha-1-antitrypsin (SERPINA1).
Inflammation, Coagulation, Complement, Immune Regulation and Cell Adhesion Are Associated with Changed Proteins in ES Plasma
The differentially expressed glycoproteins between ES and the other three groups, including HIV- and non-ES, suggests that these proteins might be involved in or a consequence of immunological suppression. To gain biological insight, we input the changed proteins into DAVID bioinformatics tool for GO analysis. The results showed that inflammatory response (p = 7E-17), complement activation (p = 5E-13), coagulation (p = 3E-12), immune response (p = 8E-10) and cell adhesion (p = 2E-6) were the representative pathways (Additional File 5: Supplementary Figure 1). The GO analysis was also performed on the basis of each comparison among the cohorts (Additional file 3: Supplementary Table 3). The changed proteins and associated pathways were summarized in Additional file 4: Supplementary Table 4. The analysis revealed that proteins IL6ST, VNN1, GPLD1, THBS1, CD163, SAA4, SERPINF1, LRG1 and SERPINA6 altered in ES versus HAART or AIDS appeared to associate with inflammation (Fig. 3). It is reported that inflammation markers IL-6 and CRP are elevated in ES [22], our analysis suggested that overlapping while distinguishable glycoprotein profiles for inflammation appeared to be present between ES and HAART cohorts suggestive of different triggers for inflammation and immune activation between natural and treatment-related viral suppression. Proteins A2M, SERPINA1, F11, MMRN1 and FGB in coagulation and C1S, C9, CFB and CFH in complement were also changed to suggest dysregulation of these pathways (Fig. 3). Finally, the diseases associated with prolonged dysregulation of the changed proteins in ES versus HIV- were revealed to be cardiovascular diseases (p = 2E-3), metabolic (p = 7E-2) and haematological (p = 9E-2).
Discussion
The HIV elite suppressors are a natural model in successfully halting or delaying disease progression. Thus, significant attention has been paid to study the host, virus, CD8+ T cells and antibody associated with this cohort [12, 13, 16, 23]. However, interesting questions remain whether the elite suppressors share similarity in the immune-relevant proteins in the plasma, even if their genetic background and particular anti-viral mechanism are likely to vary. Whether such proteins play critical roles in achieving functional cure or pose pathological risk acting like a double-edged sword. To bridge this missing gap of knowledge, we used glycoproteomics to investigate glycoproteins in plasma from individuals in an ES group compared with those in HIV-, HAART and AIDS groups. We found a significant difference existed between the plasma glycoproteins of ES and those of HIV-, HAART and AIDS. The difference could be caused, separately or in combination, by changes in glycoprotein expression, glycosylation occupancy or glycosylation isoforms. Our method did not distinguish the causes of changes rather the collective effects of all changes at each glycosylation site. However, change of glycopeptides due to any of the causes suggests dysregulation for the glycoproteins and the major cause of changes is from glycoprotein expression [9]. In this study, we identified the changed glycoproteins and used the corresponding glycoproteins to perform bioinformatic analysis. Inflammation, complement, coagulation, immune regulation and cell adhesion were strongly associated with the differentially expressed glycoproteins between ES and other sample groups. Changes in these areas suggest the ES individuals might have extensive and different multifaceted immunological regulation. Low level but ongoing viral replication might be one of the reasons causing changed plasma glycoproteins [24, 25]. Recently, evidence has emerged that individuals with ES have increasing risk of coronary atherosclerosis and immune activation [4, 22, 26]. Our finding recapitulated these studies by detecting the differentially changed proteins associated with these pathways. In an extension, we were able to delineate a more comprehensive picture of the immunological status to support understanding of the pathological mechanism in these cohorts.
