13.3
Impact Factor
Theranostics 2014; 4(5):487-497. doi:10.7150/thno.8263 This issue Cite
Research Paper
Cyclodextrin and Polyethylenimine Functionalized Mesoporous Silica Nanoparticles for Delivery of siRNA Cancer Therapeutics
1. MOE Key Laboratory of Bioinorganic and Synthetic Chemistry, School of Chemistry and Chemical Engineering, Sun Yat-sen University, Guangzhou, 510275, China,
2. Department of Nanomedicine, Houston Methodist Research Institute, Houston, Texas 77030, United States,
3. Department of Gastroenterology, The Tenth People's Hospital of Shanghai, Tongji University, Shanghai, 20072, China
4. CAS Key Laboratory for Biomedical Effects of Nanomaterials & Nanosafety, National Center for Nanoscience & Technology of China, Beijing 100190, China.
5. Department of Cell and Developmental Biology, Weill Cornell Medical College, New York, NY10065, United States
* These authors contributed equally.
Received 2013-12-2; Accepted 2014-1-20; Published 2014-2-15
Abstract
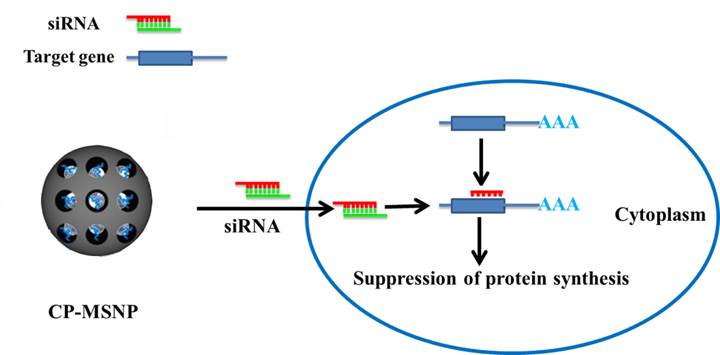
Effective delivery holds the key to successful in vivo application of therapeutic small interfering RNA (siRNA). In this work, we have developed a universal siRNA carrier consisting of a mesoporous silica nanoparticle (MSNP) functionalized with cyclodextrin-grafted polyethylenimine (CP). CP provides positive charge for loading of siRNA through electrostatic interaction and enables effective endosomal escape of siRNA. Using intravital microscopy we were able to monitor tumor enrichment of CP-MSNP/siRNA particles in live mice bearing orthotopic MDA-MB-231 xenograft tumors. CP-MSNP delivery of siRNA targeting the M2 isoform of the glycolytic enzyme pyruvate kinase (PKM2) resulted in effective knockdown of gene expression in vitro and in vivo. Suppression of PKM2 led to inhibition of tumor cell growth, invasion, and migration.
Keywords: mesoporous silica nanoparticle, siRNA, delivery, cancer therapy, gene silencing
1. Introduction
Since the discovery of RNA interference (RNAi) in 1998, there has been an impetus towards the development of siRNA therapeutics [1] . RNAi offers the potential for silencing any gene that contributes to pathogenesis, thereby providing a new avenue for treatment that does not require specific small molecule inhibitors [2, 3]. Consequently, multiple siRNA-based agents have reached various stages of clinical application for localized therapy and for liver-related diseases [4]. However, systemic delivery of siRNA therapeutics to organs outside the liver remains a considerable hurdle, since most delivery vectors tend to accumulate in organs of the reticulo-endothelial system (RES) [5-8]. In order for the siRNA to reach tumor cells in other organs, the delivery system will need to not only evade the RES, but also pass other biological barriers such as vascular endothelium, unfavorable interstitial osmotic pressure, and attack from tissue and cellular enzymes [9]. So an ideal nanocarrier for siRNA should be a multi-component delivery system with i) long blood circulation time to take advantage of the enhanced permeability and retention effect (EPR), ii) a protective environment against enzymatic degradation, iii) minimal RES uptake, and iv) effective cellular internalization and endosomal escape of siRNA [9].
Mesoporous silica nanoparticles (MSNPs), which were first applied for drug delivery in 2001 [10], can serve as the building block for such a delivery system [11]. A major advantage of these nanoparticles is that the size, shape, and surface properties can be tailored to achieve efficient loading of therapeutics, tissue-specific accumulation, high levels of intracellular uptake, favorable intracellular localization, and appropriate release of therapeutics [10]. Consequently, on the preclinical level, MSNPs have been utilized for the delivery of drugs and nucleic acids to combat diseases, such as diabetes, inflammation, and cancer [10, 12-13]. Herein, we designed a MSNP-based siRNA delivery vehicle. The MSNPs have been functionalized with cyclodextrin-grafted polyethylenimine (CP). Polyethylenimine (PEI) forms an electrostatic bond with siRNA oligos, while cyclodextrin eliminates charge-induced toxicity of PEI. The pyruvate kinase M2 isoform (PKM2), which is frequently overexpressed in several cancer types [14], was used as a target gene to evaluate the effectiveness of this delivery system. Cellular internalization, subcellular localization, gene silencing capability and anticancer activity of siRNA-loaded CP-MSNP were assessed with MDA-MB-231 human breast cancer cells. In addition, tumor accumulation and in vivo gene silencing efficiency were determined using an orthotropic mouse model of MDA-MB-231 breast cancer.
