13.3
Impact Factor
Theranostics 2014; 4(10):1039-1051. doi:10.7150/thno.7866 This issue Cite
Research Paper
Megalin-Mediated Specific Uptake of Chitosan/siRNA Nanoparticles in Mouse Kidney Proximal Tubule Epithelial Cells Enables AQP1 Gene Silencing
1. The Interdisciplinary Nanoscience Center (iNANO) and Department of Molecular Biology and Genetics, Aarhus University, Aarhus C, Denmark.
2. Department of Biomedicine, Aarhus University, Aarhus C, Denmark;
3. MEMBRANES, Aarhus University, Aarhus C, Denmark;
4. Center for Interactions of Proteins in Epithelial Transport, Aarhus University, Aarhus C, Denmark.
*Present address: Kunshan RNAi Institute, Kunshan, Jiangsu 215300, China.
#Present address: Water Security Corporation, 1455 Kleppe Lane, Sparks NV 89501, USA.
Received 2013-10-11; Accepted 2014-2-28; Published 2014-8-13
Abstract
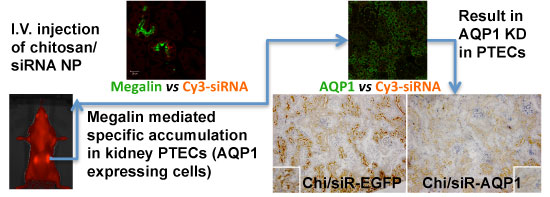
RNAi-based strategies provide a great therapeutic potential for treatment of various human diseases including kidney disorders, but face the challenge of in vivo delivery and specific targeting. The chitosan delivery system has previously been shown to target siRNA specifically to the kidneys in mice when administered intravenously. Here we confirm by 2D and 3D bioimaging that chitosan formulated siRNA is retained in the kidney for more than 48 hours where it accumulates in proximal tubule epithelial cells (PTECs), a process that was strongly dependent on the molecular weight of chitosan. Chitosan/siRNA nanoparticles, administered to chimeric mice with conditional knockout of the megalin gene, distributed almost exclusively in cells that expressed megalin, implying that the chitosan/siRNA particle uptake was mediated by a megalin-dependent endocytotic pathway. Knockdown of the water channel aquaporin 1 (AQP1) by up to 50% in PTECs was achieved utilizing the systemic i.v. delivery of chitosan/AQP1 siRNA in mice. In conclusion, specific targeting PTECs with the chitosan nanoparticle system may prove to be a useful strategy for knockdown of specific genes in PTECs, and provides a potential therapeutic strategy for treating various kidney diseases.
Keywords: siRNA, chitosan, renal targeting, megalin, aquaporin 1, optical imaging.
Introduction
Chronic kidney disease (CKD) affects 10-15% of the population and the incidence is increasing. CKD often arises as a complication from diabetes, high blood pressure, or inflammatory diseases. Without effective treatment CKD develops to renal fibrosis, which represents the main pathologically characterized progression of end-stage renal disease (ESRD) [1;2]. Currently, there is no cure for ESRD, and development of new strategies to target specific genes responsible for development of CKD may prove to be an effectively clinical treatment in patients.
RNA interference (RNAi) provides a gene silencing tool with broad applicability, high efficiency and specificity. Thus, RNAi is a potentially powerful technique for disease treatment by reducing the causative gene expression [3;4]. RNAi is mediated by small interfering RNAs (siRNA) that, upon entering the cells, provides the RNA induced silencing complex (RISC) with the guide strand complementary to the mRNA targeted for degradation. However, low stability in vivo, poor biodistribution, immunogenicity and inefficient penetration into tissue and cells are key problems for systemic administration of siRNA [5;6]. Based on our previous experience, stability [7;8] and immunogenicity issues [9] can be addressed by introducing chemical modifications, such as locked nucleic acid (LNA) and unlocked nucleic acid (UNA), and circulation time and biodistribution are heavily influenced by encapsulation of the oligonucleotides into non-viral carriers [7;8].
Accumulation of siRNA in the kidneys is dependent on the siRNA being freely filterable by the glomerulus, the rate of glomerular filtration, and the uptake of the siRNA by renal tubule epithelial cells. Using a hydrodynamic silencing approach, a technique involving rapid tail vein injection of large volumes of buffer containing siRNA or small hairpin RNA expressing DNA, it is feasible to deliver siRNA to the tubular epithelium due to transient capillary “leakage” [10]. However, this approach is accompanied with liver damage and local dysfunction of the kidneys due to the high pressure associated with the administration and, therefore, non-applicable to larger animal models and humans [11]. Local administration of siRNA directly to the kidney has been performed via the renal artery [12] or the renal vein [13], but the invasiveness of these techniques also limits their use for clinical application.
The chitosan delivery system for siRNA and related drugs has, in part, been developed in our laboratory [14;15]. Our previous biodistribution studies demonstrated that formulating the siRNA with specific chitosan particles resulted in distinct accumulation of siRNA in the kidney for at least 24 hrs post i.v. injection [7]. Here we show by real time in vivo fluorescence bioimaging and immunohistological analysis that chitosan/siRNA nanoparticles reside in the kidney for at least 48 hrs, specifically in the proximal tubule epithelial cells (PTECs). Studying the uptake of siRNA in chimeric mice with conditional knockout of the megalin gene implies that PTEC uptake of low molecular weight chitosan is mediated by the 'scavenging' receptor megalin as it has been suggested for its water soluble derivative glycol-chitosan [16]. For the first time we demonstrate delivery of non-water-soluble chitosan polyplex nanoparticles specifically into PTEC in the kidney. We furthermore exploited this technique to partially knock down the water channel aquaporin 1 (AQP1) in PTECs, providing proof-of-principle that this approach can be utilized as a novel strategy for specific targeting of genes expressed in the PTECs and thus it is a potential instrument for treating many kidney diseases [1].
Materials and Methods
siRNA Duplex Preparation
siRNA duplexes were prepared by annealing equimolar concentrations (100µM) of the sense and antisense siRNA in 5x annealing buffer (150mM Hepes, pH7.6, 500mM KC, 0.05mM MgCl2) at 95oC for 1 min and at 37oC for 1 h. The sequences used in the current study are shown in Table 1, including the siRNA against EGFP with or without fluorescence labelling, and three siRNA sequences against murine AQP1 (Ambion siRNA ID s62520, s62521 and s62562, Ambion, Copenhagen, Denmark). Chemical modification of the siRNA is also shown in Table 1.
Nanoparticle Formulation
Chitosan: Various chitosans were supplied from Novamatrix (Norway). Low molecular weight (LMW) chitosan was a gift from Bioprocess Technology, Asian Institute of Technology (Thailand).
siRNA sequences used in present study.
| Name | Sense Strand (5' to 3') | Antisense strand (5' to 3') |
|---|---|---|
| EGFP | GACGUAAACGGCCACAAGUTC | ACUUGUGGCCGUUUACGUCGC |
| LNA EGFP | GACGUAAACGGCCACAAGUTLNACLNA | ACUUGUGGCCGUUUACGUCGLNACLNA |
| F/OMe -LNA | GFAOMeCFGOMeUFAOMeAFAOMeCFGOMeGFCOMeCFAOMeCFA OMeAFGOMeUFUOMeCF | ACUUGUGGCCGUUUACGUCGLNACLNA |
| UNA | GACGUAAACGGCCACAAGUUUUNAU | ACUUGUGGCCGUUUACGUCGCUNAU |
| HNA/DNA | GACGUHNAAAAHNACGGHNACCAHNACAAGUHNAUHNATDNA | ACUUGUGGCCGUUUACGUHNACHNAGHNATDNA |
| Cy5 labeled EGFP | Cy5-GACGUAAACGGCCACAAGUTLNACLNA | ACUUGUGGCCGUUUACGUCGLNACLNA |
| Cy3 labeled EGFP | GACGUAAACGGCCACAAGUTLNACLNA | ACUUGUGGCCGUUUACGUCGLNA Cy3CLNAU |
| AQP1 s62520 (A-siR20) | CAGCAUUGGUUCUGCCCUAtt | UAGGGCAGAACCAAUGCUGat |
| AQP1 s62521 (A-siR21) | CCUCCUCCCUAGUCGACAAtt | UUGUCGACUAGGGAGGAGGtg |
| AQP1 s62522 (A-siR22) | CGCAACUUCUCAAACCACUtt | AGUGGUUUGAGAAGUUGCGgg |
| LNA A-siR22 | CGCAACUUCUCAAACCACUULNAULNA | AGUGGUUUGAGAAGUUGCGGLNAGLNAGG |
Note: The following abbreviations for modification are used: 2′-deoxy (DNA), 2′-fluoro (F), 2′-O-methyl (OMe), hexitol nucleic acid (HNA), locked nucleic acid (LNA), and unlocked nucleic acid (UNA). The position of the modification is shown in bold in the oligo sequence [27].