Inflammation was evident but distinctive between ES and non-ES. One of the elevated glycoprotein in ES cohort, IL6ST (also named gp130), binds to interleukin-6 receptor (IL-6R) and modulates the signalling of IL-6 the essential node to inflammatory response [22, 27]. A higher level of IL-6 has been shown in ES than HAART than in HIV- [22]. The soluble gp130 (sgp130) in the plasma may function as an endogenous antagonist to IL-6 [28]. Increasing evidence suggests that sgp130 is a potential therapeutic to reduce inflammation [28-31]. In addition, gp130 is required for production of IL-21 primarily from CD4+ and CD8+ T cells for viral control during chronic infection [31-33]. However, adversely, the sgp130 is significantly associated with coronary artery disease (CAD) in humans [34]. We observed a significant elevation of gp130 in ES (Fig. 2&3). This elevation indicated, on a positive side, that ES has significantly more potential to buffer systemic inflammation mediated by IL-6 and provide excellent support for IL-21 producing CD4+ and CD8+ T cells to suppress HIV. On a negative side, its elevation strongly correlated with ongoing inflammation, and previous studies report an even higher incidence of atherosclerosis in ES than that in non-ES and HIV- populations making sgp130 a potential target to manage and stratify risk of atherosclerosis in ES individuals [26]. The therapeutic potential of sgp130 for inflammation remains a novel option for clinical investigation. In addition, VNN1, protein named vanin-1, is positively associated with human inflammatory bowel diseases [35]. The protein antagonises peroxisome proliferator-activated receptor γ (PPARγ) to mediate production of inflammatory mediator by gut epithelial cells [36]. A dramatic reduction in inflammation was seen in vanin-1(-/-) mice under stimuli highlighting the role of the intestinal epithelium in the regulation of mucosal inflammation [37]. A significant elevation of vanin-1 in ES than non-ES and HIV- suggests that ES is prone to bowel inflammation (Fig. 3). Lowering vanin-1 might restore inhibition of bowel inflammation. GPLD1 cleaves GPI and GPI-anchored proteins to regulate inflammation in atherosclerosis [38, 39]. Both ES and HAART appear to have higher levels of GPLD1 to that of AIDS and HIV- (Fig. 3). The association of atherosclerosis with HIV infection suggests that GPLD1 has a regulatory role in HIV-related atherosclerosis [26]. THBS1 has anti-inflammatory and pro-inflammatory activities [40] and is found to be elevated in inflammatory processes and activates leukocytes. Therefore its increase in ES and decrease in non-ES compared to HIV- might contribute to a differential profile of inflammation and leukocyte activation (Fig. 2) [40]. Indeed, a monocyte activation marker, soluble CD163 (sCD163), has been reported to be higher in ES than non-ES and associated with coronary atherosclerosis [4]. We also observed an increase of sCD163 in ES than AIDS, supporting the observation of a higher monocyte/macrophage activation in this cohort. Four proteins i.e. SAA4, SERPINF1, LRG1, SERPINA6, were significantly lower in ES than non-ES (Fig. 3). SAA4, plasma acute phase protein, directly links to inflammation and regulation of leukocyte activation [41]. SERPINF1, a protein named PEDF, can suppress inflammation by regulating macrophage activation [42, 43]. The anti-inflammatory activity of PEDF is via induction of IL-10 expression in human monocyte-derived macrophages through an ERK2 and p38 MAPK-dependent PPARγ overexpression pathway [44]. LRG1, LRG is proposed to be a novel inflammatory biomarker in autoimmune disease [45]. SERPINA6, protein named CBG, is reported to decrease during inflammation [46]. The link of a significant decrease of SAA4 and LRG1 to inflammation in ES was observed and may implicate regulation of leukocytes. Reduction of SERPINF1 and SERPINA6 underpinned the inflammatory response in ES. Thus it was clear that, compared to non-ES, ES had a distinctive inflammatory profile that involved a sophisticated cascade of immune response to chronic infection of HIV.
Complement pathway was down-regulated in ES over other cohorts. This conclusion was inferred from the higher level of negative regulator CFH and reduced levels of positively participating proteins i.e. C1S, CFB and C9 in ES (Fig. 3). The reason for down-regulation of complement in ES was not clear. One possibility is that complement might not be essential to attack HIV and HIV infected cells directly.