2. Experimental section
2.1 Materials
Branched PEI (MW 1.8 kDa), β-cyclodextrin, N-cetyltrimethylammonium bromide (CTAB), tetraethoxysilane (TEOS), 4',6-diamidino-2-phenylindole (DAPI) and 3-isocyanatopropyltriethoxysilane (ICP) were from Sigma-Aldrich. Dulbecco's Modified Eagle Medium (DMEM) high glucose and fetal bovine serum (FBS) were obtained from Fisher Scientific. LysoTracker® Green and fluorescein isothiocyanate (FITC)-labeled dextran dyes (70-kDa) were purchased from Invitrogen. Small interfering RNA (siRNA) conjugated with Alexa555 was ordered from Qiagen. Scrambled siRNA, and PKM2 siRNA were from Sigma. PKM2 siRNA sense and anti-sense sequences are CCAUAAUCGUCCUCACCAA[dT][dT] and UUGGUGAGGACGAUUAUGG[dT][dT], respectively. Female athymic nude mice were acquired from Charles River.
2.2 Cell culture
The MDA-MB-231 cell line was obtained from ATCC (Rockville, MD). Cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin under 5% CO2 and 95% humidity at 37°C.
2.3 Preparation of β-cyclodextrin-grafted PEI
Beta-cyclodextrin-grafted PEI was synthesized according to a previous report with modifications [15]. Briefly, 6-mono-tosylated cyclodextrin (65 mg, 0.05 mmol) was added drop-by-drop into a solution containing PEI (12 mg, 0.093 mmol) and triethylamine (4.72 mg, 0.047 mmol) in 3 mL of dimethyl sulfoxide (DMSO). The reaction was allowed to proceed at 70°C in a nitrogen atmosphere for 3 days. The resulting mixture was dialyzed against water in a Spectra/Por MWCO 1000 bag, and then separated with a Sephadex-G25 column.
2.4 Synthesis of CP-MSNP
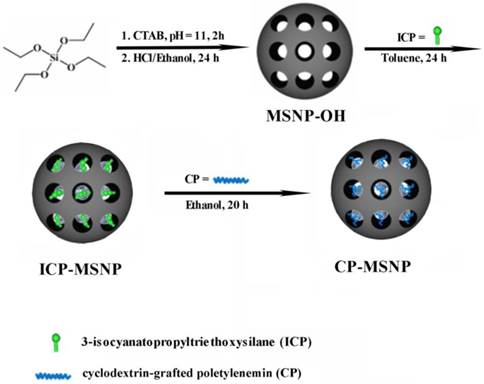
Fabrication of MSNP was based on a previously reported protocol [16]. MSNP-OH was synthesized by mixing TEOS with CTAB in a basic aqueous solution (pH = 11). The remaining CTAB was removed by suspending the particles in a solution containing methanol/hydrochloric acid (20/1, v/v) for 24 h, followed by centrifugation. MSNP-OH (1.0 g) was then resuspended in 80 mL anhydrous toluene, and mixed with 0.25 mL ICP (1.0 mmol). The reaction was allowed to proceed for 20 h to yield MSNP-ICP particles. To conjugate MSNP-ICP with CP, MSNP-ICP particles (0.3 g) were dispersed in anhydrous ethanol (5.0 mL), and 30 mg CP was added. The reaction was maintained at 20°C for 20 h. The final product was filtered and washed thoroughly with ethanol and methanol before vacuum dry overnight.
2.6. Optimization of the CP-MSNP/siRNA delivery system
CP-MSNP particles were mixed with scrambled siRNA in nuclease free water at various weight ratios (CP-MSNP/siRNA: 20:1, 40:1, 60:1, 80:1, 100:1), and incubated for 15 min at 4°C. Binding of siRNA to CP-MSNP was determined by electrophoresis in a 2% agarose gel containing ethidium bromide. Electrophoresis was carried out at a constant voltage of 120 V for 20 min in TAE running buffer (2 M Tris, 250 mM sodium acetate, 50 mM EDTA, pH 7.8). To evaluate whether CP-MSNP could protect siRNA from degradation, 0.2 μg siRNA loaded in CP-MSNP was incubated with 1 μg RNase (1 μg/μL) at 37°C for 1 h. RNase was then inactivated with ethylenediaminetetraacetic acid (EDTA, 0.25 M). siRNA was then dissociated from CP-MSNP in NaOH, and analyzed by electrophoresis.
To measure the release profile of siRNA from MSNP, CP-MSNP particles loaded with Alexa555 siRNA (w/w: 60/1) were incubated in 10 mM 2-[4-(2-hydroxyethyl) piperazin-1-yl]ethanesulfonic acid (HEPES) buffer at pH = 5.2 or 7.4, and released siRNA in supernatant was collected at different time points, and the fluorescence intensity was measured with a BioTek H4 synergy hybrid plate reader.