Each chitosan was dissolved in 1% acetic acid to give a 2% (w/v) chitosan solution. Chitosan was reprecipitated from the solution by adding 4% (w/v) NaOH dropwise until pH = 9.5. Finally, it was deacetylated with 47.7% NaOH at 60oC for 24h. The degree of deacetylation for all chitosan was around 98% and the average molecular weights were around 40, 190, 250 and 270 kDa, and represented as A, B, C and D, respectively. The detail analyses of chitosan were reported in the Supplementary Material: Table S1.
Preparation of chitosan/siRNA nanoparticles: Chitosan/siRNA nanoparticles were formed by ionic complexation between polycationic chitosan- and polyanionic siRNA solutions. RNAse free water was used for all preparations. Stock chitosan solution (1mg/ml) was prepared in 300 mM, pH 5.5 acetate buffer and was filtered through 0.45µm membrane filter. Different concentrations of 20 µL siRNA solutions were added into 1 mL chitosan solutions while stirring and left for 1h. Prior to the mixing, the ratio between chitosan and siRNA were calculated based on the molar ratio of chitosan amino groups and siRNA phosphate groups (N/P ratio).
The hydrodynamic radii of the chitosan/siRNA nanoparticles were measured by photon correlation spectroscopy using a Zetasizer Nano ZS (Malvern Instruments, Malvern, UK) prior to i.v. injection. The mean particle size and polydispersity of each formulation was recorded in the Supplementary Material: Table S2. The polydispersity index indicates the width of the particle size distribution; 0 being monodisperse and 1 being polydisperse. All measurements were carried out in triplicate with a temperature equilibration time of 1 minute at 25oC. Water (refractive index of 1.330 and a viscosity of 0.8872 cP) was chosen as dispersant.
Serum stability analysis: Twenty microliters of Chitosan/siRNA nanoparticle solution at N/P ratio of 60 was incubated with 380 µL 100% FBS (95% final serum concentration) for 0 hr (measured immediately after mixing) and 24 hr at 25oC. The particle sizes were measured by a Zetasizer Nano ZS (Malvern Instruments, Malvern, UK). The viscosity and refractive index of the resulting 95% FBS was set to 1.5 cP (Supplementary Material: Fig.S4) and 1.330 for the measurement.
Cell Lines and in vitro transfection
Madine Darby Canine Kidney (MDCK) cells stably expressing murine AQP1 were used for validation of siRNA efficiency. Cells were maintained in DMEM media containing GlutaMAX™ I, 4500 mg/L D-Glucose, Sodium Pyruvate, supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and Hygromycin B (200 μg/ml) (Invitrogen, Copenhagen, Denmark) at 37 °C in 5% CO2 and 100% humidity.
Three siRNA sequences (Ambion, Copenhagen, Denmark) were transfected via commercial reagents LipofectamineTM2000 (Invitrogen, Copenhagen, Denmark) according to standard protocols at 50 nM final siRNA concentration in 6 well plate After overnight incubation, the media was replaced with fresh cell culturing media. The cells were harvested at 48 hr post transfection for RNA and protein analysis.
Nanoparticle Injection i.v. in Mice
BALB/cJBomTac female mice (Taconic Europe, Ll.Skensved, Denmark) were used for most in vivo experiments, except conditional megalin knockout mice (megalin lox/lox cre+; B6; 129S7-Lpr2tm1Her-Wnt4tm2(EGFP/cre)Svo ). For the biodistribution experiments, 10 µg siRNA formulated with chitosan in 200 µl solution (corresponding to ~ 400 µg/kg body weight) were administered by tail vein injection. For knockdown experiments, 30 µg siRNA was formulated with chitosan in acetate buffer and concentrated using BioDesignDialysis TubingTM (14.000 MWCO, BioDesign Inc. of New York, Carmel, NY) and then was injected i.v. (corresponding to ~ 1.2 mg/kg body weight).
All procedures of animal work were approved by "The Experimental Animal Inspectorate in Denmark" under The Danish Veterinary and Food Administration, Ministry of Food, Agriculture and Fisheries.
1, Impact of Molecular Weight of chitosan on biodistribution: Ten µg Cy3 labelled EGFP siRNA duplex was formulated with chitosan A, B, C and D (N/P=10), respectively, and was injected into mice (n=2) for pharmacokinetic studies. Mice were sacrificed 24 hours post-injection and kidneys were collected.
The left kidney was dissected vertically, one half was preserved in 4% paraformaldehyde in PBS buffer, the remaining was frozen in liquid nitrogen. The fixed tissue was processed for paraffin and the frozen tissue was used for cryo-sections (see below).
The right kidney was dissected into cortex and medulla after freezing at -20oC for 10 mins. The tissue was stored in RNAlater and RNA was isolated for northern blotting (See below).
A similar experiment was performed for chitosan A/siRNA (N/P=60) (n=2), and kidneys analysed by northern blotting.
2, Fluorescence Imaging Study: To avoid autofluorescence during imaging, fur was removed from the back and the abdomen of mice by shaving. A single dose of 200 μl chitosan A/Cy5 labeled siRNA (containing 10μg siRNA duplex) nanoparticles (N/P=60) was injected via the tail vein (n=4). Both Cy5 labelled siRNA alone and buffer were injected as controls (n=3). The mice were scanned using an IVIS® 200 imaging system (Xenogen, Caliper Life Sciences, Hopkinton, MA) under anesthesia with 2.5-3.75% isoflurane (Forene®, Abbott, Copenhagen). Cy5 excitation (λex = 640 nm) and emission (λem = 700 nm) filters were used. Identical illumination settings, including exposure time (1s), binning factor (medium), f-stop (2), and fields of view (13 × 13 cm), were used for all image acquisitions. The mice were scanned at 2 min, 1, 2, 3, 4, 6, 24 and/or 48 hrs (siRNA alone and buffer were scanned at 2 min, 1, 2, 3, 24 hrs). At the end point, the mice were killed, and the individual organs removed and scanned.
Total emission from affected areas (Region of Interest, ROI) from each mouse was quantified with Living Image 4.0 software package (Caliper Life Sciences, Hopkinton, MA). The radiant efficiency of the kidney was measured ((photons/sec/cm2/sr)/(μW/cm2)) and is presented as radiance/illumination power density. Background fluorescence was subtracted prior to analysis.
3, Cellular Uptake of Chitosan/siRNA Nanoparticles in Conditional Megalin KO Mice: Chitosan/Cy3 labeled siRNA nanoparticles, Cy3 labeled chitosan/siRNA nanoparticles (N/P=60), Cy3 labeled siRNA alone or buffer were injected i.v. into an already established mouse model [17] with conditional knockout of megalin (n=3 for each group). In this model, megalin is expressed in ~ 10% of kidney proximal tubule epithelial cells. The experiment was terminated 24 hr post-injection and the kidneys were fixed by retrograde perfusion through the abdominal aorta with 2% paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.4. The tissue was dehydrated and embedded in paraffin by standard methods.