Coagulation cascades were more active in ES than non-ES. The cascades start from F12, which activates downstream F11 to convert FGB to become fibrin, the essential component for fibrin clot formation. MMRN1 supports thrombus formation [47]. In a different pathway, an excess level of F12 will catalyze the conversion of plasminogen to active plasmin that breaks down fibrin clot. Presence of the clot pieces in the blood circulation increases the risk of venous thrombosis and pulmonary embolism. Collectively, the elevated levels of F11, FGB and MMRN1 and down-regulation of negative regulators, i.e. A2M and SERPINA1 prevent production of fibrin and plasmin respectively, suggesting that individuals with ES are more prone to hypercoagulability than non-ES whose incidence of thromboembolic events has already been reported to be higher than the general population (Fig. 3) [48, 49]. It is not clear whether up-regulation of coagulation is a result or a necessity for ES or probably T cell immune-suppression. This hypercoagulability definitely affects health-management and merits use of therapeutic options.
We also observed a number of changed proteins, which have implications for immune regulation and function in cell adhesion (Additional File 4: Supplementary Table 4). The functions of GOLM1, LAMP2 and HPX in ES were obscure. IL-18BP, ICOSLG and CSF1R, however, are clearly involved in T cell function. IL-18BP is an antagonist of IL-18, which is involved in the defence against pathogens [50]. ICOS ligand and the associated ICOS pathway play a critical role in the generation of follicular helper T cells, which are the main CD4 T cell compartment for HIV [51, 52]. CSF1R is the receptor of macrophage colony-stimulating factor, a cytokine that regulates the production, differentiation and functions of the macrophage [53]. A significant decrease of the three proteins was seen in ES suggesting their down-regulation might be a consequence or be part of immunological suppression of HIV.
Conclusion
Involvement of plasma glycoproteins in ES, HAART and AIDS to HIV- was investigated through the use of glycoproteomics and mass spectrometry. We observed moderate changes in glycoprotein levels after anti-viral drug treatment indicating the profile of plasma glycoproteins might be subject to modest disturbance upon treatment. Striking change of glycoproteins was observed for ES. These changed proteins clarified that inflammation existed in ES. This finding raises the concern that neither anti-viral treatment nor natural suppression is able to normalize the health status of infected individuals. Thus, therapeutics regulating these adverse effects may be required for long-term disease management. We found that the inflammatory profile appears to be distinct between ES and non-ES suggesting that different triggers for inflammation may be present for these two cohorts. Glycoproteins involved in complement, coagulation, immune regulation and cell adhesion were also revealed to be part of the ES phenotype although their roles are unclear. It was not clear that those affected pathways and glycoproteins were an essential component for effective ES or merely a consequence of immune cell function and chronic infection. The specific glycoproteins associated with the diseases were theoretically discernable favoring translational application in HIV therapeutics.
Acknowledgements
This research was supported by National Institutes of Health (NIH), National Heart Lung and Blood Institute (NHLBI) grant, Programs of excellence in glycosciences (PEG, P01HL107153) and National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIAID) grant for JHU-Guangxi, China Clinical Trials Unit (1U01 AI69482-01) to Dr. J. Brooks Jackson. Dr. Laeyendecker and Ms. Wendel are supported by the Division of Intramural Research NIAID. We also acknowledge the support of Dr. Susan Eshleman and the HIV Prevention Trials Network (HPTN) sponsored by the NIAID, National Institute of Child Health and Human Development, National Institute of Mental Health, and the Office of AIDS research of the NIH, DHHS (UM1 AI068613). This work was also supported in part by the National Institutes of Health under grants and contracts of National Cancer Institute, Clinical Proteomics Tumor Analysis Consortium (U24CA160036), the Early Detection Research Network (EDRN, U01CA152813 and U24CA115102), and R01CA112314 and NHLBI Proteomic Center (N01-HV-00240). Publication of this article was funded in part by the Open Access Promotion Fund of the Johns Hopkins University Libraries.
Abbreviations
HIV: human immunodeficiency virus; ES: elite suppressor; HAART: highly active antiretroviral therapy; AIDS: acquired immunodeficiency syndrome.