2.7. siRNA stability in serum
Naked scrambled siRNA and CP-MSNP/scrambled siRNA were incubated in 50% FBS at 37 °C. Samples were taken at 30 min, 1 h, 3 h, 6 h, 12 h, 36 h and 60 h, and immediately mixed with gel loading buffer containing 1% SDS. After the final aliquote was collected, the samples were applied to electrophoresis in a 2% agarose gel and the presence of siRNA oligo was visualized by ethidium bromide staining.
2.8. Cellular uptake and intracellular trafficking of CP-MSNP/Alexa555 siRNA
Cellular uptake of CP-MSNP/siRNA was examined by confocal microscopy and flow cytometry. Briefly, MDA-MB-231 cells were seeded in 6-well plates with a density of 3×105 cells/well, and incubated with CP-MSNP/Alexa555 siRNA. Distribution of siRNA inside the cells was examined on days 1, 3, and 5 by confocal microscopy. For flow cytometry analysis, cells were incubated with CP-MSNP/Alexa555 siRNA complexes (50 nM siRNA final concentration). Samples were collected at different time periods (0.5, 1, 4, and 24 h), and Alexa555-positive cells were determined using an LSRII Flow Cytometer (BD Bioscience, USA).
To monitor intracellular trafficking, CP-MSNP/Alexa555 siRNA particles were incubated with MDA-MB-231 cells for various time periods (0.5, 2, and 6 h). Cells were harvested and fixed with 4% paraformaldehyde. Samples were then blocked with 1% bovine serum albumin (BSA) in phosphate buffered saline containing 0.1% Tween-20 (PBST), and stained with LysoTracker® Green and DAPI. Confocal microscopic images were obtained using a Fluo View TM 1000 confocal microscope.
2.9. Gene silencing in vitro
For expression analysis by Western blot, MDA-MB-231 cells were seeded in 6-well plates at a density of 2 × 105 cells/well in 2 mL DMEM containing 10% FBS. Cells were incubated with CP-MSNP/siRNA for 72 h before they were harvested. Protein samples were extracted using a mammalian protein extraction reagent (M-PER, Pierce Inc., USA), separated on a 12% gel by SDS-polyacrylamide gel electrophoresis (PAGE), and transferred to a nitrocellulose membrane (Bio-Rad Inc., USA) for detection with the Amersham ECL Western blot detection reagent.
2.10. Evaluation of cell viability, migration and invasion in vitro
Proliferation of MDA-MB-231 cells in the presence of CP-MSNP/siRNA was determined using a MTS-based colorimetric assay (CellTiter 96 cell proliferation assay kit; Promega, Madison, WI, USA) according to the manufacturer's instructions. Briefly, cells were seeded in 96-well culture plates (3 × 103 cells/well). They were treated with CP-MSNP/siRNA for 72 h, and cell growth was measured.
To assess cell migration and invasion, MDA-MB-231 cells were seeded in 6 mm tissue culture dishes at a density of 5 × 105 cells/well in 2 mL DMEM containing 10% FBS. Cells were incubated with empty CP-MSNP or CP-MSNP/siRNA (scrambled siRNA or PKM2 siRNA) for 48 h and then resuspended in FBS-free medium at a density of 1 × 106 cells/mL. For the migration assay, 300 μL of cell suspension was added to the upper chamber of the transwell inserts (BD Biosciences, Sparks, MD). 500 μL of medium containing 10% FBS was added into the bottom plate to act as a chemo-attractant. After 24 h incubation, cells in the upper chamber were removed with cotton swabs, and cells on the lower surface of the inserts were fixed and stained using Crystal Violet (Sigma-Aldrich). The number of migratory cells was calculated by counting ten separate fields under a phase-contrast microscope in three independent inserts. To examine cell invasion, the upper chamber was pre-coated with Matrigel (BD Biosciences), and 250 μL of cell suspension was added. Growth medium in the bottom chamber contained 20% FBS. Cells were allowed to invade for 48 h before fixation and staining.
For evaluation of cell migration by scratch assay, MDA-MB-231 cells were seeded in 6-well plates, and kept at 37°C until the cells reached a density of 85%. Cells were incubated with empty CP-MSNP or CP-MSNP/siRNA (scrambled siRNA or PKM2 siRNA) for 48 h and then scratched with a pipette tip. Microscopy images were taken immediately after scratching and 24 h later.
2.11. Evaluation of CP-MSNP delivery in vivo
All experimental procedures for animal studies were approved by the committee for the care and use of laboratory animals at Houston Methodist Research Institute. To determine particle biodistribution, CP-MSNP/Alexa555 siRNA particles were administered into female athymic nu/nu nude mice by tail vein injection (n=3, 15 μg siRNA/mouse). Major organs including heart, liver, spleen, lung, kidney, and tumor were collected 1 h post injection. Fluorescent images of the tissues were captured using the IVIS 200 imaging system (PerkinElmer, USA).
In order to visualize particle flow in live animals, mice were also inoculated i.v. with fluorescein isothiocyanate (FITC)-labeled dextran dyes (70-kDa), which delineated the tumor vasculature. Movement of the red fluorescent CP-MSNP/Alexa555 siRNA particles inside tumor blood vessels was visualized by intravital microscopy (RS 2000TM, Newport, USA), and images were captured.
