13.3
Impact Factor
Theranostics 2014; 4(9):931-944. doi:10.7150/thno.9663 This issue Cite
Review
Progress in Aptamer-Mediated Drug Delivery Vehicles for Cancer Targeting and Its Implications in Addressing Chemotherapeutic Challenges
1. Department of Neurology, Daping Hospital, Third Military Medical University, Chongqing, 400042, China.
2. Department of Anesthesia, Xinqiao Hospital, Third Military Medical University, Chongqing, 400037, China.
3. Department of Biomedical Materials Science, School of Biomedical Engineering, Third Military Medical University, Chongqing, 400038, China
4. Department of Orthopedics, Xinqiao Hospital, Third Military Medical University, Chongqing, 400037, China.
5. Department of Orthopedics, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 02130, USA.
* These two authors contributed equally.
Received 2014-5-15; Accepted 2014-6-23; Published 2014-7-13
Abstract
Aptamers are novel oligonucleotides with flexible three-dimensional configurations that recognize and bind to their cognate targets, including tumor surface receptors, in a high-affinity and highly specific manner. Because of their unique intrinsic properties, a variety of aptamer-mediated nanovehicles have been developed to directionally transport anti-cancer drugs to tumor sites to minimize systemic cytotoxicity and to enhance permeation by these tumoricidal agents. Despite advances in the selection and synthesis of aptamers and in the conjugation and self-assembly of nanotechnologies, current chemotherapy and drug delivery systems face great challenges. These challenges are due to the limitations of aptamers and vehicles and because of complicated tumor mechanisms, including heterogeneity, anti-cancer drug resistance, and hypoxia-induced aberrances. In this review, we will summarize current approaches utilizing tumor surface hallmarks and aptamers and their roles and mechanisms in therapeutic nanovehicles targeting tumors. Delivery forms include nanoparticles, nanotubes, nanogels, aptamer-drug conjugates, and novel molecular trains. Moreover, the obstacles posed by the aforementioned issues will be highlighted, and possible solutions will be acknowledged. Furthermore, future perspectives will be presented, including cutting-edge integration with RNA interference nanotechnology and personalized chemotherapy, which will facilitate innovative approaches to aptamer-based therapeutics.
Keywords: aptamer, biomarker, chemotherapy, drug delivery, nanomedicine.
Introduction
Cancer is the top public health problem all over the world. It was reported to be the second major cause of mortality in the U.S. in 2013, with 1.66 million new cases and 0.58 million deaths [1]. Moreover, it is estimated that developing countries contribute 56% of new cases and 64% of deaths to the worldwide total [2]. In China, annual statistics showed 3.09 million new cases and 1.96 million deaths [3]. Skyrocketing investment have been devoted to the battle against cancer (with an economic cost of over $1.2 trillion in the U.S. in 2013 [4]), which has become one of the top priorities for biotechnological research and pharmaceutical innovation worldwide.
At present, the most commonly adopted anti-cancer treatments are chemotherapeutics. Although current chemotherapy regimens present modest survival benefits, they still encounter drastic limitations in terms of associated poor tissue selectivity, rapid systemic clearance, low intratumoral accumulation and drug resistance. Thus, many patients succumb to the adverse effects of myelosuppression and cardiotoxicity far earlier than elevation of the tumor burden [5]. Therefore, the accurate delivery of chemotherapeutic drugs to tumor sites while sparing normal tissue is of utmost importance. Current approaches in nanotechnology have inspired most aspects of delivery vehicles, such as optimized encapsulation, improved targeting, and controlled release, among others [6, 7]. In many cases, nanoparticles (NPs) have shown great potential for targeted drug delivery due to their enhanced permeability and retention (EPR) effect [8]. Regarding directional agents, antibodies were previously the primary tools, but novel targeting agents, including aptamers, short peptides, and other small molecules, have recently become the new generation of targeting molecules [9-11].
Correspondingly, one of the most promising drug delivery strategies is aptamer-mediated nanovehicles (AMNVs). Aptamers are newly emerging small nucleic-acid ligands that target various disease markers, and they also represent an interesting class of novel pharmaceuticals due to their advantageous properties for research, diagnostics and therapeutics [12]. One milestone achieved in the translation of aptamer nanotechnology is the development of Macugen (also known as pegaptanib; Fig. 1A), which is used for the treatment of human macular degeneration. More specifically, Macugen is an RNA aptamer that antagonizes the binding of vascular endothelial growth factor (VEGF) to its receptor [13]. This pioneering attempt greatly inspired the exploration of AMNVs for cancer therapy. Additionally, a variety of aptamers targeting different cancer markers are currently under preclinical and clinical investigation. Our recent search for “aptamer AND drug delivery” returned over 2480 publications, which accounted for 5% of all results obtained when searching for “targeted drug delivery AND cancer/tumor” in the Medline database. This review aims to summarize the evolution of aptamers and their tumor counterparts, the current applications of AMNVs in major tumor models and their associated limitations, as well as the therapeutic implications for nanomedicine. In particular, we discuss the inspirations for the development of AMNVs and future perspectives from the point of view of recently emerging tumor concepts, including tumor heterogeneity, multiple drug resistance (MDR) and personalized nanomedicine.
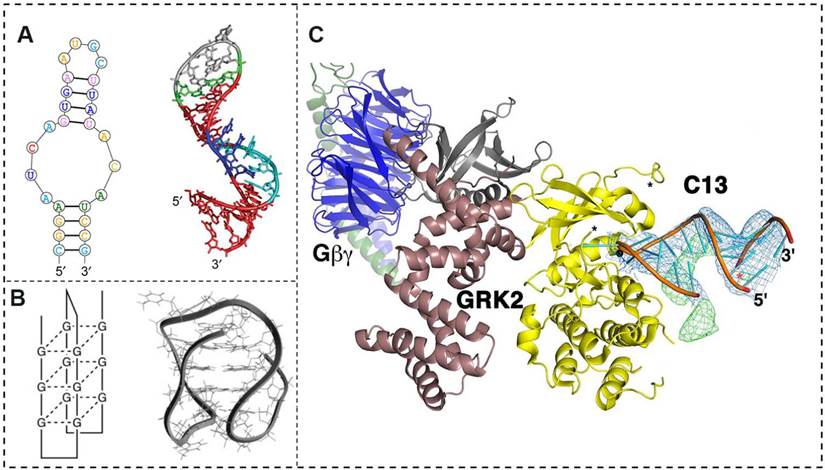
A conformational and interactional overview of aptamers and their receptors. (A) The sequential conformation (left) and secondary structure (right) of the commercialized aptamer product Macugen. (B) Schematic illustrations (left) and molecular models (right) of the quadruplex DNA for the aptamer AS1411. (C) The overall structure of the RNA aptamer C13 and its receptor, G protein-coupled receptor kinase 2 (GRK2). C13 positions an adenine nucleotide in the ATP-binding pocket of GRK2 (shown as yellow and burgundy ribbons), which stabilizes GRK2 in a unique and remodeled conformation. The terminal stem of the aptamer indirectly contributes to its affinity. Adapted from [18, 29].
General profile of aptamers and relevant significance
In contrast to linear oligonucleotides, such as micro-RNAs (miRNAs) and small interfering RNAs (siRNAs), which contain genetic sequences or antisense oligonucleotides that interrupt gene transcription, aptamers are short (20-80 bp), single-stranded, and well-formed 3D-configured RNA or DNA ligands (Fig. 1A). Aptamers bind with high affinity and specificity to their target molecules, which include proteins, phospholipids, iron channels, nucleic acids, and whole cells, among others [14]. These oligonucleotides are also called “smart ligands” due to their superior advantages in terms of small size, flexible structure, and selective internalization and intratumoral penetration. Aptamers are also easy to inexpensively synthesize, show low antigenic and immunogenic potential, and enable chemical modifications for various applications [15]. Aptamers are typically isolated and synthesized from large randomized oligonucleotide libraries by a process known as in vitro systematic evolution of ligands by exponential enrichment (SELEX), which is a combinatorial biochemical technique for producing oligonucleotides that specifically bind to a particular ligand via multiple rounds of selection [16].
Given exonuclease degradation, various methods have been adopted to synthesize aptamers with enhanced stability, such as by replacing the phosphate in the backbone with phosphorothionate or by derivatizing the 2'-ribose in pyrimidines and the 3'-inverted deoxythymidine cap [17]. Moreover, aptamers can be readily modified by adding poly (ethylene glycol) (PEG) and other moieties to reinforce the oligonucleotides' bioavailability and pharmacokinetic properties.
Interaction of aptamers with tumors
Because of their unique properties, aptamers have emerged as novel and promising molecules that target specific cancer epitopes for clinical diagnosis and therapy, comparable to antibodies. Aptamers' role in directionally guiding drugs to specific tumor lesions is achieved by working as a molecular probe to recognize and bind to corresponding receptors. In contrast to antigen-antibody interactions, an aptamer remodels its three-dimensional structure spontaneously and caters to the individual configuration of a receptor [18] (Fig. 1B, C). Human tumors show intrinsic pathological characteristics, and aptamers are sensitive in identifying these tumor-specific signatures, with the goal of improving diagnostic and therapeutic responses and reducing unnecessary toxicity in oncology patients. In this respect, a major challenge is the identification of specific cell signatures among low-content molecular targets in the presence of abundant, nonspecific plasma-membrane antigens. Here, we summarize most of the currently known biomarkers expressed on tumor cell surfaces that potentially interact with the aptamers in Table 1.