Supplementary Material
Additional File 1Supplementary Table 1 - Identified glycosite-containing peptides from all groups of plasma samples.
Supplementary Table 2 - Quantified glycosite-containing peptides from individual plasma sample.
Supplementary Table 3 - GO analysis of changed proteins in comparisons.
Supplementary Table 4 - GO analysis of biological processes for changed proteins in ES.
Supplementary Figure 1.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Global report. UNAIDS report on the global AIDS epidemic. Joint United Nations Programme on HIV/AIDS (UNAIDS). 2013
2. Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N. et al. Prevention of HIV-1 infection with early antiretroviral therapy. The New England journal of medicine. 2011;365:493-505 doi:10.1056/NEJMoa1105243
3. Blankson JN. Control of HIV-1 replication in elite suppressors. Discovery medicine. 2010;9:261-6
4. Pereyra F, Lo J, Triant VA, Wei J, Buzon MJ, Fitch KV. et al. Increased coronary atherosclerosis and immune activation in HIV-1 elite controllers. AIDS (London, England). 2012;26:2409-12 doi:10.1097/QAD.0b013e32835a9950
5. Lake JE, Currier JS. Metabolic disease in HIV infection. The Lancet infectious diseases. 2013;13:964-75 doi:10.1016/s1473-3099(13)70271-8
6. Kolarich D, Lepenies B, Seeberger PH. Glycomics, glycoproteomics and the immune system. Current opinion in chemical biology. 2012;16:214-20 doi:10.1016/j.cbpa.2011.12.006
7. Yang W, Zhou JY, Chen L, Ao M, Sun S, Aiyetan P. et al. Glycoproteomic analysis identifies human glycoproteins secreted from HIV latently infected T cells and reveals their presence in HIV+ plasma. Clinical proteomics. 2014;11:9. doi:10.1186/1559-0275-11-9
8. Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nature biotechnology. 2003;21:660-6 doi:10.1038/nbt827
9. Tian Y, Esteva FJ, Song J, Zhang H. Altered expression of sialylated glycoproteins in breast cancer using hydrazide chemistry and mass spectrometry. Molecular & cellular proteomics. 2012;11:M111_011403. doi:10.1074/mcp.M111.011403
10. Zhang H, Yi EC, Li XJ, Mallick P, Kelly-Spratt KS, Masselon CD. et al. High throughput quantitative analysis of serum proteins using glycopeptide capture and liquid chromatography mass spectrometry. Molecular & cellular proteomics: MCP. 2005;4:144-55 doi:10.1074/mcp.M400090-MCP200
11. Bailey JR, Lassen KG, Yang HC, Quinn TC, Ray SC, Blankson JN. et al. Neutralizing antibodies do not mediate suppression of human immunodeficiency virus type 1 in elite suppressors or selection of plasma virus variants in patients on highly active antiretroviral therapy. Journal of virology. 2006;80:4758-70 doi:10.1128/jvi.80.10.4758-4770.2006
12. Blankson JN, Bailey JR, Thayil S, Yang HC, Lassen K, Lai J. et al. Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. Journal of virology. 2007;81:2508-18 doi:10.1128/jvi.02165-06
13. Bailey JR, Brennan TP, O'Connell KA, Siliciano RF, Blankson JN. Evidence of CD8+ T-cell-mediated selective pressure on human immunodeficiency virus type 1 nef in HLA-B*57+ elite suppressors. Journal of virology. 2009;83:88-97 doi:10.1128/jvi.01958-08
14. Moore RD. Understanding the clinical and economic outcomes of HIV therapy: the Johns Hopkins HIV clinical practice cohort. Journal of acquired immune deficiency syndromes and human retrovirology: official publication of the International Retrovirology Association. 1998;17(Suppl 1):S38-41
15. Han Y, Lai J, Barditch-Crovo P, Gallant JE, Williams TM, Siliciano RF. et al. The role of protective HCP5 and HLA-C associated polymorphisms in the control of HIV-1 replication in a subset of elite suppressors. AIDS (London, England). 2008;22:541-4 doi:10.1097/QAD.0b013e3282f470e4