To determine knockdown of gene expression in vivo, nude mice were inoculated with MDA-MB-231 tumor cells in the mammary fat pad. When the average tumor size reached 150-200 mm3, mice were divided into three groups and treated on day 1 and day 3 with: i) CP-MSNP/scrambled siRNA (15 μg siRNA/injection), ii) CP-MSNP/PKM2 siRNA (15 μg siRNA/injection), and CP-MSNP (900 μg/injection). Mice were sacrificed on day 5 and tumor samples were collected for expression analysis by Western blot.
2.12. Statistical Analysis
All quantitative data are presented as mean ± standard deviation. Statistical analysis was performed with the Student's t-test. Differences were considered statistically significant at p < 0.05 (*) and p < 0.01 (**).
3. Results and discussion
3.1. Synthesis and characterization of CP and CP-MSNPs
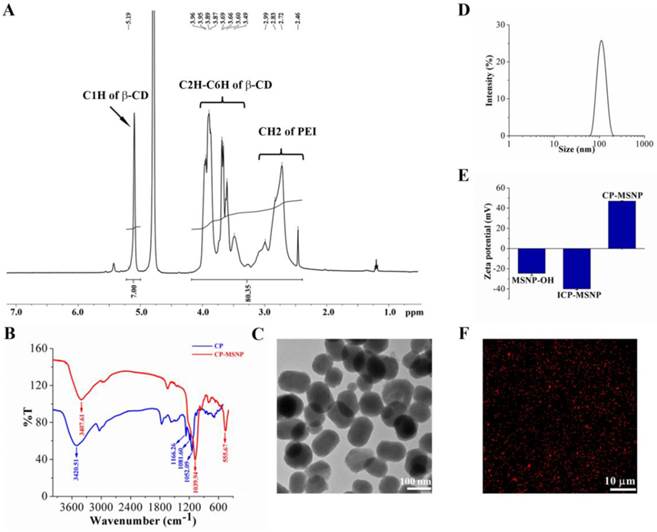
The extent of cyclodextrin grafting to polyethylenimine was determined based on proton integration of 1H-nuclear magnetic resonance (NMR) (400 MHz, D2O): δ5.19 ppm (s, C1H of beta-CD), 3.3-4.2 ppm (m br., C2H-C6H of beta-CD), 2.4-3.2 ppm (m br., CH2 of PEI). The integration yielded a calculated beta-cyclodextrin/PEI ratio of approximately 5 (Fig. 1A), which is very similar to that reported by Davis and co-workers [15]. As shown in Figure 1B, the strong band at 1166.26 cm-1 can be assigned to the asymmetric glycoside vibration (C-O-C) and the bands at 1052.09 and 1081.60 cm-1 correspond to the coupled stretch vibrations (C-C/C-O), which are typical for cyclodextrin. The broad band at 3420.51 cm-1 corresponds to the O-H vibration and the N-H vibration arising from PEI.
The two-step procedure for fabrication of MSNPs, illustrated in Scheme 1/Figure A, is based on a protocol similar to that previously reported [16]. Fourier transform infrared spectroscopy (FTIR) confirmed that the CP was successfully conjugated with CP-MSNP (Fig. 1B). The MSNP peaks at 555.67 and 1039.94 cm-1 are in accordance with previously reported results [17]. The CP peak around 1039.34 is almost covered by the stronger MSNP peak. Moreover, the peak at 3407.61 with a slight blue shift corresponds to the CP-MSNP complex. Transmission electron microscopy (TEM) was applied to analyze the morphology of CP-MSNP (Fig. 1C). The size of CP-MSNPs was in the range of 80-140 nm with a mean size of 105 nm as determined by dynamic light scattering (Fig. 1D). Surface chemical modification of the particles was confirmed by changes in surface charge (Fig. 1E). CP modification changed zeta potential of the particles from negative to positive. The zeta potentials were -24.6 mV for MSNP-OH, -39.97 mV for ICP-MSNP, and +46.97 mV for CP-MSNP. Alexa555 siRNA was used to visualize siRNA loading to CP-MSNP. Confocal microscopy confirmed successful loading of siRNA to the particles (Fig. 1F).
(Scheme 1) Schematic illustration on fabrication of the CP-MSNP delivery carrier for gene silencing agents.
Characterization of CP-MSNP as a delivery carrier for gene silencing agents. (A) 1H nuclear magnetic resonance (NMR) analysis of cyclodextrin-grafted polyethylenimine (CP) in D2O (400 MHz). (B) Fourier transform infrared spectroscopy (FTIR) spectrums of CP and CP-MSNPs. (C) Transmission electron microscopy (TEM) image of CP-MSNP. (D) Size distribution of CP-MSNPs measured by dynamic light scattering. (E) Changes in zeta potential of particles at various stages of fabrication. Results are presented as the mean of five measurements ± standard deviation. (F) Confocal microscopic image of CP-MSNP/siRNA. The red fluorescence stems from Alexa555-conjugated siRNA.