Verified hallmarks on tumor cell surface for potential tumoricidal targeting
| Biomarker | Classification | Known expressing tumors | Role in Tumorigenesis | Reference |
|---|---|---|---|---|
| Prostate specific mem- brane antigen (PSMA) | Type II transmembrane proteins | Prostate, kidney , bladder cancers, etc. | Catalyzing the hydrolysis of N-acetylaspartyl- glutamate for cancer cell over-proliferation. | Benjamin T, et al.2013. |
| V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) | Ras family member and GTPase proteins | Pancreatic, colorectal, lung cancers, mucinous adenoma, ductal carcinoma, leukemia, etc. | Product of Kirsten ras proto-oncogene switching the propagating of growth factors and receptor's signal (c-Raf, PI3KCG, RALGDS, RASSF2). | Miles KA, et al. 2014. |
| RET receptor tyrosine kinase (RTK) | Flial cell-derived neurotrophic factor family members | Thyroid, breast, lung cancers, etc. | RET proto-oncogene signaling by oncogenic mutation, gene rearrangement, overexpression or transcriptional up-regulation. | Plaza-Menachol I, et al. 2014. |
| Human epidermal growth factor 2 (HER2) | Membrane tyrosine kinases | Breast, gastric, lung, bladder, colorectal, esophageal, ovarian cancers, etc. | Tumorigenic signaling via MAPK, PI3K/Akt, PKC and STAT pathways. | Krishnamurti U, et al. 2014. |
| Epithelial cell adhesion molecule (EpCAM) | Type I transmembrane glycoproteins | Bladder, breast, colon, esophagus, lung, hepatocellular, ovarian, pancreas, prostate, etc. | Regulating gene expression of c-myc, e-fabp, cyclin, etc. and modulating EMT. | Patriarca C, 2012. |
| Nestin | Class VI intermediate filament family members | Prostate, breast, pancreatic, ovarian, cervical cancers, glioblastoma, lung adeonocarcinoma, multiple myeloma, etc. | Influencing FAK and integrin localization and modulating Akt/Sox-2 for tumor invasion. | Hyder CL et al. 2014; Chen ZG, et al. 2014; Narita K, et al. 2014. |
| Annexin A2 (p36) | Calcium dependent phospholipid-binding proteins | Leukemia, breast, gastric, lung, pancreatic cancers, hepatocellular, colorectal, squamous cell carcinomas, multiple myeloma, etc. | Regulating tumor behaviors such as neoangiogenesis, migration and invasion, and chemoresistance | Cibiel A, et al. 2014; Wang CY, et al. 2014. |
| Tumor endothelial marker 1 (TEM 1) | C-type lectin trans- membrane receptors | Most sarcomas, brain tumors, etc. | Promoting angiogenesis via HIF-2α pathway | Facciponte JG, et al. 2014; Bagley RG. 2009. |
| Mucin 1 (MUC1) | Members of the mucin family | Ovarian, breast, lung, pancreatic cancers, pros- tate adenocarcinoma, multiple myeloma, etc. | Promoting cancer cell invasion through beta-catenin and initiating EMT. | Dalziel M, et al.1999. |
| Nucleolin (NCL) | Multi-functional proteins translocated from nucleic | Gastric, lung, colorectal, prostate, breast cancers, melanomas, glioblastoma, gliomas, osteosarcoma , leukemia, etc. | Activating CXCR4 signaling to promote EMT, inhibiting Fas-mediated apoptosis and IL-9 dependent apoptosis. | Yang X, et al. 2014; Wise JF, et al. 2013; Shang Y, et al. 2012. |
| Endoglin (CD105) | Type III TGF-β super- family receptors | Renal cell, hepatocellular carcinoma, gastric, prostate cancers , sarcoma, leukemia, etc. | Development of resistance VEGF inhibition; Modulating tumor growth and metastasis via TGF-B receptor signaling. | Rosen LS, et al. 2014; Breen MJ, et al. 2013. |
| CD24, 30, 44, 133, 166. | Surface transmembrane glycoproteins | Breast, brain, colon, gastric, lung, prostate cancers, leukemia, Hodgkin's lymphoma, et al. | Regulating tumor cell proliferation, differentiation, migration, angiogenesis. | Schmitt F, et al. 2014, Ni C, et al. 2013; Wen L, et al. 2013; Takaishi K, et al. 2009. |
| Integrin α3βv, α2β1, α5β1 | Cell surface adhesion molecules | Breast, pancreatic, leukemia, prostate, colorectal cancers, sarcoma, etc. | Promoting tumorigenesis by transducting stimuli from ECM to oncogenic signal transducers of ErbB, Ras, and Src, etc. | Chen JC, et al. 2013. |
| CXCR4 | Chemokine (C-X-C motif) receptors | Gliomas, renal cell carcinoma, lymphoma, bre- ast, leukemia, gastric, ovarian, colorectal, etc. | Modulating tumor proliferation, invasion, angiogenesis and immune response. | Gagliardi F, et al.2014. |
| CXCR3 | Chemokine (C-X-C motif) receptors | Breast, gastric, cancers, melanoma, renal cell carcinoma, etc. | Inducing cell growth, mobility and invasion and angiogenesis. | Billottet C, et al.2013. |
| Vascular endothelial growth factor receptor (VEGF-R) | Type V receptor tyrosine kinases | Ovarian, breast, cervical, lung cancers, thyroid, renal cell carcinoma, etc. | Key regulator in tumor angiogenesis and angiogenesis. | Shibuya M, et al.2014; Stuttfeld E, et al.2009. |
| Platelet derived growth factor receptor (PDGF-R) | Cell surface tyrosine kinase receptors | Gastrointestinal stromal tumors, leukemia, multiple myeloma, dermatofibrosarcoma, melanoma, glioblastoma, etc. | Modulating epithelial-mesenchymal transition (EMT) and tumor addiction by over-expression or mutation. | Heldin CH, et al.2013. |
| Hepatocyte growth factor receptor (HGFR/MET) | Transmembrane receptor tyrosine kinases | Colorectal, lung, breast, pancreatic, ovarian, gastric, endometrial cancers, etc. | Product of c-Met proto-oncogene modulating oncogenic pathways (RAS, PI3K, STAT3, beta-catenin) and angiogenesis. | Peng Z, et al. 2014; Harshman LC, et al. 2013; Mahlknecht G,2013 |
| Ephrin type-B receptor 4 (EphB4) | Eph receptor tyrosine kinases | Breast, , prostate, lung cancers, mesothelioma, glioblastoma, etc. | Regulating tissue homeostasis including tissue boundaries formation, intricate neuronal circuits assembly, blood vessels remodeling. | Pasquale EB. 2010. |
| IL-6 receptor (CD126) | Type I cytokine receptors | Colon, ovarian and pancreatic cancers, mul- tiple myeloma, hepatocellular carcinoma, etc. | Promoting EMT via IL-6R/STAT3, GP130, Cyclin D1, Bcl-xl, etc. | Rokavec M, et al.2014; Cui X, et al. 2014 |
| Transforming growth factor β receptor (TGF-β R) | Single pass serine/ threonine kinase receptors | Breast, lung, colon, prostate, pancreatic cancers, ductal adenocarcinoma, hepatocellular carcinoma, etc. | Promoting EMT via interaction with autocrine or paracrine growth factors and inducting microenvironment. | Giannelli G, et al. 2014; Hiemer SE, et al. 2014. |
| Transcobalamin II receptor (TCII-R) | Plasma membrane receptors | Breast, pancreatic, ovarian, lung cancers , renal adenocarcinoma, Ewing's sarcoma, glioblasto- ma, leukemia, Burkitt's lymphoma, etc. | Vitamin B12-dependent cellular replication for tumor proliferation. | Sysel AM, et al. 2013; Amagasaki T, et al. 1999; Seetharam B, et al. 1999. |
| Protein tyrosine kinase 7 (PTK7) | Tyrosine kinase-like transmembrane receptors | T-cell acute lymphoblastic leukemia, lung, gastric cancers, colon carcinoma, etc. | Regulating planar cell polarity, cell migration and invasion via non-canonical Wnt signaling. | Shangguan D, et al. 2008; Peradziryi H, et al. 2011 Na HW, et al. 2012 |
Delivery formulations of AMNVs to tumors
The current paradigm of an aptamer-mediated tumoricidal therapeutic approach is to develop dual-functional complexes consisting of an outstanding aptamer guide that targets the extracellular region of a tumor-specific surface biomarker and a cargo portion that carries a cytotoxic agent. To date, the most commonly employed vehicles are aptamer-linked NPs encapsulating a hydrophobic chemotherapeutic agent (NPs; Fig. 2A). These NPs show great diversity based on their drug-loading characteristics, and they may be classified primarily as polymeric NPs (e.g., poly (lactide-co-glycolide), or PLGA), metallic NPs (e.g., gold NPs, or GNPs), magnetic NPs (e.g., Fe3O4), silica NPs, liposomes, micelles, and quantum dots (QDs), among others [19-22]. These vehicles are advantageous for drug loading and an extended period of chemotherapeutic drug administration because the drug cargo is independent. Most studies have shown over 5% loading capability, an encapsulation efficacy of up to 90%, and in vitro release profiles over 48 hours (Table 2).
Another derivation from nanoscaled drug delivery system, termed as “aptamer-drug conjugate,” has been generated to overcome the complicated assembly techniques and low controllability of site-specific drug conjugation [23]. The simple assembly largely relies on the structural property of the drug and its noncovalent association with specific DNA sequences. One classical example is the anthracycline class of anti-cancer drugs, such as doxorubicin (Dox), which prefer to bind to a repetitive sequence (5'-CG-3') in aptamers and to intercalate within the GC pairs to form physical conjugates at certain molar ratios [24]. Under these circumstances, the aptamer acts both as a guide and as a cargo for drug delivery (Fig. 2B and C). The superiority of these agents is achieved via their improved binding affinity (Kd=1-10 nM), their ability to be efficiently internalized by many cancer cells, and their ability to pass through the blood-brain/blood-tumor barriers [25]. However, the majority of these approaches have suffered from low synthesis yield, reduced drug loading capacity and rapid elimination by systemic clearance.