4, AQP1 Gene Silencing in Mice: Chitosan/AQP1 siRNA, chitosan/control siRNA (against EGFP), or buffer were injected i.v. in mice (n=5). SiRNA was LNA modified in the 3′ overhangs for both strands and nanoparticles were made at N/P ratio = 60. Three total injections were made at day 0, day 3 and day 5, with dose of 30 µg siRNA (corresponding to approximately ~ 1.2 mg/kg body weight). The experiment was terminated at day 7. The kidney was dissected vertically, one half was preserved in 4% paraformaldehyde in PBS buffer; the remaining was further separated into two equal portions, one for RNA purification (stored in RNA later) and one for protein purification (quick frozen in liquid nitrogen).
Evaluation Methods for siRNA Integrity, Cellular Localization and Gene Expression
siRNA Integrity Analysis by Northern Blotting: Total RNA was purified by Trizol® reagent (Invitrogen, Copenhagen, Denmark) from kidney tissues in RNAlater. Four µg total RNA was run on 15% denaturing polyacrylamide gels and transferred onto Hybond-N+ membrane (Amersham Biosciences, Copenhagen, Denmark). After UV cross-linking, the membranes were probed with [γ-32P] ATP labelled sense strand LNA modified siRNA according to standard procedures. The membranes were analyzed on a Typhoon phosphoimager to visualize the antisense strand signal that bound with 32P labelled sense strand of the siRNA [7].
Quantitative real time RT PCR (qRT-PCR): Total RNA was used for reverse transcription using the SuperScript® II Reverse Transcriptase (RT) kit according to the manufacturer's protocol (Invitrogen, Copenhagen, Denmark). Quantitative real-time RT PCR was performed as described previously [18] on an Mx4000® Multiplex Quantitative PCR System (Stratagene, Copenhagen, Denmark). The comparative CT (threshold cycle) method was used to quantitate the relative AQP1 mRNA expression levels, comparing treated samples to concordant non-treated controls. The beta-actin gene mRNA was amplified as an internal control to normalise the data of AQP1 mRNA level. The primer sequences used for AQP1 gene are, Forward: 5'-TGAGATCATTGGCACTCTGC, Reverse: 5'-TGATACCGCAGCCAGTGTAG; the primer sequences for beta-actin gene are, Forward: 5'-ACACAGTGCTGTCTGGTGGT, Reverse: 5'-CTGGAAGGTGGACAGTGAGG.
Western Blotting: Affinity-purified antibodies used were against AQP1 [19] AQP2 [20], and beta-actin (Sigma, Copenhagen, Denmark). Kidney tissues were homogenized in 0.3M sucrose, 25 mM imidazol, 1 mM EDTA in ddH2O, pH 7.2) containing 8.4 mM leupeptin and 0.4 mM pefabloc. The tissue samples were homogenized using an ultra Turrax T8 homogenizer (IKA Labortechnik; Staufen, Germany) and centrifuged at 4000 3 g for 15 min at 4C. The supernatant (or cell pellets from MDCK cell studies) were diluted in SDS buffer containing a final concentration of 62 mM Tris (hydroxymethyl)-aminomethane, 0.1 M SDS, 8.7% glycerol, 0.09 mM bromophenol blue, and 0.04 M dithiothreitol (DTT), pH 6.8. The protein samples were heated for 5 min at 90C. SDS-PAGE was performed on 4-15% gradient polyacrylamide gels (Ready Gels, Bio-Rad, Copenhagen, Denmark). The proteins were transferred electrophoretically to immobilon PVDF membranes. Membranes were dried and blocked for 1 h at room temperature in Odyssey blocking buffer:PBS, 1:1 (blocking buffer), and probed overnight at 4°C with primary antibody in blocking buffer. After washing, blots were incubated with species-specific fluorescence secondary antibodies (Alexa 688) and visualized using the Odyssey Infrared Imaging System (LiCor, Lincoln, NE). Quantification was performed using Odyssey software.
Fluorescence Microscopy and immunohistochemical (IHC) Staining: The renal tubule and cell type location of chitosan /Cy3 siRNA nanoparticles were assessed according to the histological structure and by fluorescence labeling with the segment specific markers AQP1 (proximal tubule, thin descending limbs of Henles loop, vasa recta), AQP2 (connecting tubule and collecting ducts). Paraffin embedded tissue was sectioned at 2µm using a Leica RM 2165 microtome (Leica, Wetzlar, Germany). Sections were dried at 60°C for 1 hour, placed xylene overnight, rehydrated in graded alcohols and blocked for endogenous peroxidise. Antigen retrieval was performed by heating the sections in TEG buffer (Tris-EGTA buffer, pH 9) at approximately 50°C for 20 min, thereafter the sections were cooled at room temperature for 30 min and incubated for 30 min in 50 nM NH4Cl in 0.01 M PBS. Permeabilization was obtained with 0.05% saponin (1% BSA, 0.2% gelatine, 0.05% saponin in 0.01 M PBS) and sections were incubated with primary antibody in 0.01 M PBS, 0.1% BSA and 0.3% Triton X-100 followed by incubation with Alexa-conjugated secondary antibody. Sheep anti megalin was kindly provided by Dr. Pierre Verroust [21]. Images were obtained using either a confocal laser-scanning microscope (LSM 510-META; Carl Zeiss, Göttingen, Germany) and Zeiss Zen software (2009, Light Edition) or a Leica TCS SL confocal microscope.
Statistical Analysis: Student's t-test was performed to compare the fluorescent signal between mice injected with Cy5-siRNA and chitosan/Cy5-siRNA at different time points, scanned by optical imaging system. Analysis of Variance (ANOVA) was performed to evaluate the significant difference among three groups for their AQP1 mRNA expression level and protein level in the in vivo gene silencing experiment. Difference between A-siR22 and EGFPsiR/Buffer was further evaluated by post Tukey's Multiple Comparison Test.
Results
Chemically modified siRNA formulated with chitosan accumulates in the kidney independent of siRNA modifications
Chitosan [16;22-24], and chitosan-formulated LNA-modified siRNA [7] have previously been shown to accumulate in the kidney. To study the effect of chemical modification on siRNA distribution, various chemical modifications including 2′-deoxy, 2′-fluoro 2′-O-methyl-LNA (F/OMe-LNA), unlocked nucleic acid (UNA) and Hexitol nucleic acid (HNA) were included in the RNA strands (Table 1) [25]. The chitosan formulated siRNA variants were injected into the tail vein of mice and their biodistribution examined. Various organs were collected 24 hrs post injection, and total RNA was purified and subjected to northern blotting analysis using the passenger strand of the siRNA as probe. All chemically modified siRNAs were found to accumulate in the kidney 24 hrs post injection (Supplementary Material: Fig. S1), whereas unmodified siRNA could not be detected in any organs except from a weak signal in spleen 24 hrs post injection (Supplementary Material: Fig. S2a) Formulation of the siRNA with chitosan provided up to 5 minutes protection to unmodified siRNA in blood circulation (Fig. S2b) compared to < 1 minute life time for naked unmodified siRNA [7]. All siRNA applied in subsequent in vivo experiments in this report contained LNA modifications.
Accumulation of siRNA in the kidneys depends on the molecular weight of chitosan
It is well known that the degree of deacetylation (DD) and the molecular weight (MW) of chitosan are crucial parameters for optimal transfection in vitro. In order to investigate the impact of the MW of chitosan on siRNA biodistribution in a mouse, we prepared highly deacetylated chitosan with 4 different molecular weights. Using these chitosan samples, chitosan-siRNA polyplex nanoparticles were formulated. The detail characteristics of chitosan and the size of the resultant chitosan-siRNA nanoparticles were reported in the Supplementary Material: Table S1 and S2.