3.2. Optimization and release kinetics of CP-MSNP/siRNA
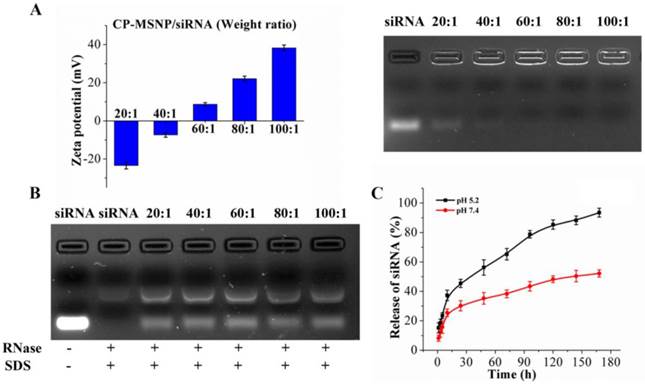
Binding affinity, stability, and release profile of siRNA are some of the most important factors to consider when developing a siRNA delivery platform [7]. In this study, agarose gel electrophoresis was performed in order to examine the condensing ability and binding affinity of CP-MSNPs. We used a wide range of CP-MSNP to siRNA ratios (20:1 to 100:1) in this study. The surface charge of the CP-MSNP/siRNA complexes ranged from approximately -23 to +38 mV depending on the weight ratio of CP-MSNP to siRNA (Fig. 2A, left panel). No significant amount of siRNA release could be detected by electrophoresis when the MSNP/siRNA ratio was above 40 (Fig. 2A, right panel), indicating that the CP-MSNP carrier was able to bind the negatively charged siRNA tightly. In addition, stability of the loaded siRNA was assessed by incubating CP-MSNP/siRNA with RNase, an enzyme that degrades RNA. As shown in Figure 2B, free siRNA was degraded after incubation with RNase, while the CP-MSNP platform effectively protected siRNA from degradation.
It is important for the delivery vehicle to display a moderately positive charge, since cellular uptake is not as effective for negatively charged particles [18]. On the other hand, particles with a very high positive charge can cause cytotoxic effects [19]. Hence, the CP-MSNP/siRNA particles with a weight ratio of 60:1 and a positive charge of ~+8 mV were selected for further studies.
To compare stability of between the naked siRNA and siRNA in CP-MSNP particles, samples were incubated in 50% FBS at 37 °C for a different amount of time, and then separated in a 2% agarose gel (Supplementary Material: Fig. S1). After 12 h of incubation, 86.5% of the naked siRNA and 45.5% of the particle-bound siRNA had degraded. The result indicates that binding of siRNA to MSNP protects siRNA from degradation by serum enzymes. Release of siRNA from the CP-MSNP at pH 5.2 and 7.4 was also evaluated. After 168 h of incubation, the cumulative release reached 93% at pH 5.5 and 52% at pH 7.4 (Fig. 2C). This result demonstrates that siRNA oligos can be effectively released from the carrier.
3.3. Uptake of CP-MSNP/siRNA by tumor cells
Cellular uptake of CP-MSNP/Alexa555 siRNA was evaluated in MDA-MB-231 human breast cancer cells. Confocal microscopy was used to monitor the intracellular accumulation of particles at different time points. Particle uptake by the breast cancer cells was apparent 1 day after incubation (Fig. 3A), and multiple particles per cell could be visualized on day 3 and 5 (Fig. 3A). Flow cytometry was also used to measure cellular uptake of Alexa555 siRNA at various time periods after incubation (0.5, 1, 4, and 24 h). The percentage of cells with internalized CP-MSNP/Alexa555 siRNA was 32.8% (0.5 h), 58.0% (1 h), 81.5% (4 h) and 99.2% (24 h) (Fig. 3B). These results indicate that the particles can be internalized rapidly and effectively by tumor cells.
Optimization of CP-MSNP/siRNA and siRNA release from CP-MSNP/siRNA. (A) Left panel: Changes in zeta potential at different ratios of CP-MSNP and siRNA (w/w, 20:1 to 100:1). Right panel: Agarose gel retardation assay of CP-MSNP/siRNA complexes at various ratios. Naked siRNA (100 μg) was used as a control. (B) Agarose gel analysis on protection of siRNA from RNase degradation by CP-MSNPs. siRNA was dissociated from the carrier in the presence of SDS. Naked siRNA (200 μg) was used as a control. (C) Release profiles of siRNA from CP-MSNP/siRNA (60:1). CP-MSNP/Alexa555 siRNA was incubated in 10% fetal bovine serum at pH 5.2 or pH 7.4. Supernatant was separated from the particles, and was used to measure fluorescent intensity at Ex543/Em590 nm. Results are presented as the mean of three independent measurements ± standard deviation.
Uptake of CP-MSNP/Alexa555 siRNA by MDA-MB-231 cells. (A) Time-dependent analysis of CP-MSNP/Alexa555 siRNA (red) inside MDA-MB-231 cells. Final siRNA concentration: 50 nM. (B) Quantitative flow cytometry analysis of Alexa555-positive cells after incubation with CP-MSNP/Alexa555 siRNA at different time points. Untreated cells were used as a negative control. Final siRNA concentration: 50 nM. (C-D) MTS assay on cell viability following incubation with CP-MSNP (C) or CP-MSNP/siRNA (D) for 72 h. Results are presented as the mean of three measurements ± standard deviation.