16. Pohlmeyer CW, Buckheit RW 3rd, Siliciano RF, Blankson JN. CD8+ T cells from HLA-B*57 elite suppressors effectively suppress replication of HIV-1 escape mutants. Retrovirology. 2013;10:152. doi:10.1186/1742-4690-10-152
17. Buckheit RW 3rd, Siliciano RF, Blankson JN. Primary CD8+ T cells from elite suppressors effectively eliminate non-productively HIV-1 infected resting and activated CD4+ T cells. Retrovirology. 2013;10:68. doi:10.1186/1742-4690-10-68
18. Buckheit RW 3rd, Allen TG, Alme A, Salgado M, O'Connell KA, Huculak S. et al. Host factors dictate control of viral replication in two HIV-1 controller/chronic progressor transmission pairs. Nature communications. 2012;3:716. doi:10.1038/ncomms1697
19. Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic acids research. 2007;35:D61-5 doi:10.1093/nar/gkl842
20. Kim N, Park H, He N, Lee HY, Yoon S. QCanvas: An Advanced Tool for Data Clustering and Visualization of Genomics Data. Genomics & informatics. 2012;10:263-5 doi:10.5808/gi.2012.10.4.263
21. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44-57 doi:10.1038/nprot.2008.211
22. Krishnan S, Wilson EM, Sheikh V, Rupert A, Mendoza D, Yang J. et al. Evidence for Innate Immune System Activation in HIV Type 1-Infected Elite Controllers. The Journal of infectious diseases. 2014;209:931-9 doi:10.1093/infdis/jit581
23. Sather DN, Carbonetti S, Kehayia J, Kraft Z, Mikell I, Scheid JF. et al. Broadly neutralizing antibodies developed by an HIV-positive elite neutralizer exact a replication fitness cost on the contemporaneous virus. Journal of virology. 2012;86:12676-85 doi:10.1128/jvi.01893-12
24. Salgado M, Swanson MD, Pohlmeyer CW, Buckheit RW 3rd, Wu J, Archin NM. et al. HLA-B*57 elite suppressor and chronic progressor HIV-1 isolates replicate vigorously and cause CD4+ T cell depletion in humanized BLT mice. Journal of virology. 2014;88:3340-52 doi:10.1128/jvi.03380-13
25. O'Connell KA, Brennan TP, Bailey JR, Ray SC, Siliciano RF, Blankson JN. Control of HIV-1 in elite suppressors despite ongoing replication and evolution in plasma virus. Journal of virology. 2010;84:7018-28 doi:10.1128/jvi.00548-10
26. Hsue PY, Hunt PW, Schnell A, Kalapus SC, Hoh R, Ganz P. et al. Role of viral replication, antiretroviral therapy, and immunodeficiency in HIV-associated atherosclerosis. AIDS (London, England). 2009;23:1059-67 doi:10.1097/QAD.0b013e32832b514b
27. Askevold ET, Gullestad L, Dahl CP, Yndestad A, Ueland T, Aukrust P. Interleukin-6 Signaling, Soluble Glycoprotein 130, and Inflammation in Heart Failure. Current heart failure reports. 2014 doi:10.1007/s11897-014-0185-9
28. Mitsuyama K, Tomiyasu N, Suzuki A, Takaki K, Takedatsu H, Masuda J. et al. A form of circulating interleukin-6 receptor component soluble gp130 as a potential interleukin-6 inhibitor in inflammatory bowel disease. Clinical and experimental immunology. 2006;143:125-31 doi:10.1111/j.1365-2249.2005.02960.x
29. Nasi M, Pinti M, Mussini C, Cossarizza A. Persistent inflammation in HIV infection: Established concepts, new perspectives. Immunology letters. 2014 doi:10.1016/j.imlet.2014.01.008
30. Garbers C, Thaiss W, Jones GW, Waetzig GH, Lorenzen I, Guilhot F. et al. Inhibition of classic signaling is a novel function of soluble glycoprotein 130 (sgp130), which is controlled by the ratio of interleukin 6 and soluble interleukin 6 receptor. The Journal of biological chemistry. 2011;286:42959-70 doi:10.1074/jbc.M111.295758
31. Montufar-Solis D, Garza T, Vigneswaran N, Klein JR. Soluble gp130 promotes intestinal epithelial hyperplasia during reovirus infection. International journal of experimental pathology. 2010;91:276-80 doi:10.1111/j.1365-2613.2010.00710.x