Publications of aptamer-mediated nano-vehicles, physico-chemical and tumoricidal characterizations in the recent years.
| Aptamer /Target | Type (bp) | Physico-chemical Property | Anti-tumor Potency | Reference | ||||
|---|---|---|---|---|---|---|---|---|
| Vehicle | Drug | Drug loading | Release profile | Cancer model(cell line) | Study type | |||
| AS1411 /NCL | DNA (28) | Triblock co- polymer NPs* | Dox | LP 3.64% | pH-responsive: 70.30% in PBS, 48h | MCF-7, PANC-1 | In vitro | Lale SV, et al. 2014 |
| DNA (28) | Carbon nanotube | Dox | DNS | DNS | Nude mice bearing xenograft (PC3 cell) | In vitro & In vivo | Zhang H, et al. 2014 | |
| DNA (38) | Liposome | Dox | LP 12.5% | Hydrolytic release: 10% in plasma, 24h | Nude mice bearing xenograft (MCF-7) | In vitro & In vivo | Xing H, et al. 2013 | |
| DNA (26) | Mesoporous silica NPs | Dox | LP 4.6% | pH-responsive: 72% in PBS, 50h | MCF-7,MDA MB-231 | In vitro | Li LL, et al. 2012 | |
| DNA (26) | PLGA-lecithin-PEG NPs | Ptx | EE 60.93% | Hydrolytic release: 94% in PBS, 120h | GI-1, MCF-7 | In vitro | Aravind A, et al. 2012 | |
| DNA (28) | PEG-PLGA NPs | Ptx | EE 44.7%, LP 1.02% | Hydrolytic release: 78.4% in PBS, 84.1% in plasma, 12d | Nude mice bearing xenograft (C6 glima cell) | In vitro & In vivo | Guo JW, et al. 2011 | |
| DNA (26) | Silica-coated gold nanorods | Dox | LP 3.8% | photothermo-responsive release | MCF-7 | In vitro | Yang XJ, et al. 2012 | |
| A10 /PSMA | RNA (57) | Unimolecular micelles NPs | Dox | LP 10.4% | pH-responsive: 91% in PBS, 45h | Nude mice bearing xenograft (CWR22Rv1 cells) | In vitro & In vivo | Xu WJ, et al. 2013 |
| RNA (57) | PLA-PLGA NPs | Platinum Docetaxel | Pt: EE95%, LP 5%; Dtx: EE80%, LP 1%. | Hydrolytic release: 98% Dxtl and 80%, Pt in PBS, 72h | LNCaP | In vitro | Kolishetti N, et al. 2010 | |
| RNA (57) | PLGA NPs | Platinum | LP 5% | DNS | Nude mice bearing xenograft (LNCaP) | In vivo | Dhar S, et al. 2010 | |
| RNA (91) | Golden-NPs | Dox | Apt-Dox ratio: 2:15 (molar) | Hydrolytic release: 52% in PBS, 24h | LNCaP | In vitro | Kim, D, et al. 2010 | |
| RNA (57) | QD-Apt-drug conjugate | Dox | QD-Apt/Dox ratio: 1/7 (molar) | DNS | LNCaP | In vitro | Bagalkot V et al. 2009 | |
| SZTI01 /PSMA | DNA (48) | Dimeric apt- drug complex | Dox | Apt-Dox ratio: 1:4 (molar) | Hydrolytic release: 40% in PBS, 6h | C4-2 cells | In vitro | Boyacioglu O, et al. 2013 |
| 5TR1 /MUC1 | DNA (25) | Apt-SPION complex | Epirubicin | LP 2.27% | DNS | BALB/c mice bearing xenograft (C26 cells) | In vitro & in vivo | Jalalian SH, et al. 2013 |
| MA3 /MUC1 | DNA (86) | Apt-drug conjugate | Dox | Apt-dox ratio: 1/10 (molar) | DNS | A549 lung cancer, MCF-7 | In vitro | Hu Y, et al. 2012 |
| Apt /MUC1 | DNA (25) | DNA Icosahedral | Dox | LP 40% | DNS | CHO-K1, MCF-7 | In vitro | Chang M, et al. 2011 |
| S2.2 /MUC1 | DNA (19) | PLGA NPs | Ptx | EE 83.6%, LP 4.2% | Hydrolytic release: 65% in PBS, 48h | MCF-7,HepG2 | In vitro | Yu CC , et al. 2011 |
| Apt /MUC1 | DNA (25) | QD-Apt-Dox conjugate | Dox | QD-dox ratio: 1/46 (molar) | pH-responsive: 35%, 5h | athymic nu/nu mice bearing xenograft (A2780/AD cells) | In vitro & in vivo | Savla R, et al. 2011 |
| Sgc8c /PTK7 | DNA (41) | PLGA hybrid NPs | Ptx, Dox | LP 3.5%, EE35%. | DNS | CEM cells and Ramos cells | In vitro | Huang FJ, et al. 2014 |
| DNA (41) | Apt-drug conjugate | Fluorouracil | Apt/drug ratio: 1/5 (molar) | Photo-controllable release | HCT116 cells | In vitro | Wang RW, et al. 2014 | |
| DNA (41) | Apt -drug Nanotrain | Dox | Apt-NTr/drug ratio: 1/50 (molar) | DNS | NOD. Cg-Prkdc mice bear- ing xenograft (CCRF-CEM) | In vitro & in vivo | Zhu GZ, et al. 2013 | |
| DNA (41) | AuNR-based Nanogel | Dox | DNS | Photothermo-responsive: 74%, 50min | CCRF-CEM cells, Ramos cells | In vitro | Kang HZ, et al. 2011 | |
| DNA (41) | Lipid-coated droplet | Dox | EE 50.6%, LP 30.4% | Ultrasound-responsive: 12% in PBS, 24h | CCRF-CEM cell lines | In vitro | Wang CH, et al. 2012 | |
| DNA (41) | Carbon nanotube | Daunorubicin | LP 157% | pH-responsive: 60%, 72h | Molt-4, U266 | In vitro | Taghdisi SM, et al. 2011 | |
| DNA (41) | Apt-drug conjugate | Dox | Dox/sgc8c ratio: 1/1 (molar) | DNS | CCRF-CEM cell lines | In vitro | Huang YF, et al. 2009 | |
| Ploy-Apt /PTK7 | DNA(41)n | Apt-drug conjugate | Dox | Apt/drug ratio: 1/10 (molar) | Nuclease-responsive | CCRF-CEM cells, Ramos cells | In vitro | Zhang ZQ, et al. 2013 |
| Apt /EpCAM | RNA (14) | PLGA NPs | Curcumin | EE: 89.98% | Hydrolytic release: 81% in PBS, 100h | HT 29, HEK293T | In vitro | Li L, et al. 2014 |
| EpDT3 /EpCAM | RNA (19) | Apt-drug conjugate | Dox | Apt-Dox ratio: 1:2 (molar) | Hydrolytic release: 37% in PBS, 6h | Y79 and WERI-Rb1 | In vitro | Subramanian N, et al. 2012 |
| DDSs /Cyt c | DNA (48) | Silica-shelled gold nanorods | Rotenone | LP 7.3% | Photothermo-responsive: 25%, 12h | Hela cells | In vitro | Ju EG, et al. 2014 |
| Apt /CD30 | RNA (39) | HAuNS | Dox | EE 90%, LP 30% | pH-responsive: 80% in PBS, 2h | Karpas 299, SUDHL-1 | In vitro | Zhao NX, et al. 2013 |
| TLS11a /LH86 | DNA (63) | Apt-drug conjugate | Dox | Apt/Dox ratio: 1/25 (molar) | DNS | NOD. Cg-Prkdc mice bearing xenograft (LH86 cells) | In vitro & in vivo | Meng L, et al. 2012 |
| HB5 /HER2 | DNA (86) | Apt-drug conjugate | Dox | Apt-Dox ratio: 1/10 (molar) | DNS | SK-BR-3,MDA-MB-231, MCF-7 | In vitro | Liu Z, et al. 2012 |
*triblock copolymer: [pPEGMA-PCL-pPEGMA= poly(polyethylene glycol methacrylate)-poly(caprolactone)-poly(polyethylene glycol methacrylate.
Abbreviation: SPIO, superparamagnetic iron oxide; Ptx, paclitaxel; HAuNS, hollow gold nanosphere; Dox, doxorubicin; Apt, aptamer; QD, Quantum dot; AuNR, gold nanorods; EE, encapsulation efficiency; LP, loading percentage; DNS, data not shown.
A schematic illustration of novel forms of advanced AMNVs and their tumoricidal response in vitro and in vivo. (A) Lipid-polymer hybrid NPs combining the positive attributes of both liposomes and polymeric NPs and comprising a hydrophobic polymeric core (PLGA, drug and fluorescent dye) and a lipid layer conjugated to aptamers. (B) Self-assembled hybrid nanoparticles for the targeted co-delivery of two different drugs to cancer cells. A lipid-PEG-aptamer loaded with Dox forms the hydrophilic shell, whereas PLGA encapsulating PTX forms the hydrophobic core. (C) A poly-aptamer-drug composition based on rolling-circle amplification that induces cooperative binding and increases the strength and frequency of interactions with target tumor cells. (D) Aptamer-tethered DNA nanotrains, self-assembled from short DNA building blocks upon initiation from a chimeric aptamer-tethered trigger probe, significantly improve the drug payload capacity and the anti-tumor efficacy. (E-G) Increased endocytosis, cisplatin-d (GpG) intrastrand cross-links and cytotoxicity to tumor cells were confirmed by confocal laser scanning microscopy using different fluorescent probes (NR dye, R-C18 antibody and tubulin marker, respectively). Left, non-targeted NPs; right, aptamer-functionalized NPs. (H) The biodistribution discrepancy between non-targeted (left) and Muc1-targeted (right) QD-Dox conjugates shows active tumor targeting by preferentially accumulating in subcutaneous ovarian tumors. Adapted from [21, 24, 26].