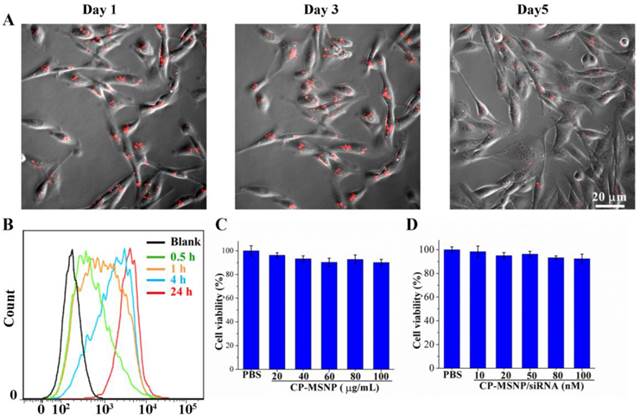
Since PEI is known to cause cytotoxicity due to the prevalence of primary amino groups in this polymer [20], we evaluated potential cell-killing effect from empty CP-MSNP and scrambled siRNA-loaded CP-MSNP. Over 95% of the cells were viable at concentrations up to 100 μg/mL after 72 h of incubation (Fig. 3C, 3D). It is likely that conjugation of PEI to ICP-MSNP has converted primary amino groups in PEI to secondary or tertiary amines, thereby reducing overall toxicity.
3.4. Intra-cellular trafficking and gene silencing in vitro
Endosomal/lysosomal escape is essential for effective delivery of gene silencing agents [21]. We examined intra-cellular location of CP-MSNP/Alexa555 siRNA by tracking the red fluorescent Alexa555 siRNA with confocal microscopy (Fig. 4A). The acidic late endosomes and lysosomes were stained with LysoTrack Green. The images indicate that the siRNA reached the endosomes/lysosome after 30 min of incubation, as is evident from the colocalization of the red and green fluorescence signal. Maximum levels of colocalization could be observed after 2 h, when almost all of the Alexa555siRNA was located in the endosome/lysosome. By 6 h, some of the siRNA was present in the cytoplasm, suggesting that the particles had escaped from the late endosome or lysosome. The potential mechanism behind this phenomenon is most likely the proton sponge effect triggered by the polyamine moiety present in the delivery vehicle [22].
Intracellular trafficking and gene silencing of CP-MSNP/siRNA in MDA-MB-231 cells. (A) Intracellular trafficking of CP-MSNP/siRNA in MDA-MB-231 cells. Cells were harvested at the indicated time points and stained with Lysotracker (green) and DAPI (blue). Fluorescent images were captured with a confocal microscope. (B) Western blot analysis of PKM2 protein expression in MDA-MB-231 cells. Beta-actin was used as an internal control. Cells were treated with 50 nM siRNA. PKM2: PKM2 siRNA; Scr: scrambled siRNA.
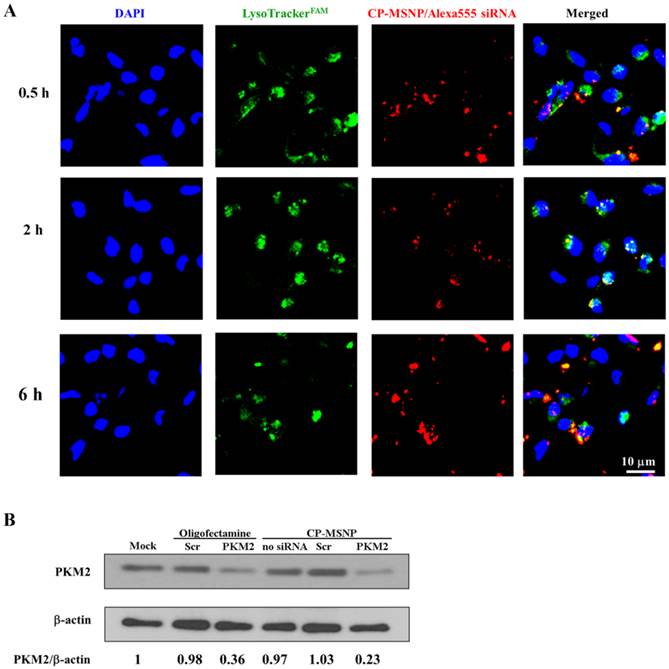
To evaluate gene knockdown efficiency of the CP-MSNP delivered siRNA, MDA-MB-231 breast cancer cells were treated with CP-MSNP carrying siRNA specific for the PKM2 gene (Fig. 4B). Expression of PKM2 in cancer cells results in aerobic glycolysis, which is believed to bestow a selective growth advantage [14]. CP-MSNP/PKM2 siRNA treatment caused significant reduction in PKM2 protein level as compared to treatment with CP-MSNP/scrambled siRNA. Importantly, the CP-MSNP delivery platform was more efficient than the commercial transfection reagent Oligofectamine.