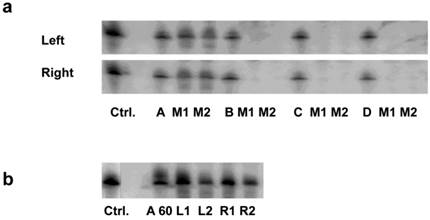
Among these formulations, we found that only the nanoparticles prepared with chitosan A (40kDa) led to significant accumulation in the cortex of kidney (Fig. 1a). The siRNA intensities between left and right kidney were, on average, very similar (Fig. 1a). A similar distribution was observed using higher N/P ratio (N/P= 60; Fig. 1b). Because an in vitro transfection assay showed the formulation with chitosan A at N/P ratio of 60 was far higher than that of 10 (Supplementary Material: Fig.S3a), and the nanoparticles in N/P=10 formulation tended to form agglomerates and later deposited in lung whereas no such occurrence was observed for N/P=60 (Fig.S3b), we, therefore, decided to use only the chitosan-A/siRNA nanoparticles with N/P ratio of 60 for further investigation.
Impact of molecular weight of chitosan on siRNA delivery to the kidney: (a). Delivery using chitosan with various molecular weights. siRNA formulated with chitosan of different molecular weights (N/P=10) were i.v. injected in mice. Kidneys were collected 24 hrs post injection. Four micrograms of RNA isolated from cortex of left and right kidneys was run on 15% denaturing polyacrylamide gels and northern analysis was performed using [γ-32P] ATP labelled siRNA sense strand. Lane 1, 1 ng siRNA duplexes (Ctrl.); Lanes 2-4/5-7/8-10/11-13, 1 ng siRNA/chitosan complex (N/P=10) (Chitosan A, B, C, D, respectively), experiments were performed in groups of two mice (M1, M2) for each formulation. (b). Delivery using chitosan A/siRNA (N/P= 60). siRNA formulated with chitosan A/siRNA (N/P= 60) was injected i.v. in mice (n=2) and kidneys were collected 24 hrs post injection. RNA was obtained from cortex and northern analysis was performed as described above. Lane 1, 1 ng siRNA duplexes (Ctrl.); Lane 2, 1 ng chitosan A/siRNA; Lanes 3-6, RNA from left (L1 and L2) and right kidney (R1 and R2) from mouse 1 (L1 and R1) and mouse 2 (L2 and R2), respectively.
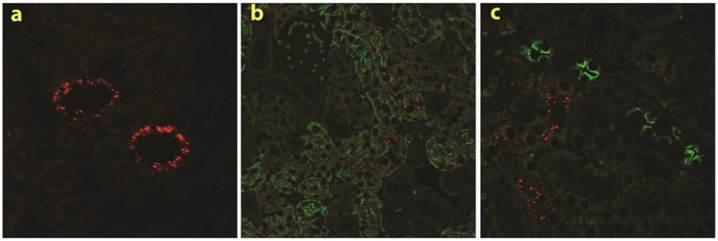
Specific accumulation of chitosan A/siRNA nanoparticles in kidney PTECs: The kidneys were collected at 24 hrs post-injection from the mice injected with polyplexes formulated with Cy3-siRNA and chitosan A (N/P=60). Sections were made and analysed by fluorescence microscopy, Cy3 signal (red dots) was observed in the kidney (a); the neighbouring sections from the same experiment were labelled with specific antibodies against AQP1 and AQP2 (both in green), clearly demonstrated that chitosan/Cy3-siRNA is co-localised with AQP1 at proximal tubular cells (b), but not with AQP2 in collecting duct cells (c).
The chitosan-A/siRNA nanoparticles were incubated in 95% FBS for 24 hrs, and the nanoparticle sizes were determined in water (Supplementary Material: Fig. S4a) and FBS as dispersant (Fig. S4c) and FBS alone (Fig. S4b), in order to assess the serum stability (Fig. S4d). Although a small shift of size distribution was observed, no particle aggregation was observed.
Chitosan/siRNA accumulates in kidney PTECs
To investigate the specific location of siRNA in the kidney in more detail, we made tissue sections from both frozen and paraffin-embedded kidneys. Cy3 conjugated siRNA was clearly observed by confocal laser-scanning microscopy in histological sections obtained from mice injected with chitosan A/Cy3-siRNA (2a). The fluorescence intensity was greatest in the proximal tubular cells (Fig. 2a). No signal was detectable in kidney sections obtained from mice injected with Cy3-siRNA formulated with B, C or D chitosan or buffer (Data not shown).
To confirm the localization of chitosan A/Cy3-siRNA in PTECs, immunohistochemical staining and confocal microscopy were performed on serial tissue sections using antibodies against markers for the proximal tubule, aquaporin 1 (AQP 1), or for the distal tubule, aquaporin 2 (AQP2). Cy3-siRNA was detected only in cells also positive for AQP1 expression (Fig. 2b), but not in cells expressing AQP2 (Fig. 2c), confirming that chitosan/siRNA nanoparticles specifically accumulated in PTECs. The consistent results obtained from both northern blotting and confocal microscopy prompted us to choose LMW chitosan A (subsequently referred to as chitosan) and an N/P ratio of 60 (conditions where we observed greatest significant knockdown in vitro for this type of chitosan; Data not shown) for all subsequent experiments.
Endocytotic uptake of chitosan/siRNA in PTECs is mediated by megalin
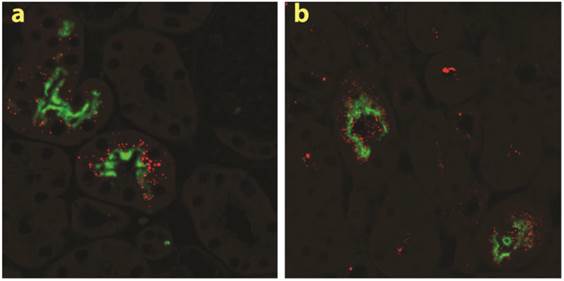
The two multiligand, endocytic receptors, megalin or cubilin are responsible for the receptor-mediated endocytotic uptake of low molecular weight (LMW) proteins from the glomerular ultrafiltrate in PTECs [26]. Since low MW chitosan/siRNA specifically accumulated in PTECs we speculated if megalin and/or cubilin could be responsible for this uptake. To examine this hypothesis we applied our delivery system to transgenic mice carrying a conditional knockout of the megalin gene. In these animals approximately 90% of the PTECs lack megalin expression [17]. In addition to Cy3-labelled siRNA we also injected chitosan that was directly labelled with Cy3. Fluorescence signal was observed in mice injected with either chitosan/Cy3-siRNA or Cy3-chitosan/siRNA in the subset of PTECs also expressing megalin, as visualized by immunolabeling using a megalin specific antibody (Fig. 3a, b). These results demonstrate that the uptake of both siRNA and chitosan in PTECs is mediated by a megalin-dependent endocytotic pathway. Furthermore, this result suggests that chitosan and siRNA remain associated when being filtered by the kidney and upon entering the PTECs. No signal was detected in mice injected with Cy3-labeled siRNA alone or buffer (n=3 for each group) after 24 hours (data not shown), suggesting that the megalin-associated uptake in the PTECs is chitosan specific.
Dynamics of chitosan/siRNA accumulation in kidney observed by optical imaging
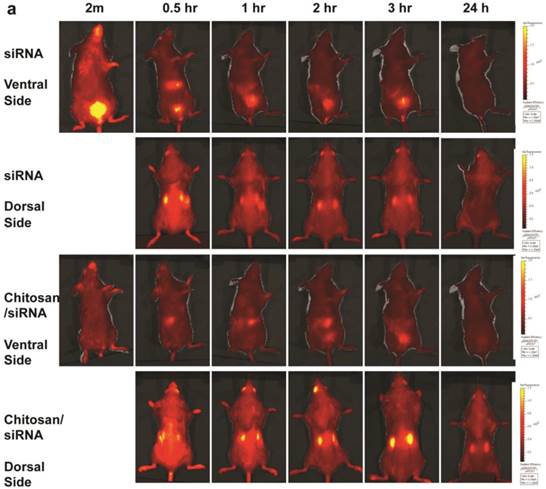
To monitor the dynamic changes in siRNA blood clearance, urine excretion and kidney accumulation, fluorescent imaging of fluorescently labelled siRNA was performed in living animals. To facilitate in vivo imaging and reduce fluorescent background, the red-shifted Cy5 dye was used as a siRNA tracer. Chitosan/Cy5-siRNA was prepared and injected via the tail vein, and fluorescent images were recorded dorsally and ventrally at 2 min, 30 min, and 1, 2, 3, 4, 6, 24, and 48 hrs post-injection using a 2D optical imaging system. Animals treated with unformulated Cy5 labelled siRNA or buffer alone were included as controls and imaged at various time points.