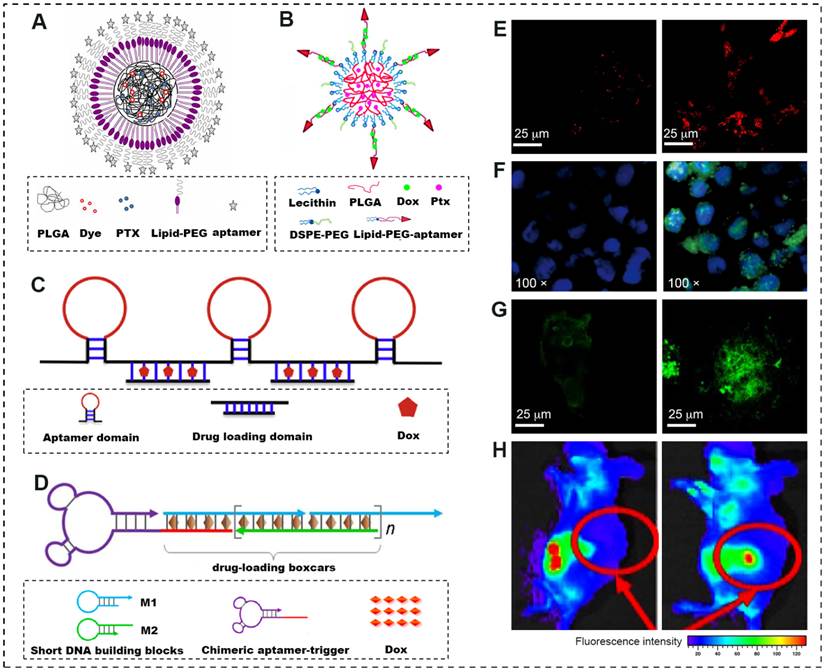
From this perspective, it is worth noting that an innovative delivery form named a “molecular train” was recently described by Tan [26]. This form can significantly improve the drug loading capacity by as much as 10-fold. In brief, a molecular train is an assembly of an aptamer moiety on one end, serving as the locomotive, and tandem modules at the other end, serving as the boxcars by integrating the drug into phosphoramidites based on a solid-phase synthesis platform (Fig. 2D). Furthermore, specific linkers can be added between the two to enable photothermal- or pH-responsive release [27].
Notably, as a progressive formulation integrating natural and artificial nanomaterials at physiological conditions, a class of aptamer-based DNA nanostructure shows greater promise as candidates for targeted drug delivery and cancer therapy with the characteristics of multiple functionality and high programmability. These novel nanoassemblies, reported as nanoflower, nanorobot, DNA origami, and aptamer-micelle self-assemblies [28-31], are capable of hybridizing the predesigned DNA building blocks from one-dimensional into two- and three-dimensional nanostructures. By precise programming, many myriad functional moieties, such as cancer-targeting aptamers, intercalated anticancer drugs, therapeutic antisense oligonucleotides, and fluorescent bioimaging components, can be readily introduced to construct a single nanoassembly, with the advantage of facile assembly, high packaging efficiency, thousand-fold loading of chemotherapeutic and bioimaging agents, along with possible solutions to chemoresistance [32].
Progress in AMNVs used in anti-cancer nanomedicine
A renowned aptamer in anti-cancer research is AS1411, a 26-nucleotide guanosine-rich DNA sequence discovered by Bates et al. in 1914 [33]. AS1411 specifically binds to overexpressed or translocated nucleolin (NCL) in many types of cancer cells [34]. A range of preclinical studies have shown significant inhibition of various tumor cell lines at very low concentrations, with good efficacy and minimal side effects (Table 2). For example, one comprehensive study reported a formulation of AS1411-functionalized liposomes encapsulating Dox. In vitro results indicated a high targeting efficiency and enhanced cytotoxicity in MCF-7 cells, and the targeted liposomes also improved anti-tumor efficacy against xenografted MCF-7 breast tumors in nude mice due to enhanced tumor tissue penetration and the early onset of tumor suppression [35]. Except for the chemotherapeutic toxicity, the anti-cancer effects of AS1411 included two specific mechanisms: blockade of the binding of NCL to the Bcl-2 oncogene, which induces the escape of cells from apoptosis, thereby enabling tumorigenicity, and inhibition of the signaling of the pro-survival molecule NF-κB, which otherwise maintains DNA replication and inhibits cell cycle arrest [36, 37].
This preclinical success inspired great interest in the clinical study of AS1411. Based on early-stage data from a phase I clinical trial supporting its safety and maximum tolerated dose in advanced solid tumors, the tumoricidal effect of its commercial version (generated by Aptamera Inc., Louisville, KY) is being investigated in phase II trials for acute myeloid leukemia (AML) and renal cell carcinoma (RC) [38]. An early phase II trial for AML demonstrated that combination with cytarabine was superior to cytarabine alone [39]. However, a recently published report including 36 cases of RC unveiled minimal responses to AS1411 (<3%) in unselected cases, except in one case with FGFR2 and mTOR mutations [40].
Compared with the diverse and even contradictory results for AS1411, another valuable discovery was aptamer A10, whose use has been focused on prostate cancer research for decades. A10 is an aptamer that binds to a hallmark antigen on prostate cancer cell surfaces, named prostate-specific membrane antigen (PSMA), which is predominantly expressed on the vasculature of many neoplasms but that shows very low expression in normal tissue [41]. Many studies have revealed encouraging results for the advancement of anti-prostate cancer nanomedicine using A10.
In one preliminary study, A10 was conjugated to PLGA NPs for the controlled release of a platinum compound (PtIV), which was reduced to cisplatin (Pt) intracellularly. A lethal dose of Pt was specifically delivered to LNCaP cells, with evidence of endocytosis and nuclear DNA cross-links (Fig. 2E and F). The A10-NPs were 80 times more toxic than free Pt was, and the IC50 was only a quarter of that of non-targeted NPs [19].
To further improve the drug encapsulation efficiency and co-delivery of multiple drugs for combined chemotherapy, another study constructed an A10-functionalized, dual-drug delivery NP platform with high loading of hydrophilic PtIV and hydrophobic Dtxl. This powerful carrier exhibited 10-fold increased toxic efficacy compared with a single PtIV carrier (Fig. 2G) [42]. The significance of this study lies in the strategy used to minimize the batch-to-batch variability of the NPs during synthesis and the potential for combination therapy. Interestingly, the aptamer density on the NP surface can be precisely tuned by adjusting the mixing ratio of the PLGA-PEG-A10 tri-block copolymer with the PLGA-PEG di-block copolymer. The optimum ligand density for PSMA-specific endocytosis was estimated to range from 10-80 nmol of aptamer per μmol of NPs. This minimum amount was considered to be an optimal ligand density that conferred the maximal targeted cellular uptake. This theory was confirmed by comparative NP biodistribution studies in vivo using an LNCaP xenograft mouse model [43].
In addition to enhanced tumor cytotoxicity, certain non-polymer-based NPs offer enhanced choices for monitoring the real-time behaviors of AMNVs by visualizing drug transportation to tumor sites. For example, a QD-mucin 1 aptamer-Dox conjugate exhibited preferential accumulation in ovarian carcinoma resulting from implantation of a subcutaneous tumor for in vivo targeting (Fig. 2H). Moreover, A9 (a predecessor of A10) and A10 PMSA aptamers were each conjugated to GNPs and superparamagnetic iron oxide (SPIO) NPs, both of which can be tracked when they specifically bind to prostate cancer using clinical CT and MRI [44,45]. This system fulfills the dual demands of both fluorescent/radiographic contrast and a drug delivery system.
One feature of the evolution of these known aptamers is that they fully embody the extension of the cancer spectrum for in vivo and in vitro evaluation. These aptamers can also be upgraded as refined aptamers, such as the updating of PMSA aptamers from the previous A9 to the current A10.3. In fact, following insight into novel tumor biomarkers, other new aptamers are still currently under selection and investigation. These studies offer a solid proof-of-concept for drug delivery strategies and tumor-targeting approaches. For example, an aptamer-equipped hollow GNP carrying Dox was fabricated to target CD30 (a cell membrane protein of the tumor necrosis factor receptor family) on lymphoma cells [46]. In another study, a PLGA-lecithin-PEG NP targeting endothelial cell adhesion molecule (EpCAM) was prepared to selectively deliver curcumin to colon cancer cells [47]. In addition, the human epidermal growth factor receptor 2 (HER2) aptamer HB5 and the mucin 1 aptamer MA3 were developed to exploit their tumoricidal effects on breast and lung cancers [48, 49]. Other parallel studies are summarized in Table 2. Of note, a non-polymeric drug transport platform called aptamer-tethered DNA nanotrains (apt-NTrs) was recently presented by Tan et al [26]. In their series of studies, sgc8-NTrs were self-assembled from two short hairpin DNA monomers upon initiation from a modified aptamer-tethered trigger probe (Fig. 2D). These nanotrains selectively transported a drug payload to target leukemia cells and offloaded the drug to induce tumor cytotoxicity in both in vitro and in vivo models. This system presents advances in terms of its simple design and preparation, high payload capacity, and optional upload of theranostic agents into the tandem repetitive cargo.
Limitations of aptamers in chemotherapy
Compared with antibody-mediated nanovehicles, aptamers are prone to unbinding from receptors, resulting in pharmacokinetic instability and rapid systemic clearance. Several discussions about the insufficient affinity of aptamers for their receptors have suggested suboptimal bioavailability and anti-cancer efficacy. Most reports have proposed that the affinity of aptamers, quantified using the value of the dissociation constant (Kd), is generally 30-300 nM [23-26, 44-49]. However, a study on 840 rabbit monoclonal antibodies (http://www.abcam.com/index.html?pageconfig=resource&rid=15749) revealed that their affinities were nearly 1000 times higher than those of aptamers (Kd: 10-1000 pM). This theoretical discrepancy was elucidated by one study aiming to evaluate the targeting selectivity, extent of internalization, and anti-cancer efficacy of liposomes loaded with the α-particle generator 225Ac to kill PSMA-expressing tumor cells. The liposomes were labeled with the PSMA J591 antibody or with the A10 aptamer [50]. The results demonstrated that the J591-labeled liposomes display significantly higher levels of conjunction intensity, total specific binding and cytotoxicity to PMSA (+) cell lines than do A10-labeled liposomes.