32. Williams LD, Bansal A, Sabbaj S, Heath SL, Song W, Tang J. et al. Interleukin-21-producing HIV-1-specific CD8 T cells are preferentially seen in elite controllers. Journal of virology. 2011;85:2316-24 doi:10.1128/jvi.01476-10
33. Yi JS, Du M, Zajac AJ. A vital role for interleukin-21 in the control of a chronic viral infection. Science. 2009;324:1572-6 doi:10.1126/science.1175194
34. Luchtefeld M, Schunkert H, Stoll M, Selle T, Lorier R, Grote K. et al. Signal transducer of inflammation gp130 modulates atherosclerosis in mice and man. The Journal of experimental medicine. 2007;204:1935-44 doi:10.1084/jem.20070120
35. Gensollen T, Bourges C, Rihet P, Rostan A, Millet V, Noguchi T. et al. Functional polymorphisms in the regulatory regions of the VNN1 gene are associated with susceptibility to inflammatory bowel diseases. Inflammatory bowel diseases. 2013;19:2315-25 doi:10.1097/MIB.0b013e3182a32b03
36. Berruyer C, Pouyet L, Millet V, Martin FM, LeGoffic A, Canonici A. et al. Vanin-1 licenses inflammatory mediator production by gut epithelial cells and controls colitis by antagonizing peroxisome proliferator-activated receptor gamma activity. The Journal of experimental medicine. 2006;203:2817-27 doi:10.1084/jem.20061640
37. Pouyet L, Roisin-Bouffay C, Clement A, Millet V, Garcia S, Chasson L. et al. Epithelial vanin-1 controls inflammation-driven carcinogenesis in the colitis-associated colon cancer model. Inflammatory bowel diseases. 2010;16:96-104 doi:10.1002/ibd.21031
38. Lindqvist M, Wallinder J, Bergstrom J, Henriksson AE. Plasma glycosylphosphatidylinositol phospholipase D (GPI-PLD) and abdominal aortic aneurysm. International journal of clinical and experimental medicine. 2012;5:306-9
39. Wallinder J, Bergstrom J, Henriksson AE. Discovery of a novel circulating biomarker in patients with abdominal aortic aneurysm: a pilot study using a proteomic approach. Clinical and translational science. 2012;5:56-9 doi:10.1111/j.1752-8062.2011.00372.x
40. Lopez-Dee Z, Pidcock K, Gutierrez LS. Thrombospondin-1: multiple paths to inflammation. Mediators of inflammation. 2011;2011:296069. doi:10.1155/2011/296069
41. El Kebir D, Jozsef L, Khreiss T, Pan W, Petasis NA, Serhan CN. et al. Aspirin-triggered lipoxins override the apoptosis-delaying action of serum amyloid A in human neutrophils: a novel mechanism for resolution of inflammation. Journal of immunology (Baltimore, Md: 1950). 2007;179:616-22
42. Zamiri P, Masli S, Streilein JW, Taylor AW. Pigment epithelial growth factor suppresses inflammation by modulating macrophage activation. Investigative ophthalmology & visual science. 2006;47:3912-8 doi:10.1167/iovs.05-1267
43. Zhang SX, Wang JJ, Gao G, Shao C, Mott R, Ma JX. Pigment epithelium-derived factor (PEDF) is an endogenous antiinflammatory factor. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2006;20:323-5 doi:10.1096/fj.05-4313fje
44. Yang SL, Chen SL, Wu JY, Ho TC, Tsao YP. Pigment epithelium-derived factor induces interleukin-10 expression in human macrophages by induction of PPAR gamma. Life sciences. 2010;87:26-35 doi:10.1016/j.lfs.2010.05.007
45. Serada S, Fujimoto M, Ogata A, Terabe F, Hirano T, Iijima H. et al. iTRAQ-based proteomic identification of leucine-rich alpha-2 glycoprotein as a novel inflammatory biomarker in autoimmune diseases. Annals of the rheumatic diseases. 2010;69:770-4 doi:10.1136/ard.2009.118919
46. Garrel DR. Corticosteroid-binding globulin during inflammation and burn injury: Nutritional modulation and clinical implications. Horm Res. 1996;45:245-51