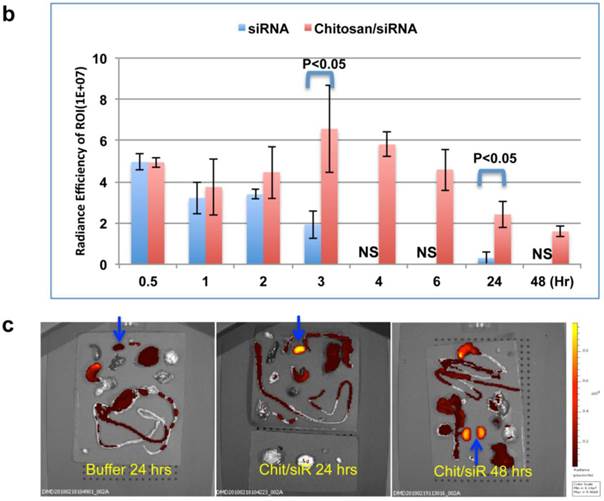
A strong fluorescent signal was observed at the nose and paws, regions of the mouse that are rich in blood capillaries, immediately following injection of chitosan/Cy5 siRNA (ventral view; Fig. 4a), whereas no clear signal was observed in the bladder. This is in contrast to the mice treated with Cy5-siRNA alone, where the bladder signal rapidly increases to become the strongest signal 2 min post injection (Fig. 4a), indicating that the majority of the naked siRNA was excreted into the urine a few minutes post injection. No Cy5 signal was observed in mice injected with buffer alone (data not shown). Signal was observed in the liver ~ 30 mins after injection both for siRNA alone and siRNA formulated with chitosan (Fig. 4a), and diminished 2 to 3 hrs post injection. When chitosan/siRNA was imaged from the dorsal side, Cy5 signals were apparent in the kidney after 20 - 30 min and peaked at 3 - 4 hrs, with some signal remaining more than 48 hrs post injection (Fig. 4a). siRNA alone peaked at an earlier time point (30 min) and disappeared almost completely 1 hr to 3 hrs post injection (Fig. 4a). A significant difference in fluorescent intensity was observed between mice treated with chitosan/siRNA and siRNA alone that scanned at 3 hrs and 24 hrs post injection (p<0.05), respectively (Fig. 4b). After 24 or 48 hrs post injection the mice were euthanized at and the organs were dissected for scanning. For the chitosan/siRNA group, the strongest signal was clearly observed in the kidneys, with the signal intensity being >10 fold higher in kidneys compared with other organs (except stomach which was strongly autofluorescent) both 24 and 48 hrs post injection (Fig. 4c). These results are in agreement with our observations obtained by northern blot analysis and fluorescence microscopy, which showed that chitosan/siRNA specifically accumulates and resides in the kidney for at least 48 hrs post injection.
Megalin-mediated endocytotic uptake of chitosan/siRNA nanoparticles in PTECs: Chitosan/Cy3-siRNA and Cy3-chitosan/siRNA polyplexes were injected via tail vein into a mouse model with conditional deletion of the megalin gene and the kidneys were collected after 24 hrs (n=3). Serial sections were labelled with specific antibodies against megalin. Both chitosan/Cy3-siRNA (a) and Cy3-chitosan/siRNA (b) signals co-localized with megalin expressing cells.
Dynamic observation of chitosan/siRNA polyplexes in the kidney using in vivo optical imaging: Whole animal fluorescence scanning was performed at the indicated time points post-injection of chitosan/Cy5 labelled siRNA. (a) Representative images of mice injected with siRNA alone or chitosan/siRNA nanoparticles at 2 min, 0.5hr, 1hr, 2hr, 3 hr, and 24 hr. Scanning was performed either from the ventral side (panel 1 & 3) showing the signals at mouth/nose, liver, and bladder or dorsal side (panel 2 & 4), showing signals from paws, ears, and kidney. (b), To quantify the signal intensity of images, the average radiant efficiency [(photons/sec/cm2/sr)/(µW/cm2)] was measured using the Living Image 4.0 software package. Radiant efficiency from the region of interest (ROI) (kidney) in mice injected with siRNA (n=3) and chitosan/siRNA (n=4) at the indicated time points is presented (NS - not scanned at this time point for siRNA injected mice). Significant difference for the fluorescent intensity was seen between the two groups at 3 hrs and 24 hrs (p<0.05 evaluated by Student t-test). (c) Ex vivo imaging of dissected organs 24 or 48 hrs after administration of chitosan/siRNA nanoparticle or buffer (only at 24 hrs), including kidneys (arrows), stomach, liver, intestine/colon, spleen, lung, and heart. Strong Cy5 signal was only observed in the kidney except a prominent auto-fluorescence signal originated from feed in the stomach.
Specific knockdown of AQP1 expression in mouse kidney
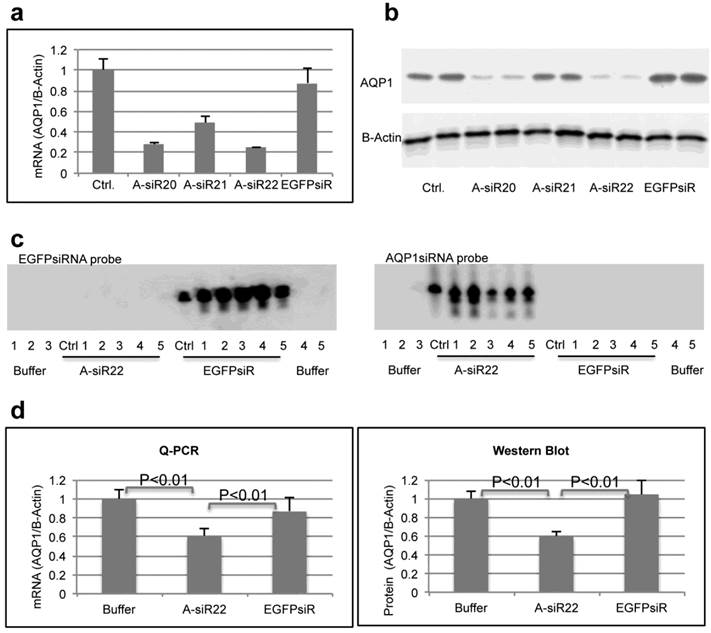
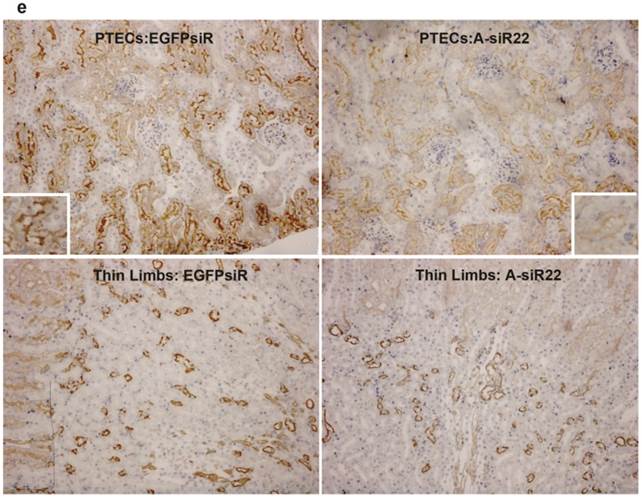
To determine if the chitosan/siRNA particles could be used to deliver functionally active siRNAs to PTECs, we examined the effectiveness of this approach to silence the gene expression of the water channel AQP1, which is predominately expressed in PTECs. Three siRNAs, designed to target mouse AQP1 (Table 1), were tested in MDCK cells expressing mouse AQP1. Two of these siRNAs showed a strong knockdown of both AQP1 mRNA and protein expression (Fig. 5a, b). A-siR22 was selected for further experiments and modified chemically with LNA in the overhangs to increase stability in vivo. Thirty micrograms of AQP1 or control EGFP siRNA (1.2 mg/kg body weight for 25g mice) were formulated with chitosan and administrated via the tail vein at days 0, 3, and 5. The experiment was terminated at day 7, and the kidneys were collected. Total RNA was isolated and northern blotting was performed. Both intact AQP1 siRNA and control EGFP siRNA could be detected in all kidney samples, indicating efficient renal delivery in a sequence independent fashion (Fig. 5c). Both AQP1 mRNA and protein levels were specifically decreased by > 40% in kidneys from animals treated with AQP1 siRNA/chitosan compared to the control groups treated with either buffer or EGFP siRNA (Fig. 5d). To further investigate the AQP1 knock down at a cellular level, AQP1 immunolabeling was performed on kidney sections from the treated animals. A clear reduction in AQP1 labelling was observed in PTECs of kidneys from animals treated with chitosan formulated AQP1 siRNA, compared with control EGFP siRNA treated animals. No apparent reduction in AQP1 expression was observed in thin limbs of Henle's loop (Fig. 5e). These data confirm that the accumulating chitosan/siRNA in PTECs is functional and capable of specific knockdown of gene expression.