Another concern is that secondary targets would be affected following aptamer treatment because of these targets' minimal or unknown mechanism in normal tissues and the multifunctional nature of aptamer-targeting receptors. For example, physiologically, NCL exerts its principal functions in rRNA synthesis, ribosome biogenesis, and cell cycle modulation [51]. Several unpredictable outcomes, such as dysregulated gene silencing and cell cycles and even the phenotypic transformation of cancer cells, might emerge following AS1411 treatment.
The third limitation is that few measures can be taken when we treat certain cancers with negative signatures using AMNVs. Triple-negative breast cancer, one pathological type that completely lacks the estrogen receptor (ER), the progesterone receptor (PR) and HER2, currently has no effective solution except for surgery and combined chemotherapy [52]. The lack of biomarkers for certain cancers makes them more difficult to treat, so most targeted therapies will be in vain.
Current concepts in tumorigenesis and implications for aptamer-related chemotherapy
Tumor heterogeneity
One issue troubling researchers is that many drug delivery vehicles appear particularly promising in preclinical studies but fail unexpectedly in clinical evaluations. Despite hundreds of preclinical confirmations, AS1411 was reported to be ineffective in its anti-cancer activity in a phase II clinical trial for RC [40]. This finding was a profound enlightenment, although this result is still not conclusive. Currently, it is recognized that tumor heterogeneity is one of the significant reasons underlying this effect, in addition to chemoresistance and cancer relapse [53, 54]. Increasing evidence continues to show that tumor heterogeneity can be manifested in at least three ways: (1) tumor entities comprising heterogeneous cells with different phenotypic properties, which may variously contribute to the progression of the malignancy; (2) pathological diversified tissue formation observable within the same tumor, with at least five subtypes categorized for breast cancer, based on the varying presence of receptors; and (3) tumor cells recovered from metastatic sites that may be a newly acquired variant compared with the original tumor [55,56]. Heterogeneity presents a great challenge in terms of targeting therapeutics because the current tumor models for cancer toxicity generally operate on the premise of homogeneity, and these models are not ideal biomimetic niches to simulate variability. Moreover, the majority of current AMNVs only specifically bind to one type of receptor, and subpopulations of other cells within the tumor will not be affected by the drug mechanism and will easily acquire MDR [57].
One possible solution is chimerization or multi-conjugation strategies, referring to combination with another aptamer, a biomacromolecule and/or a small molecular compound, in which the functional capability of the chimera is the superposition of the aptamer and these molecules [58]. The aim is to broaden the range of recognition of different cell subpopulations within a tumor. This goal was substantiated by several preliminary studies, including a study on multi-specific and self-assembled drug vehicles generated based on the monovalent aptamers sgc8c and sgd5a. These aptamers are capable of recognizing and killing different leukemia cell lines, including CEM, Ramos, and Toledo cells. Other studies have examined an anti-HER2 antibody and S6 RNA aptamer-equipped GNPs capable of detecting different breast cancer cells and a dual-aptamer SPIO-NP system co-conjugated with the A10 and DUP-1 peptide aptamers, which induce selective cell uptake and effective drug delivery to both PSMA (+) and PSMA (-) cancer cells [59-61] (Fig. 3).
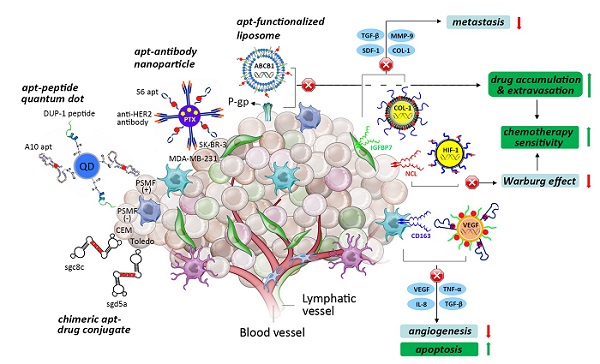
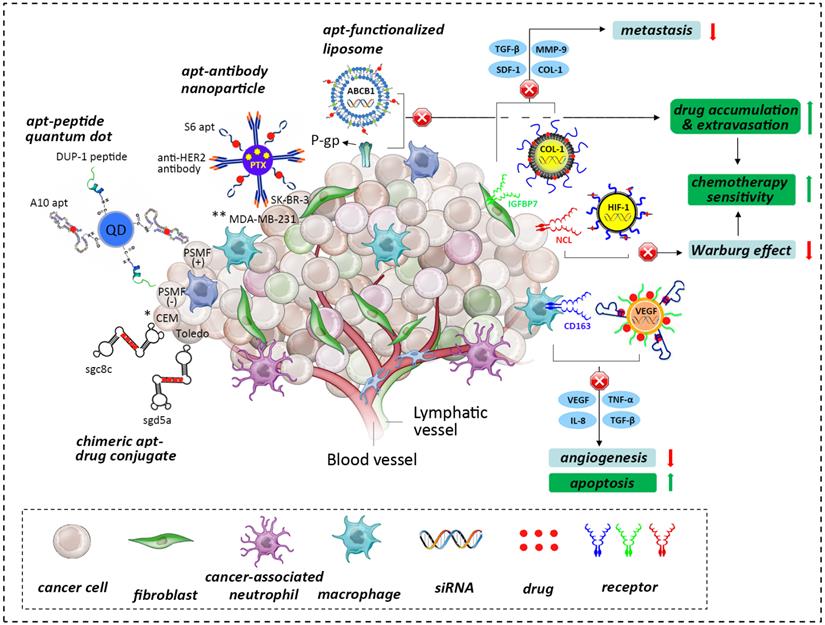
The role of the tumor niche in the origin and influence of tumor heterogeneity, hypoxic aberrancy and the tumor-stroma interactome. Tumorigenesis involves the co-evolution of tumor cells with the ECM and vascular endothelial, immune and stromal cells (adapted from [52]). Various anti-tumor strategies can be used to develop advanced AMNVs to overcome current limitations in chemotherapy, such as chimerization, co-delivery with siRNA, and stroma-targeted nanomedicine, among others. Aptamer is abbreviated as apt in this figure. * CEM and Toledo are different hemopoietic cancer cell lines; **MDA-MB-231 and SK-BR-3 are different breast cancer cell lines.
Another possible route is a cancer stem cell (CSC)-based strategy. CSCs are tumor cells showing self-renewal and multi-lineage differentiation properties that are believed to be the central echelon of the cellular hierarchy present within tumors and to drive further heterogeneity [62]. Despite continuing controversy about their existence and their proposed role in tumor maintenance, anti-cancer strategies targeting CSCs are widely accepted as a path-breaking approach. To date, dozens of reports have paved the way for CSC-targeting therapies, such as those seeking biomarkers of CSCs and targeting genes that regulate the malignant activities of CSCs. These example suggest that aptamers designed to target specific hallmarks of CSCs within brain and colon cancers (such as CD133), as well as AMNVs carrying cytotoxins or siRNAs, should be theoretically possible [63, 64].
Above all, a prerequisite for the tactics discussed here is knowledge of the specific pathological and immunohistological evidence provided by tumor biopsies. The identification of these important characteristics will permit a better definition of tumor heterogeneity, which may ultimately lead to better treatment strategies. However, currently, the complicated genetic, epigenetic, phenotypic and morphologic mechanisms underlying heterogeneity present a huge challenge for these investigations.
Hypoxia-induced aberrant EPR and MDR
One of the most validated issues in oncology is that most growing, solid tumors contain regions experiencing hypoxia [65]. Hypoxia inhibits standard anti-cancer therapies by reducing drug inflow into the cell, increasing drug outflow from the cell, enhancing anti-cancer-drug metabolism, increasing DNA repair, and obstructing programmed cell death [66]. In addition to tumor heterogeneity and stemness, the cellular hypoxia machinery drives most aspects of impaired EPR and MDR, which may inspire the improvement of AMNVs based on the following mechanisms.
First, the poorly formed vasculature and diffusion barrier hamper the transport of the drug-carrying nanovehicle into the center of the tumor, thereby impairing EPR and reducing cytotoxicity in hypoxic regions [67, 68]. Because the majority of AMNVs are administered systemically, such as by intravenous injections, they reach the tumor site via the systemic circulation. The drug administration route can be optimized by localized delivery and regional release using a controllable vehicle, such as in situ-forming gels [69]. It would be beneficial to reduce systemic distribution and to increase the intratumoral concentration by using a strong aptamer-receptor binding moiety. Chemotherapeutic killing could also be increased by targeted injection into the central hypoxic area of the tumor mass with ultrasonic or radiographic assistance; by locoregional regimens, such as aerosols for pulmonary cancer; or by transcatheter arterial administration for hepatocellular carcinoma.
Second, cancer cells can maintain a high glycolytic rate, even under oxygen-saturated conditions [70]. This metabolic shift, defined as the Warburg effect, is considered to be a major contributor to chemoresistance [71]. It is known that the process is controlled by two key regulators: hypoxia-inducible factors 1 and 2 (HIF-1, 2), which primarily modulate tumor glycolysis and lipid metabolism and further affect the processes of angiogenesis and vascularization [72, 73]. Thus, AMNVs carrying a hypoxic cytotoxin/antagonist or a specific inhibitor targeted against HIF-1 and HIF-2 and their target genes, such as pyruvate dehydrogenase kinase (PDK) [74], may be beneficial for reversing the intracellular hypoxic metabolism and increasing the effectiveness of hypoxia-targeted therapies (Fig. 3). In particular, a series of studies has suggested that it is possible to reduce metabolic flexibility and to reverse the Warburg effect in cancer cells to improve chemotherapeutic sensitivity by inhibiting HIF using NSC-134754, cetuximab, tirapazamine and echinomycin, among others [75-77].