3.5. Migration and invasion in vitro
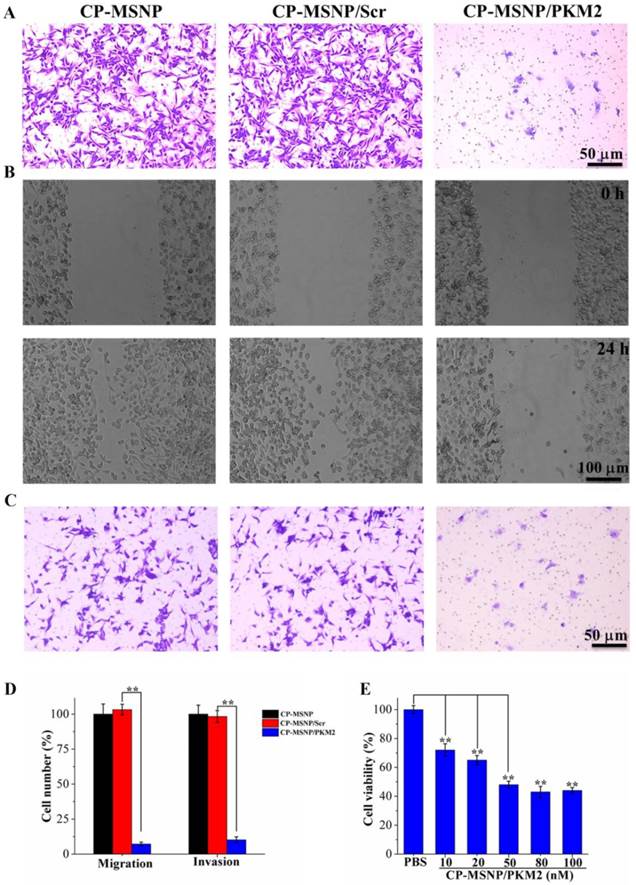
Since inhibition of PKM2 is known to result in suppression of cancer cell proliferation and migration [23], we assessed cell migration in response to CP-MSNP/PKM2 siRNA treatment in a two-chamber assay. Compared to the negative control groups (CP-MSNP and CP-MSNP/scrambled siRNA, 50 nM), cells treated with CP-MSNP/PKM2 siRNA exhibited a 93% decrease in migration (Fig. 5A, 5D). We also examined cell migration in a scratch assay. While cells treated with empty CP-MSNP or CP-MSNP/scrambled siRNA migrated rapidly, the cells with suppressed PKM2 did not show apparent migration (Fig. 5B). We observed similar results in the invasion assay, where the number of invading cells was reduced by ~90% in cells treated with CP-MSNP/PKM2 siRNA (Fig. 5C, 5D). Besides preventing migration and invasions, treatment with CP-MSNP/PKM2 siRNA platform also significantly reduced viability of MDA-MB-231 cells (Fig. 5E). Taken together, these results confirm that CP-MSNP particles are very effective in delivering siRNA into tumor cells to achieve knockdown of gene expression.
3.6. Evaluation of the CP-MSNP delivery system in vivo
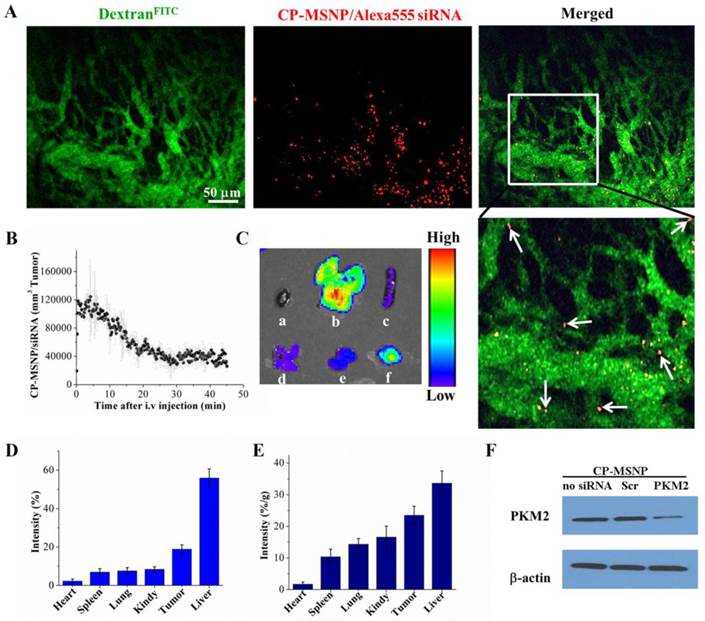
Athymic nude mice bearing orthotopic MDA-MB-231 tumors were used to test the ability of CP-MSNPs to deliver gene-silencing agents to solid tumors in vivo. In order to track CP-MSNP/Alexa555 siRNA particles (in red) inside tumors, mice were intravenously administered with FITC-labeled dextran dyes (in green) to delineate the tumor vasculature. Images and videos were captured during the first hour after particle administration by intravital microscopy. As shown in Figure 6A, particles were successfully delivered to the tumor tissue. While most particles passed through the tumor vasculature, many of them remained stationary within the tumor (video S1), indicating successful particle retention. Time-dependent particle flow-through and retention was analyzed and shown in Figure 6B. There was a surge of particles that passed through tumor blood vessels in the first 10 minutes following administration. The number of particles leveled off after 20 minutes, and approximately 40,000 CP-MSNP/siRNA particles per cubic millimeter were detected in the tumor 30 minutes after administration. Mice were subsequently sacrificed and major organs (heart, spleen, kidney, liver, and lung) and tumor tissues were visualized using an IVIS-200 imaging system (Fig. 6C). While most particles accumulated in the liver, as much as 19% of the total combined fluorescent intensity from these organs/tissues was detected in the tumor tissue, (Fig. 6D), which was equivalent to 24% of the measured fluorescence/gram tissue (Fig. 6E). Although the result does not reflect a whole-body or quantitative analysis, the biodistributional pattern suggests that the EPR effect may have played a significant role in tumor accumulation of CP-MSNP/siRNA. Indeed nano-scale objects are known to preferentially accumulate in tumors due to the presence of leakier vasculature [24].