Efficient gene silencing of AQP1 gene in vitro and in vivo: Three siRNA targeted against murine AQP1 (A-siR20, A-siR21 and A-siR22) were transfected in duplicate into AQP1-MDCK cells using Lipofectamine 2000. Non-transfected cells (Ctrl) or cells transfected with siRNA against EGFP (EGFPsiR) were included as controls. Cells were harvested 48 hrs post transfection and AQP1 mRNA (a) and protein levels (b) were evaluated by quantitative RT-PCR and western blotting, respectively. (c) A-siR22 or EGFPsiR was LNA modified in the 2-nucleotides overhangs for in vivo experiments and formulated with chitosan. Thirty micrograms of A-siR22 or EGFPsiR (equal to 1.2mg/kg body weight for 25g mice) were administrated i.v. at day 0, 3, and 5 (n=5). Buffer was injected as control (n=5). The experiment was terminated at day 7, and the kidneys were collected. Total RNA was isolated and northern blotting was performed using sense strands from siRNA targeted against EGFP or AQP1 as probes. A major band migrating as intact guide strands was detected for both probes whereas no signal was observed in mice injected with buffer. One nanogram of siRNA duplex was loaded as control (Ctrl). (d), AQP1 mRNA and protein levels were evaluated by quantitative RT-PCR and western blotting, respectively. Both AQP1 mRNA and protein expression were normalized with β-actin and presented as mean±SD (n=5). A significant reduction (p<0.01 evaluated by One way Analysis of Variance) in AQP1 mRNA and protein levels compared to either buffer alone or EGFPsiR/buffer was detected in mice injected with A-siR22. (e), Immunohistochemical staining of AQP1 demonstrated that AQP1 protein levels are decreased in PTECs, but not at thin limbs of Henle's loop. The knockdown in PTECs was only observed in mice injected with A-siR22 and not in the control group injected with EGFPsiR, a representative region was magnified within in a square at the corner.
Discussion
In this report we have demonstrated specific renal delivery of chitosan/siRNA nanoparticles into kidney PTECs in a living animal. We have utilized the technique to obtain gene silencing of AQP1 in PTECs, and as such provided “proof-of-principle” that this approach could be utilized for knockdown of various other genes in PTECs. Compared to previous delivery methods, chitosan mediated siRNA targeting to the PTEC avoids high pressure hydrodynamic injection [10], which may led to disruption of the cellular membrane and undesired side effects [27]. Furthermore, the use of chitosan can potentially avoid surgical injury due to direct administration into the renal artery or veins [12;28], which may hamper clinical use.
Other systemic delivery systems targeting the kidney have been previously reported [11]. For example, siRNA conjugated with specific antibodies were successfully delivered into podocytes [29]; nanoparticles with defined size (~75 ± 25-nm diameters) could target the mesangium [30]; and cationic cyclodextrin-containing polymer (CDP)-based siRNA nanoparticles were able to accumulate and disassemble in the glomerular basement membrane [31]. Furthermore, subcutaneous injection of cholesterol-conjugated siRNA led to accumulation of siRNA predominantly in the kidney cortex and glomeruli [32]. This approach was successful for ameliorating diabetic nephropathy in mice, but required injections of a high dose (16-20 mg/kg siRNA per injection) of siRNA biweekly for 7 weeks. Naked delivery of functional siRNA to renal PTECs has also been shown, but, again, requiring relatively high doses, i.e.12-24mg/kg of siRNA by bolus i.v. injection, and with a relative short lifetime in tissues ~1 hr [33]. Additionally, it was reported that both free 3H-labeled antisense oligonucleotides (ODNs) and ODNs encapsulated in a PLGA polymer (Mw 3000 Da) could accumulate in PTECs 24 hrs post subcutaneous injection [34]. However the mechanism for specific accumulation of ODNs and the integrity of ODNs were not investigated.
Chitosan, a β (1-4) linked copolymer of D-glucosamine and N-acetyl-D-glucosamine derived from chitin, is orally non-toxic in animals [35] and humans [36] and has been intensively investigated for drug and gene delivery [37]. Low molecular weight chitosan and its water soluble derivative, glycol-chitosan, were reported to selectively accumulate in the mouse kidney renal tubules following i.v. injection and thus could potentially be utilized to deliver drugs or other compounds to the kidneys [16;22-24;38]. In present study we utilized non-water-soluble chitosan polyplex nanoparticles to deliver siRNA specific into the PTECs.
Several techniques, including optical live imaging, fluorescence microscopy, immunohistochemical staining and RNAi, were applied to demonstrate that chitosan siRNA particles can be functionally targeted to PTECs. In agreement with previous observations, only LMW chitosan formulated siRNA localized in the kidney [7;22], with 40 kDa chitosan being most efficient for kidney siRNA delivery. In contrast, the N/P ratio had only little impact on siRNA accumulation in the kidney, implying that the molecular weight of chitosan, rather than the N/P ratio, plays a dominant role in kidney specific accumulation of siRNA.
In the current study we also investigated the mechanism for chitosan/siRNA delivery into PTECs. In principle, antisense oligonucleotides can be taken up both at the capillary and tubular lumen sides of the kidney [39;40]. Uptake of solutes from the tubular lumen side of PTECs is often mediated by the two receptors, megalin and cubilin, that are responsible for the receptor-mediated endocytotic uptake of various substances from the glomerular ultrafiltrate. Notably, a natural substrate for megalin, aminoglycosides [41], shares a strong resemblance to chitosan; in particular the glucosamine unit. Additionally, the aminoglycoside gentamycin effectively competes with chitosan for the association with PTECs in mice [16], suggesting that the specific accumulation of LMW chitosan in kidney proximal tubules may be mediated by megalin. To confirm that the renal delivery of chitosan/siRNA is mediated by megalin, we administrated our delivery system to a transgenic mouse with conditional knockout of the megalin gene [17]. In this model, megalin deletion occurs in a mosaic pattern and allows a direct comparison of PTECs with and without megalin expression in the same nephron. The clear overlap between siRNA uptake and megalin-expressing cells implies that megalin is indeed the mediator of chitosan/siRNA uptake. Thus, we conclude that our siRNA delivery system using non-water-soluble chitosan shares a megalin-dependent endocytotic pathway with water soluble chitosan, as observed previously [16] [17].