Third, it has been illustrated that hypoxia also upregulates the expression of certain ATP-binding cassette genes (ABCB1, 2); increases the activity of efflux transporters, such as P-glycoprotein (P-gp), a protein encoded by multidrug resistance protein 1 (MRP1); and finally leads to an ATP-dependent decrease in cellular drug accumulation, resulting in treatment failure [78]. Recent studies in nanomedicine have presented several inspired approaches to overcome MDR by inhibiting the translation of the mRNAs of MDR efflux pump proteins, such as MDR1 or MRP1, by using siRNA or antisense oligonucleotides [32, 79, 80], or by blocking MDR efflux proteins, such as P-gp/ABCB1 or MRP1/ABCC1, using chemosensitizers (e.g., verapamil) or small-molecule compounds and/or antibodies [81-83]. Therefore, silencing these target genes using integrated AMNVs that deliver a construct of single or multiple siRNAs or a pharmacologic antagonist that improves the sensitivity of chemotherapy is plausible (Fig. 3). This strategy was verified in a preliminary attempt that used a dual minicell system in which sequential delivery of si/shRNA-encoding plasmids was used to knock down MDR1/P-gp and cytotoxic drugs were used to eliminate tumor xenografts [84]. Additionally, a recent advanced study used azobenzene as a hypoxia-responsive bioreductive linker for the hypoxia-targeted delivery of siRNA to downregulate GFP-expressing tumors [85].
Tumor-stroma interactome in microenvironment
It is apparent that the genetic and phenotypic properties of cancer cells alone are not enough to enable cancer progression without a supporting microenvironment [86]. In fact, cancer cells and their stromal partners coordinate in many processes that control tumorigenesis. This bidirectional crosstalk was fully replicated in a prostate cancer model that provided evidence that the progression from original growth to distant bone metastasis was further propelled by the cancer-associated stroma (CAS) [87]. In particular, permanent genetic or phenotypic changes were noted in cancer cells when they were co-cultured with CAS cells, such as fibroblasts and macrophages [88]. Moreover, cancer-associated fibroblasts enhance the secretion of SDF-1, TGF-β1, interleukin-6 (IL-6), and matrix metalloproteinase 9 (MMP-9) to facilitate the detachment and invasion of cancer cells [89, 90]. Additionally, hypoxia recruits macrophages that provide trophic function to stimulate angiogenesis by expressing VEGF and angiopoietin, these cells also produce epidermal growth factor (EGF), tumor necrosis factor (TNF), TGF- β and IL-8, contributing to matrix breakdown and tumor cell motility and providing proliferative and anti-apoptotic support[91, 92]. These findings suggest that the previously underestimated CAS is an important therapeutic target for AMNVs. Along these lines, a specific aptamer that uses IGFBP7 or CD163 could be designed to enhance tumor ablation by identifying the tumor stromal markers of epithelial cancers and mesenchyme-derived sarcomas [93, 94] (Fig. 3).
Additionally, the inclination of prostate cancer to metastasize into bone tissue encouraged researchers to unveil the participation of extracellular matrix (ECM) components in cancer progression. In one study, integrin α2β1-overexpressing LNCaP prostate cancer cells derived from successive plating on type I collagen (COL-I) displayed enhanced chemotactic migration toward COL-I, the most abundant bone protein[95], by inhibiting the activation of the suppressor gene BRCA2, thereby reducing E-cadherin expression and upregulating the PI3K/Akt pathway [96,97]. It is highly plausible that the ECM plays a pivotal role in the epithelial-mesenchymal transition (EMT). Thus, whole-cell-recognizing AMNVs binding to COL-I-secreting cells, such as cancer-associated fibroblasts, and aptamers targeting integrin α2β1-overexpressing cancer cells may be a therapeutic alternative for intervening in the progression of the EMT.
Conclusions and Perspectives
An imminent human disaster was forecasted by the WHO, based on the prediction that cancer cases are expected to surge by 57% in the coming 20 years and to exceed 13.1 million [69]. Therefore, advanced weaponry will definitely be required to conquer cancer. However, current chemotherapy lacks specificity, so the majority of chemotherapeutics agents induce deleterious side effects and genetic and phenotypic transformations resulting in tumor chemoresistance and regression. The emergence of aptamers imbues nanoscale vehicles with the property of specificity to minimize “off-target” effects, and a variety of tumoricidal agents (e.g., chemotherapy drugs, cytotoxins, and siRNAs) have been directionally delivered to cancer cell lines associated with nearly all clinical cancers using the modalities of NPs, QDs, liposomes, hydrogels and other self-assembled constructs [35, 42, 44, 45, 98, 99]. Although these AMNVs have displayed advantageous enhanced selective killing via EPR effects to suppress malignant progression, to our knowledge, numerous limitations should be addressed prior to entering clinical trials with these nanovehicles and permitting their use in the daily management of cancer.
The requirement to optimize the intrinsic parameters of aptamer-nanovehicle entities continues, and the detailed internalization mechanisms of AMNVs are still elusive. After endocytosis by a cancer cell, AMNVs undergo very complicated cytoplasmic processes, such as protein binding, transporter delivery, oxidation and ubiquitylation, and infiltration through multiple barriers or compartments [7, 100-102]. Further understanding of the intracellular interactions will significantly improve the flexibility and bioavailability of nanovehicles in tumors, such as by enhancing their accumulation and extravasation, thereby damaging the tumor vasculature [103, 104].
The spectrum of recognition targets should also be significantly extended. Previous aptamers have been primarily used to target transmembrane proteins, but their use in modulating the function of intracellular proteins has been limited due to a poor understanding of the mechanisms. In the coming years, many proteins/antigens contributing to the progression of tumors, either in the cytoplasm or in the nucleus, may be worthy of therapeutic targeting instead, using specially designed aptamers. Furthermore, using this strategy, many of the cancer-related transcriptional factors and oncogenes, such as the transcription factor NF-κB and its responsive genes Bcl-xl, HIF-1α and VEGF [105-107], which are critical for the establishment of malignancies, could possibly be modulated by aptamers. This theory is supported by a study that blocked the activation of NF-κB and reversed Dox-induced chemoresistance using a novel adenoviral expression system to deliver an RNA aptamer directly targeted to the p50 subunit of NF-κB [108].
Another approach is the use of genetic and epigenetic modulations based on whole-genome sequences at the single-cell level and on tumor auto-biopsies [53]. Various miRNAs have been shown to possess tumor-suppressing potency in breast, colon, and hematological malignancies, among others [109]. Therefore, these miRNAs can be targeted to cancer cell signaling components or EMT regulators, and aptamers can be utilized to direct the application of miRNAs to the malignant machinery [110]. It is anticipated that the interdisciplinary combination of aptamer chemistry, chemo-pharmaceutics and RNA interference-based techniques will potentiate the manipulation of key molecular events associated with cancer initiation and progression at both the transcriptional and the posttranscriptional levels [111].
With expanding knowledge of the genome, epigenome, transcriptome, proteome and metabolome [112], another prospective avenue will be personalized oncology. Instead of the traditional “one treatment fits all” paradigm, more attention will be paid to determining the genetic, epigenetic, phenotypic, and morphologic characteristics of a tumor in individual samples using whole-genome sequences, mass spectroscopy-based proteomic analyses, and next-generation sequencing [113] to uncover specific tumor biomarkers/antigens, oncogenes, and non-coding regulators, with very high resolution. Based on these personalized data, an ideal profile of an advanced AMNV may be sketched to generate a selective and integrative nanoplatform that incorporates multiple or chimeric aptamers for targeting, tumoricidal chemotherapeutics/cytotoxins, RNA interference that blocks the mechanisms of oncogene activation and chemoresistance, an epigenetic regulator targeting tumor progression, and a biocompatible nanocarrier [114].