47. Reheman A, Tasneem S, Ni H, Hayward CP. Mice with deleted multimerin 1 and alpha-synuclein genes have impaired platelet adhesion and impaired thrombus formation that is corrected by multimerin 1. Thrombosis research. 2010;125:e177-83 doi:10.1016/j.thromres.2010.01.009
48. Sule AA, Pandit N, Handa P, Chadachan V, Tan E, Sum FN. et al. Risk of Venous Thromboembolism in Patients Infected with HIV: A Cohort Study. The International journal of angiology: official publication of the International College of Angiology, Inc. 2013;22:95-100 doi:10.1055/s-0033-1333866
49. Funderburg NT. Markers of coagulation and inflammation often remain elevated in ART-treated HIV-infected patients. Current opinion in HIV and AIDS. 2014;9:80-6 doi:10.1097/coh.0000000000000019
50. Carbotti G, Barisione G, Orengo AM, Brizzolara A, Airoldi I, Bagnoli M. et al. The IL-18 antagonist IL-18-binding protein is produced in the human ovarian cancer microenvironment. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:4611-20 doi:10.1158/1078-0432.ccr-13-0568
51. de la Fuente H, Cibrian D, Sanchez-Madrid F. Immunoregulatory molecules are master regulators of inflammation during the immune response. FEBS letters. 2012;586:2897-905 doi:10.1016/j.febslet.2012.07.032
52. Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK. et al. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. The Journal of experimental medicine. 2012;210:143-56 doi:10.1084/jem.20121932 [doi]
53. Luo J, Elwood F, Britschgi M, Villeda S, Zhang H, Ding Z. et al. Colony-stimulating factor 1 receptor (CSF1R) signaling in injured neurons facilitates protection and survival. The Journal of experimental medicine. 2013;210:157-72 doi:10.1084/jem.20120412
Author biography
 Dr. Weiming Yang obtained his Ph.D. degree in virology from the University of Warwick, United Kingdom. He is currently a postdoctoral fellow in Dr. Hui Zhang's group that emphasizes innovation and application of glycoproteomics and glycomics. He has a special interest in developing and using novel proteomic technologies to approach critical questions in HIV research.
Dr. Weiming Yang obtained his Ph.D. degree in virology from the University of Warwick, United Kingdom. He is currently a postdoctoral fellow in Dr. Hui Zhang's group that emphasizes innovation and application of glycoproteomics and glycomics. He has a special interest in developing and using novel proteomic technologies to approach critical questions in HIV research.
 Dr. Hui Zhang is an associate professor of Pathology at Johns Hopkins University. Her primary focus is to study protein modification on the proteome scale and the effects of modification on protein function and disease progression. She has developed high-throughput technologies to study two of the most abundant protein modifications - phosphorylation and glycosylation.
Dr. Hui Zhang is an associate professor of Pathology at Johns Hopkins University. Her primary focus is to study protein modification on the proteome scale and the effects of modification on protein function and disease progression. She has developed high-throughput technologies to study two of the most abundant protein modifications - phosphorylation and glycosylation.
![]() Corresponding author: Department of Pathology, Johns Hopkins University, 4011 Smith Building, 400 N. Broadway, Baltimore, MD 21231, USA. E-mail: hzhang32edu
Corresponding author: Department of Pathology, Johns Hopkins University, 4011 Smith Building, 400 N. Broadway, Baltimore, MD 21231, USA. E-mail: hzhang32edu
 Global reach, higher impact
Global reach, higher impact