However, the suggested role of chitosan for delivery of siRNA to the PTECs raises an interesting question: How are the siRNA/chitosan nanocomplexes filtered in view of the glomerular sieving threshold? One possibility is that a fraction of chitosan/siRNA nanoparticles comprising with very small nanoparticles could pass through the endothelial layer and dissembled at the glomerular basement membrane (GBM) as recently described by Zuckerman,J.E., et al. for cationic cyclodextrin-containing polymer based siRNA nanoparticles [31] and siRNA loaded dextran nanogels [42] . After filtration into the urinary space the chitosan could re-associate with the siRNA in accordance with the observation reported for cationic cyclodextrin-containing polymer based siRNA nanoparticles [31] and facilitate uptake of siRNA in the PTEC. Although Thompton et al. [43] have demonstrated that naked siRNA can enter the PTEC, our data clearly indicate a role for chitosan in increasing the efficiency of this event. Thompton et al. only observed a short transient uptake in PTEC of naked siRNA, whereas we find a significant amount of siRNA still present after 48 hours. An alternative model is that the siRNA-chitosan complex only becomes partially disassembled at the GBM. A siRNA associated with just one molecule of chitosan has a molecular weight of approximately 40 KDa, which is considered to be within the molecular weight cut-off (MWCO) for glomerular filtration. Also we cannot exclude that certain mechanisms may allow even larger complexes to pass the GBM. It has for instance been reported that a carbonanotube of 200-300nm, which are much larger than the MWCO, can be cleared through glomerular filtration, which cannot be explained by traditional theory [44].
PTECs play an important role in the reabsorption of a variety of substances following their glomerular filtration, for example, albumin [45;46] A prominent group of PTECs express the aquaporin (AQP) membrane protein family that mediates transport of water or small solutes in a concentration dependent manner. One family member, AQP1, is localized to both the apical and basolateral membranes of the kidney proximal tubules, the descending thin limb of long loop nephrons and the vasa recta. AQP1 plays a critical role in transcellular water reabsorption across PTECs and body water balance [47]. Consistent with PTEC delivery, chitosan/Cy3-siRNA co-localized with AQP1 expressing cells, but not with AQP2, which is expressed exclusively in the distal portion of the kidney tubule - the collecting duct [48]. We confirmed that the efficient delivery of siRNA also correlated with a significant knock down of AQP-1 expression in PTECs, highlighting that at least part of the siRNA escapes endosomes and becomes incorporated into RISC complexes in the PTECs, a process highly inefficient for naked siRNA.
Unlike total gene deletion of AQP1 [49], our 40-50% knock down was insufficient to produce any overt phenotype [49] and it may be necessary to increase the current dose (1.2mg/kg body weight per injection) in future studies. Alternatively, as our gene silencing was only observed in PTECs and not in other AQP1 expressing tubule segments such as the descending thin limb of Henle's loop or in vasa recta, it is conceivable, that AQP-1 function in these segments is sufficient to maintain water balance and account for lack of a robust phenotype in the treated animals [49].
Conclusion
Chitosan/siRNA could be specifically delivered into kidney PTECs, where it persisted for more than 48 hrs. The specific uptake was mediated by a megalin-dependent endocytotic pathway and proved to be a viable approach for gene silencing in PTECs. Our demonstration of long lasting siRNA accumulation in PTECs and gene knockdown may be beneficial for treatment of certain chronic diseases e.g. kidney fibrosis. Our findings provide a potential generic platform for renal therapy via specifically targeting of drugs or siRNAs to PTECs, which play essential roles in the onset of various kidney diseases [1]. Furthermore, the approach will allow clarification of the physiological or patho-physiological roles of a variety of proteins expressed in PTECs and a strategy to provide target validation for novel pharmacological inhibitors.
Supplementary Material
Figures S1-S4, Tables S1-S2.
Acknowledgements
This work was supported by Lundbeck Foundation Nanomedicine Center for Individualized Management of Tissue Damage and Regeneration and the EU-FP6 RIGHT project (LSHB-CT-2004-005276). Funding to RAF, RN, and EIC is provided by the Danish Medical Research Council and the Lundbeck Foundation. The authors would like to thank Claus Bus, Rita Rosendahl, Inger Blenker Kristoffersen, Hanne Siedelmann and Inger Merete Paulsen for their excellent technical assistance, as well as Jesper B. Bramsen for siRNA selection.
Competing Interests
NO conflict of interest to disclose.
References
1. Dolman ME, Harmsen S, Storm G, Hennink WE, Kok RJ. Drug targeting to the kidney: Advances in the active targeting of therapeutics to proximal tubular cells. Adv Drug Deliv Rev. 2010;62:1344-1357
2. Abbate M, Zoja C, Remuzzi G. How does proteinuria cause progressive renal damage? J Am Soc Nephrol. 2006;17:2974-2984
3. Kanasty RL, Whitehead KA, Vegas AJ, Anderson DG. Action and reaction: the biological response to siRNA and its delivery vehicles. Mol Ther. 2012;20:513-524
4. Bramsen JB, Kjems J. Development of Therapeutic-Grade Small Interfering RNAs by Chemical Engineering. Front Genet. 2012;3:154
5. Zhang X, Shan P, Jiang D, Noble PW, Abraham NG, Kappas A. et al. Small interfering RNA targeting heme oxygenase-1 enhances ischemia-reperfusion-induced lung apoptosis. J Biol Chem. 2004;279:10677-10684
6. Lu PY, Xie F, Woodle MC. In vivo application of RNA interference: from functional genomics to therapeutics. Adv Genet. 2005;54:117-142
7. Gao S, Dagnaes-Hansen F, Nielsen EJ, Wengel J, Besenbacher F, Howard KA. et al. The effect of chemical modification and nanoparticle formulation on stability and biodistribution of siRNA in mice. Mol Ther. 2009;17:1225-1233
8. Laursen MB, Pakula MM, Gao S, Fluiter K, Mook OR, Baas F. et al. Utilization of unlocked nucleic acid (UNA) to enhance siRNA performance in vitro and in vivo. Mol Biosyst. 2010;6:862-870
9. Schyth BD, Bramsen JB, Pakula MM, Larashati S, Kjems J, Wengel J. et al. In vivo screening of modified siRNAs for non-specific antiviral effect in a small fish model: number and localization in the strands are important. Nucleic Acids Res. 2012;40:4653-4665
10. Lewis DL, Wolff JA. Systemic siRNA delivery via hydrodynamic intravascular injection. Adv Drug Deliv Rev. 2007;59:115-123
11. Stokman G, Qin Y, Racz Z, Hamar P, Price LS. Application of siRNA in targeting protein expression in kidney disease. Adv Drug Deliv Rev. 2010;62:1378-1389
12. Takabatake Y, Isaka Y, Mizui M, Kawachi H, Takahara S, Imai E. Chemically modified siRNA prolonged RNA interference in renal disease. Biochem Biophys Res Commun. 2007;363:432-437
13. Hamar P, Song E, Kokeny G, Chen A, Ouyang N, Lieberman J. Small interfering RNA targeting Fas protects mice against renal ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2004;101:14883-14888
14. Nawroth I, Alsner J, Behlke MA, Besenbacher F, Overgaard J, Howard KA. et al. Intraperitoneal administration of chitosan/DsiRNA nanoparticles targeting TNFalpha prevents radiation-induced fibrosis. Radiother Oncol. 2010;97:143-148