Acknowledgements
The authors acknowledge Myron Spector from Harvard Medical School and Ali Khademhosseini from the Harvard-MIT Division of Health Sciences and Technology for their help in preparing the manuscript. This work was supported by the National Natural Science Foundation of China (grant no. 81271980).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11-30
2. Zhang Y, Hong H, Cai W. Tumor-targeted drug delivery with aptamers. Curr Med Chem. 2011;18:4185-94
3. Chen W, Zheng R, Zhang S. et al. Annual report on status of cancer in China, 2010. Chin J Cancer Res. 2014;26:48-58
4. Zheng W, McLerran DF, Rolland BA. et al. Burden of Total and Cause-Specific Mortality Related to Tobacco Smoking among Adults Aged ≥45 Years in Asia: A Pooled Analysis of 21 Cohorts. PLoS Med. 2014;11:e1001631
5. Shapira A, Livney YD, Broxterman HJ. et al. Nanomedicine for targeted cancer therapy: towards the overcoming of drug resistance. Drug Resist Updat. 2011;14:150-63
6. Hubbell JA, Langer R. Translating materials design to the clinic. Nat Mater. 2013;12:963-6
7. Zhang XQ, Xu X, Bertrand N. et al. Interactions of nanomaterials and biological systems: Implications to personalized nanomedicine. Adv Drug Deliv Rev. 2012;64:1363-84
8. Kobayashi H, Watanabe R, Choyke PL. Improving Conventional Enhanced Permeability and Retention (EPR) Effects; What Is the Appropriate Target? Theranostics. 2014;4:81-9
9. Hughes B. Antibody-drug conjugates for cancer: poised to deliver? Nat Rev Drug Discov. 2010;9:665-7
10. You J, Zhang R, Xiong C. et al. Effective photothermal chemotherapy using doxorubicin-loaded gold nanospheres that target EphB4 receptors in tumors. Cancer Res. 2012;72:4777-86
11. D'Huyvetter M, Vincke C, Xavier C. et al. Targeted Radionuclide Therapy with A 177Lu-labeled Anti-HER2 Nanobody. Theranostics. 2014;4:708-20
12. Sundaram P, Kurniawan H, Byrne ME. et al. Therapeutic RNA aptamers in clinical trials. Eur J Pharm Sci. 2013;48:259-71
13. Ng EW, Shima DT, Calias P. et al. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov. 2006;5:123-32
14. Bunka DH, Stockley PG. Aptamers come of age - at last. Nat Rev Microbiol. 2006;4:588-96
15. Levy-Nissenbaum E, Radovic-Moreno AF, Wang AZ. et al. Nanotechnology and aptamers: applications in drug delivery. Trends Biotechnol. 2008;26:442-9
16. Sefah K, Shangguan D, Xiong X. et al. Development of DNA aptamers using Cell-SELEX. Nat Protoc. 2010;5:1169-85
17. Yang X, Bassett SE, Li X. et al. Construction and selection of bead-bound combinatorial oligonucleoside phosphorothioate and phosphorodithioate aptamer libraries designed for rapid PCR-based sequencing. Nucleic Acids Res. 2002;30:e132
18. Tesmer VM, Lennarz S, Mayer G. et al. Molecular mechanism for inhibition of g protein-coupled receptor kinase 2 by a selective RNA aptamer. Structure. 2012;20:1300-9
19. Dhar S, Gu FX, Langer R. et al. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA-PEG nanoparticles. Proc Natl Acad Sci U S A. 2008;105:17356-61
20. Lu W, Arumugam SR, Senapati D. et al. Multifunctional oval-shaped gold-nanoparticle-based selective detection of breast cancer cells using simple colorimetric and highly sensitive two-photon scattering assay. ACS Nano. 2010;4:1739-49
21. Yen SK, Padmanabhan P, Selvan ST. Multifunctional Iron Oxide Nanoparticles for Diagnostics, Therapy and Macromolecule Delivery. Theranostics. 2013;3:986-1003
22. Ding K, Alemdaroglu FE, Börsch M. et al. Engineering the structural properties of DNA block copolymer micelles by molecular recognition. Angew Chem Int Ed Engl. 2007;46:1172-5
23. Bagalkot V, Farokhzad OC, Langer R. et al. An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew Chem Int Ed Engl. 2006;45:8149-52
24. Zhang Z, Ali MM, Eckert MA. et al. A polyvalent aptamer system for targeted drug delivery. Biomaterials. 2013;34:9728-35
25. Huang YF, Shangguan D, Liu H. et al. Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumor cells. Chembiochem. 2009;10:862-8
26. Zhu G, Zheng J, Song E. et al. Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc Natl Acad Sci U S A. 2013;110:7998-8003
27. Han D, Zhu G, Wu C. et al. Engineering a cell-surface aptamer circuit for targeted and amplified photodynamic cancer therapy. ACS Nano. 2013;7:2312-9
28. Zhu G, Hu R, Zhao Z. et al. Noncanonical self-assembly of multifunctional DNA nanoflowers for biomedical applications. J Am Chem Soc. 2013;135:16438-45
29. Douglas SM, Bachelet I, Church GM. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012;335:831-4
30. Jiang Q, Song C, Nangreave J. et al. DNA origami as a carrier for circumvention of drug resistance. J Am Chem Soc. 2012;134:13396-403
31. Wu Y, Sefah K, Liu H. et al. DNA aptamer-micelle as an efficient detection/delivery vehicle toward cancer cells. Proc Natl Acad Sci U S A. 2010;107:5-10
32. Wu C, Han D, Chen T. et al. Building a multifunctional aptamer-based DNA nanoassembly for targeted cancer therapy. J Am Chem Soc. 2013;135:18644-50
33. Mongelard F, Bouvet P. AS-1411, a guanosine-rich oligonucleotide aptamer targeting nucleolin for the potential treatment of cancer, including acute myeloid leukemia. Curr Opin Mol Ther. 2010;12:107-14
34. Hovanessian AG, Soundaramourty C, El Khoury D. et al. Surface expressed nucleolin is constantly induced in tumor cells to mediate calcium-dependent ligand internalization. PLoS One. 2010;5:e15787
35. Xing H, Tang L, Yang X. et al. Selective Delivery of an Anticancer Drug with Aptamer-Functionalized Liposomes to Breast Cancer Cells in Vitro and in Vivo. J Mater Chem B Mater Biol Med. 2013;1:5288-5297
36. Soundararajan S, Chen W, Spicer EK. et al. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008;68:2358-65
37. Girvan AC, Teng Y, Casson LK. et al. AGRO100 inhibits activation of nuclear factor-kappaB (NF-kappaB) by forming a complex with NF-kappaB essential modulator (NEMO) and nucleolin. Mol Cancer Ther. 2006;5:1790-9
38. Laber DA, Sharma VR, Bhupalam L. et al. Update on the first phase I study of AGRO100 in advanced cancer. J Clin Oncol. 2005;23:3064
39. Chen W, Sridharan V, Soundararajan S. et al. Activity and mechanism of action of AS1411 in acute myeloid leukemia cells. Blood. 2007;110:479-84
40. Rosenberg JE, Bambury RM, Van Allen EM. et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Invest New Drugs. 2014;32:178-87
41. Ristau BT, O'Keefe DS, Bacich DJ. The prostate-specific membrane antigen: Lessons and current clinical implications from 20 years of research. Urol Oncol. 2014;32:272-79
42. Kolishetti N, Dhar S, Valencia PM. et al. Engineering of self-assembled nanoparticle platform for precisely controlled combination drug therapy. Proc Natl Acad Sci U S A. 2010;107:17939-44
43. Gu F, Zhang L, Teply BA, Mann N. et al. Precise engineering of targeted nanoparticles by using self-assembled biointegrated block copolymers. Proc Natl Acad Sci U S A. 2008;105:2586-91
44. Kim D, Jeong YY, Jon S. A drug-loaded aptamer-gold nanoparticle bioconjugate for combined CT imaging and therapy of prostate cancer. ACS Nano. 2010;4:3689-96
45. Jalalian SH, Taghdisi SM, Shahidi Hamedani N. et al. Epirubicin loaded super paramagnetic iron oxide nanoparticle-aptamer bioconjugate for combined colon cancer therapy and imaging in vivo. Eur J Pharm Sci. 2013;50:191-7
46. Zhao N, You J, Zeng Z. et al. An ultra pH-sensitive and aptamer-equipped nanoscale drug-delivery system for selective killing of tumor cells. Small. 2013;9:3477-84
47. Li L, Xiang D, Shigdar S. et al. Epithelial cell adhesion molecule aptamer functionalized PLGA-lecithin-curcumin-PEG nanoparticles for targeted drug delivery to human colorectal adenocarcinoma cells. Int J Nanomedicine. 2014;9:1083-96
48. Liu Z, Duan JH, Song YM. et al. Novel HER2 aptamer selectively delivers cytotoxic drug to HER2-positive breast cancer cells in vitro. J Transl Med. 2012;10:148
49. Hu Y, Duan J, Zhan Q. et al. Novel MUC1 aptamer selectively delivers cytotoxic agent to cancer cells in vitro. PLoS One. 2012;7:e31970
50. Bandekar A, Zhu C, Jindal R. et al. Anti-prostate-specific membrane antigen liposomes loaded with 225Ac for potential targeted antivascular α-particle therapy of cancer. J Nucl Med. 2014;55:107-14
51. Farin K, Schokoroy S, Haklai R. et al. Oncogenic synergism between ErbB1, nucleolin, and mutant Ras. Cancer Res. 2011;71:2140-51
52. Sekar TV, Foygel K, Ilovich O. et al. Noninvasive Theranostic Imaging of HSV1-sr39TK-NTR/GCV-CB1954 Dual-Prodrug Therapy in Metastatic Lung Lesions of MDA-MB-231 Triple Negative Breast Cancer in Mice. Theranostics. 2014;4:460-74
53. Bhatia S, Frangioni JV, Hoffman RM. et al. The challenges posed by cancer heterogeneity. Nat Biotechnol. 2012;30:604-10
54. Denison TA, Bae YH. Tumor heterogeneity and its implication for drug delivery. J Control Release. 2012;164:187-91
55. Hoey T. Drug resistance, epigenetics, and tumor cell heterogeneity. Sci Transl Med. 2010;2:28ps19
56. Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346-54
57. Wilting RH, Dannenberg JH. Epigenetic mechanisms in tumorigenesis, tumor cell heterogeneity and drug resistance. Drug Resist Updat. 2012;15:21-38
58. Burke DH, Willis JH. Recombination, RNA evolution, and bifunctional RNA molecules isolated through chimeric SELEX. RNA. 1998;4:1165-75