15. Howard KA, Paludan SR, Behlke MA, Besenbacher F, Deleuran B, Kjems J. Chitosan/siRNA nanoparticle-mediated TNF-alpha knockdown in peritoneal macrophages for anti-inflammatory treatment in a murine arthritis model. Mol Ther. 2009;17:162-168
16. Yuan ZX, Zhang ZR, Zhu D, Sun X, Gong T, Liu J. et al. Specific renal uptake of randomly 50% N-acetylated low molecular weight chitosan. Mol Pharm. 2009;6:305-314
17. Weyer K, Storm T, Shan J, Vainio S, Kozyraki R, Verroust PJ. et al. Mouse model of proximal tubule endocytic dysfunction. Nephrol Dial Transplant. 2011;26:3446-3451
18. Gao S, Nielsen BS, Krogdahl A, Sorensen JA, Tagesen J, Dabelsteen S. et al. Epigenetic alterations of the SERPINE1 gene in oral squamous cell carcinomas and normal oral mucosa. Genes Chromosomes Cancer. 2010;49:526-538
19. Terris J, Ecelbarger CA, Nielsen S, Knepper MA. Long-term regulation of four renal aquaporins in rats. Am J Physiol. 1996;271:F414-F422
20. Hoffert JD, Fenton RA, Moeller HB, Simons B, Tchapyjnikov D, McDill BW. et al. Vasopressin-stimulated increase in phosphorylation at Ser269 potentiates plasma membrane retention of aquaporin-2. J Biol Chem. 2008;283:24617-24627
21. Moestrup SK, Nielsen S, Andreasen P, Jorgensen KE, Nykjaer A, Roigaard H. et al. Epithelial glycoprotein-330 mediates endocytosis of plasminogen activator-plasminogen activator inhibitor type-1 complexes. J Biol Chem. 1993;268:16564-16570
22. Yuan ZX, Sun X, Gong T, Ding H, Fu Y, Zhang ZR. Randomly 50% N-acetylated low molecular weight chitosan as a novel renal targeting carrier. J Drug Target. 2007;15:269-278
23. Yuan ZX, Li JJ, Zhu D, Sun X, Gong T, Zhang ZR. Enhanced accumulation of low-molecular-weight chitosan in kidneys: a study on the influence of N-acetylation of chitosan on the renal targeting. J Drug Target. 2011;19:540-551
24. Park K, Kim JH, Nam YS, Lee S, Nam HY, Kim K. et al. Effect of polymer molecular weight on the tumor targeting characteristics of self-assembled glycol chitosan nanoparticles. J Control Release. 2007;122:305-314
25. Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjaer N. et al. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37:2867-2881
26. Christensen EI, Birn H, Storm T, Weyer K, Nielsen R. Endocytic receptors in the renal proximal tubule. Physiology (Bethesda ). 2012;27:223-236
27. Shimizu H, Fujita T. New short interfering RNA-based therapies for glomerulonephritis. Nat Rev Nephrol. 2011;7:407-415
28. Wan X, Fan L, Hu B, Yang J, Li X, Chen X. et al. Small interfering RNA targeting IKKbeta prevents renal ischemia-reperfusion injury in rats. Am J Physiol Renal Physiol. 2011;300:F857-F863
29. Hauser PV, Pippin JW, Kaiser C, Krofft RD, Brinkkoetter PT, Hudkins KL. et al. Novel siRNA delivery system to target podocytes in vivo. PLoS One. 2010;5:e9463
30. Choi CH, Zuckerman JE, Webster P, Davis ME. Targeting kidney mesangium by nanoparticles of defined size. Proc Natl Acad Sci U S A. 2011;108:6656-6661
31. Zuckerman JE, Choi CH, Han H, Davis ME. Polycation-siRNA nanoparticles can disassemble at the kidney glomerular basement membrane. Proc Natl Acad Sci U S A. 2012;109:3137-3142
32. Yuan H, Lanting L, Xu ZG, Li SL, Swiderski P, Putta S. et al. Effects of cholesterol-tagged small interfering RNAs targeting 12/15-lipoxygenase on parameters of diabetic nephropathy in a mouse model of type 1 diabetes. Am J Physiol Renal Physiol. 2008;295:F605-F617
33. Molitoris BA, Dagher PC, Sandoval RM, Campos SB, Ashush H, Fridman E. et al. siRNA targeted to p53 attenuates ischemic and cisplatin-induced acute kidney injury. J Am Soc Nephrol. 2009;20:1754-1764
34. Khan A, Benboubetra M, Sayyed PZ, Ng KW, Fox S, Beck G. et al. Sustained polymeric delivery of gene silencing antisense ODNs, siRNA, DNAzymes and ribozymes: in vitro and in vivo studies. J Drug Target. 2004;12:393-404
35. Rao SB, Sharma CP. Use of chitosan as a biomaterial: studies on its safety and hemostatic potential. J Biomed Mater Res. 1997;34:21-28
36. Aspden TJ, Mason JD, Jones NS, Lowe J, Skaugrud O, Illum L. Chitosan as a nasal delivery system: the effect of chitosan solutions on in vitro and in vivo mucociliary transport rates in human turbinates and volunteers. J Pharm Sci. 1997;86:509-513
37. Duceppe N, Tabrizian M. Advances in using chitosan-based nanoparticles for in vitro and in vivo drug and gene delivery. Expert Opin Drug Deliv. 2010;7:1191-1207
38. Kim JH, Kim YS, Park K, Lee S, Nam HY, Min KH. et al. Antitumor efficacy of cisplatin-loaded glycol chitosan nanoparticles in tumor-bearing mice. J Control Release. 2008;127:41-49
39. Sawai K, Mahato RI, Oka Y, Takakura Y, Hashida M. Disposition of oligonucleotides in isolated perfused rat kidney: involvement of scavenger receptors in their renal uptake. J Pharmacol Exp Ther. 1996;279:284-290
40. Zhai XY, Nielsen R, Birn H, Drumm K, Mildenberger S, Freudinger R. et al. Cubilin- and megalin-mediated uptake of albumin in cultured proximal tubule cells of opossum kidney. Kidney Int. 2000;58:1523-1533
41. Moestrup SK, Cui S, Vorum H, Bregengard C, Bjorn SE, Norris K. et al. Evidence that epithelial glycoprotein 330/megalin mediates uptake of polybasic drugs. J Clin Invest. 1995;96:1404-1413
42. Naeye B, Deschount H, Caveliers V, Descamps B, Braeckmans K, Vanhove C. et al. In vivo disassembly of IV administered siRNA matrix nanoparticles at the renal filtration barrier. Biomaterials. 2013;34:2350-2358
43. Thompson JD, Kornbrust DJ, Foy JW, Solano EC, Schneider DJ, Feinstein E. et al. Toxicological and pharmacokinetic properties of chemically modified siRNAs targeting p53 RNA following intravenous administration. Nucleic Acid Ther. 2012;22:255-264
44. Ruggiero A, Villa CH, Bander DA, Rey M, Bergkvist M, Batt CA. et al. Paradoxical glomerular filtration of carbon nanotubes. Proc.Natl.Acad.Sci.U.S.A. 2010;107:12369-12374
45. Cui S, Verroust PJ, Moestrup SK, Christensen EI. Megalin/gp330 mediates uptake of albumin in renal proximal tubule. Am J Physiol. 1996;271:F900-F907
46. Birn H, Fyfe JC, Jacobsen C, Mounier F, Verroust PJ, Orskov H. et al. Cubilin is an albumin binding protein important for renal tubular albumin reabsorption. J Clin Invest. 2000;105:1353-1361
47. Tamma G, Procino G, Svelto M, Valenti G. Cell culture models and animal models for studying the patho-physiological role of renal aquaporins. Cell Mol Life Sci. 2011
48. Moeller HB, Fenton RA. Cell biology of vasopressin-regulated aquaporin-2 trafficking. Pflugers Arch. 2012;464:133-144
49. Fenton RA, Knepper MA. Mouse models and the urinary concentrating mechanism in the new millennium. Physiol Rev. 2007;87:1083-1112
Author contact
![]() Corresponding authors: J.K.: Interdisciplinary Nanoscience Center (iNANO) and Department of Molecular Biology and Genetics, Aarhus University, Gustav Wieds Vej 14, 8000 Aarhus C, Denmark. Email: jkau.dk; Telephone: 0045 87155494; Fax: 0045 86196500. OR R.A.F.: Department of Biomedicine - Anatomy, Aarhus University, Wilhelm Meyers Allé 3, building 1233, Aarhus C, Denmark. Email: rofeau.dk; Telephone: 0045 87167671; Fax: 0045 871 67102.
Corresponding authors: J.K.: Interdisciplinary Nanoscience Center (iNANO) and Department of Molecular Biology and Genetics, Aarhus University, Gustav Wieds Vej 14, 8000 Aarhus C, Denmark. Email: jkau.dk; Telephone: 0045 87155494; Fax: 0045 86196500. OR R.A.F.: Department of Biomedicine - Anatomy, Aarhus University, Wilhelm Meyers Allé 3, building 1233, Aarhus C, Denmark. Email: rofeau.dk; Telephone: 0045 87167671; Fax: 0045 871 67102.
 Global reach, higher impact
Global reach, higher impact