59. Zhu G, Meng L, Ye M. et al. Self-assembled aptamer-based drug carriers for bispecific cytotoxicity to cancer cells. Chem Asian J. 2012;7:1630-6
60. Lu W, Arumugam SR, Senapati D. et al. Multifunctional oval-shaped gold-nanoparticle-based selective detection of breast cancer cells using simple colorimetric and highly sensitive two-photon scattering assay. ACS Nano. 2010;4:1739-49
61. Min K, Jo H, Song K. et al. Dual-aptamer-based delivery vehicle of doxorubicin to both PSMA (+) and PSMA (-) prostate cancers. Biomaterials. 2011;32:2124-32
62. Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21:283-96
63. Qu H, Li R, Liu Z. et al. Prognostic value of cancer stem cell marker CD133 expression in non-small cell lung cancer: a systematic review. Int J Clin Exp Pathol. 2013;6:2644-50
64. Wang R, Liu W, Helfer CM. et al. Activation of SOX2 expression by BRD4-NUT oncogenic fusion drives neoplastic transformation in NUT midline carcinoma. Cancer Res. 2014;74:3332-43
65. Kwan JJ, Kaya M, Borden MA. et al. Theranostic Oxygen Delivery Using Ultrasound and Microbubbles. Theranostics. 2012;2:1174-84
66. Shapira A, Livney YD, Broxterman HJ. et al. Nanomedicine for targeted cancer therapy: towards the overcoming of drug resistance. Drug Resist Updat. 2011;14:150-63
67. Denison TA, Bae YH. Tumor heterogeneity and its implication for drug delivery. J Control Release. 2012;164:187-91
68. Chung AS, Lee J, Ferrara N. Targeting the tumour vasculature: insights from physiological angiogenesis. Nat Rev Cancer. 2010;10:505-14
69. Wu W, Chen H, Shan F. et al. A Novel Doxorubicin-Loaded in Situ Forming Gel Based High Concentration of Phospholipid for Intratumoral Drug Delivery. Mol Pharm. 2014 [Epub ahead of print]
70. Kaelin WG Jr. Cancer and altered metabolism: potential importance of hypoxia-inducible factor and 2-oxoglutarate-dependent dioxygenases. Cold Spring Harb Symp Quant Biol. 2011;76:335-45
71. Guarente L.The many faces of sirtuins. Sirtuins and the Warburg effect. Nat Med. 2014;20:24-5
72. Lum JJ, Bui T, Gruber M. et al. The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev. 2007;21:1037-49
73. Rankin EB, Rha J, Selak MA. et al. Hypoxia-inducible factor 2 regulates hepatic lipid metabolism. Mol Cell Biol. 2009;29:4527-38
74. Greco O, Marples B, Joiner MC. et al. How to overcome (and exploit) tumor hypoxia for targeted gene therapy. J Cell Physiol. 2003;197:312-25
75. Baker LC, Boult JK, Walker-Samuel S. et al. The HIF-pathway inhibitor NSC-134754 induces metabolic changes and anti-tumour activity while maintaining vascular function. Br J Cancer. 2012;106:1638-47
76. Lu H, Li X, Luo Z. et al. Cetuximab reverses the Warburg effect by inhibiting HIF-1-regulated LDH-A. Mol Cancer Ther. 2013;12:2187-99
77. Zhang J, Cao J, Weng Q. et al. Suppression of hypoxia-inducible factor 1α (HIF-1α) by tirapazamine is dependent on eIF2α phosphorylation rather than the mTORC1/4E-BP1 pathway. PLoS One. 2010;5:e13910
78. Chen ZS, Tiwari AK. Multidrug resistance proteins (MRPs/ABCCs) in cancer chemotherapy and genetic diseases. FEBS J. 2011;278:3226-45
79. Chen Y, Bathula SR, Li J. et al. Multifunctional nanoparticles delivering small interfering RNA and doxorubicin overcome drug resistance in cancer. J Biol Chem. 2010;285:22639-50
80. Patil YB, Swaminathan SK, Sadhukha T. et al. The use of nanoparticle-mediated targeted gene silencing and drug delivery to overcome tumor drug resistance. Biomaterials. 2010;31:358-65
81. Wu J, Lu Y, Lee A. et al. Reversal of multidrug resistance by transferrin-conjugated liposomes co-encapsulating doxorubicin and verapamil. J Pharm Pharm Sci. 2007;10:350-7
82. Chen Y, Zhang L, Lu X. et al. Sinomenine reverses multidrug resistance in bladder cancer cells via P-glycoprotein-dependent and independent manners. Pharmazie. 2014;69:48-54
83. Liu Y, Li LL, Qi GB. et al. Dynamic disordering of liposomal cocktails and the spatio-temporal favorable release of cargoes to circumvent drug resistance. Biomaterials. 2014;35:3406-15
84. Zhang H, Wang J, Cai K. et al. Downregulation of gene MDR1 by shRNA to reverse multidrug-resistance of ovarian cancer A2780 cells. J Cancer Res Ther. 2012;8:226-31
85. Perche F, Biswas S, Wang T. et al. Hypoxia-Targeted siRNA Delivery. Angew Chem Int Ed Engl. 2014;53:3362-6
86. Chung LW, Baseman A, Assikis V. et al. Molecular insights into prostate cancer progression: the missing link of tumor microenvironment. J Urol. 2005;173:10-20
87. Sanità P, Capulli M, Teti A. et al. Tumor-stroma metabolic relationship based on lactate shuttle can sustain prostate cancer progression. BMC Cancer. 2014;14:154
88. Yu Y, Xiao CH, Tan LD. et al. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of breast cancer cells through paracrine TGF-β signalling. Br J Cancer. 2014;110:724-32
89. Hale MD, Hayden JD, Grabsch HI. Tumour-microenvironment interactions: role of tumour stroma and proteins produced by cancer-associated fibroblasts in chemotherapy response. Cell Oncol (Dordr). 2013;36:95-112
90. Choi YP, Lee JH, Gao MQ. et al. Cancer-associated fibroblast promote transmigration through endothelial brain cells in three-dimensional in vitro models. Int J Cancer. 2014 [Epub ahead of print]
91. Obeid E, Nanda R, Fu YX. et al. The role of tumor-associated macrophages in breast cancer progression. Int J Oncol. 2013;43:5-12
92. Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4:71-8
93. Rupp C, Scherzer M, Rudisch A. et al. IGFBP7, a novel tumor stroma marker, with growth-promoting effects in colon cancer through a paracrine tumor-stroma interaction. Oncogene. 2014 [Epub ahead of print]
94. Andersen MN, Abildgaard N, Maniecki MB. et al. Monocyte/macrophage-derived soluble CD163: a novel biomarker in multiple myeloma. Eur J Haematol. 2014 [Epub ahead of print]
95. Hall CL, Dai J, van Golen KL. et al. Type I collagen receptor (alpha 2 beta 1) signaling promotes the growth of human prostate cancer cells within the bone. Cancer Res. 2006;66:8648-54
96. Moro L, Arbini AA, Marra E. et al. Down-regulation of BRCA2 expression by collagen type I promotes prostate cancer cell proliferation. J Biol Chem. 2005;280:22482-91
97. Kiefer JA, Farach-Carson MC.Type I collagen-mediated proliferation of PC3 prostate carcinoma cell line. implications for enhanced growth in the bone microenvironment. Matrix Biol. 2001;20:429-37
98. Yang H, Liu H, Kang H. et al. Engineering target-responsive hydrogels based on aptamer-target interactions. J Am Chem Soc. 2008;130:6320-1
99. Yang L, Zhang X, Ye M. et al. Aptamer-conjugated nanomaterials and their applications. Adv Drug Deliv Rev. 2011;63:1361-70
100. Nel AE, Mädler L, Velegol D. et al. Understanding biophysicochemical interactions at the nano-bio interface. Nat. Mater. 2009;8:543-57
101. Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8:129-38
102. Bareford LM, Swaan PW. Endocytic mechanisms for targeted drug delivery. Adv. Drug Deliv. Rev. 2007;59:748-58
103. Jain RK. Delivery of molecular and cellular medicine to solid tumors. Adv Drug Deliv Rev. 2012;64:353-65
104. Guo J, Gao X, Su L. et al. Aptamer-functionalized PEG-PLGA nanoparticles for enhanced anti-glioma drug delivery. Biomaterials. 2011;32:8010-20
105. Buggins AG, Pepper C, Patten PE. et al. Interaction with vascular endothelium enhances survival in primary chronic lymphocytic leukemia cells via NF-kappaB activation and de novo gene transcription. Cancer Res. 2010;70:7523-33
106. Kim HJ, Yim GW, Nam EJ. et al. Synergistic Effect of COX-2 Inhibitor on Paclitaxel-Induced Apoptosis in the Human Ovarian Cancer Cell Line OVCAR-3. Cancer Res Treat. 2014;46:81-92
107. Li Z, Guo Y, Jiang H. et al. Differential regulation of MMPs by E2F1, Sp1 and NF-kappa B controls the small cell lung cancer invasive phenotype. BMC Cancer. 2014;14:276
108. Mi J, Zhang X, Rabbani ZN. et al. RNA aptamer-targeted inhibition of NF-kappa B suppresses non-small cell lung cancer resistance to doxorubicin. Mol Ther. 2008;16:66-73
109. Srinivasan S, Selvan ST, Archunan G. et al. MicroRNAs -the Next Generation Therapeutic Targets in Human Diseases. Theranostics. 2013;3:930-42
110. Xu X, Xie K, Zhang XQ. et al. Enhancing tumor cell response to chemotherapy through nanoparticle-mediated codelivery of siRNA and cisplatin prodrug. Proc Natl Acad Sci U S A. 2013;110:18638-43
111. Draz MS, Fang BA, Zhang P. et al. Nanoparticle-Mediated Systemic Delivery of siRNA for Treatment of Cancers and Viral Infections. Theranostics. 2014;4:872-92
112. Rosenblum D, Peer D. Omics-based nanomedicine: The future of personalized oncology. Cancer Lett. 2013 doi: 10.1016/j.canlet.2013.07.029
113. Vogelstein B, Papadopoulos N, Velculescu VE. et al. Cancer genome landscapes. Science. 2013;339:1546-58
114. Zhang Y, Hong H, Cai W. Tumor-targeted drug delivery with aptamers. Curr Med Chem. 2011;18:4185-94
Author contact
![]() Corresponding author: Yuan Zhang, yzhang75org or joint_chuengedu.cn. Tel: +86 23 68755618; Fax: +86 23 68755608.
Corresponding author: Yuan Zhang, yzhang75org or joint_chuengedu.cn. Tel: +86 23 68755618; Fax: +86 23 68755608.
 Global reach, higher impact
Global reach, higher impact