In a separate study, mice bearing MDA-MD-231 orthotopic tumors were treated i.v. with two doses of CP-MSNP/PKM2 siRNA (15 μg siRNA in 900 μg CP-MSNP particles/injection). Tumor samples were collected 5 days after the first treatment, and PKM2 expression was measured by Western blot. The CP-MSNP/PKM2 siRNA treatment knocked down PKM2 expression, while the expression level was unaltered in the control group that received CP-MSNP/scrambled siRNA (Fig. 6F). These in vivo results demonstrated that CP-MSNP can effectively deliver gene-silencing agents to tumor tissues and mediate suppression of gene expression.
4. Conclusion
In summary, we have developed an efficient multi-component system for delivery of siRNA therapeutics. Each component is design to perform one or more unique functions in order for the whole system to overcome biological barriers and achieve tumor enrichment of therapeutics. This system is capable of not only protecting siRNA oligos from attack by plasma RNases, but also enriching the gene-silencing agents in tumor tissues. Overcoming multiple biological barriers and tumor tissue enrichment are perhaps the two most important properties of a good delivery system. In contrast to most other delivery systems where complicated chemical conjugation steps are needed to package siRNA [25], loading of siRNA into the CP-MSNP carrier is achieved by simple mixing. In the in vivo studies, we have demonstrated that siRNA accumulation inside the primary tumor is more profound than in most major organs. By loading the delivery system with siRNA oligos specific for the PKM2 gene, we have also shown effective knockdown of gene expression in vitro and in vivo, and subsequent inhibition of cancer cell growth, migration, and invasion. Taken together, this system has the potential for broad applications both in basic research and in clinical treatments.
The current study is the first step towards development of a powerful system for systemic delivery of siRNA therapeutics. Future efforts should be devoted to fully evaluation of particle biodistribution at the whole body level, such as using whole body imaging techniques, and functional validation on the therapeutic efficacy of siRNA.
Evaluation of migration, invasion, and cell viability in MDA-MB-231 cells following CP-MSNP/siRNA treatment. (A) Migration assay. Cells were fixed with crystal violet. (B) Scratch assay. (C) Invasion assay. Cells were fixed with crystal violet. (D) Quantitative analysis of migration and invasion results. The number of cells that had migrated across the transwell insert was measured by counting ten separate fields/well under a phase-contrast microscope. Percentage of cell number was presented as the average of measurements in three independent wells after normalized against the control. Data are presented as the mean of thirty measurements ± standard deviation. Final siRNA concentration: 50 nM. (E) MTS assay to measure cell viability following CP-MSNP/siRNA treatment for 72 h. Cell viability was quantified by measuring the absorbance at 490 nm, and normalized against the control.
In vivo evalution of CP-MSNP/siRNA tumor accumulation, biodistribution, and suppression of gene expression. (A) Intravital microscopic images captured one hour after administration of CP-MSNP/Alexa555. CP-MSNP/Alexa555 siRNA particles are shown in red, and tumor blood vessels are displayed in green (FITC-dextran). White arrows point to selected particles that have left the tumor vasculature. (B) Measurement of CP-MSNP/Alexa555 siRNA in tumor blood vessels at various time points by intravital microscopy. (C-E) Biodistribution of CP-MSNP/Alexa555 siRNA in tumor tissue and major organs. (C) Fluorescent intensity from tumor and major organs. Samples are presented as a: heart, b: liver, c: spleen, d: lung, e: kidney, and f: tumor. (D) Percentage of fluorescent intensity/sample and (E) percentage of fluorescent intensity/gram sample are presented. Percentage was calculated as fluorescent intensity in the sample/total combined intensity. (F) Western blot analysis on knockdown of PKM2 expression 5 days after treatment with CP-MSNP/siRNA (n = 3 mice/group).
Supplementary Material
Fig.S1.
Acknowledgements
The authors acknowledge financial support from Houston Methodist Research Institute, the National Natural Science Foundation of China (Nos., 21231007 and 21121061), the Ministry of Education of China (Nos., 20100171110013 and 313058), the National Basic Research Program of China (973 Program No. 2014CB845604), and the Fundamental Research Funds for the Central Universities.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806-11
2. Hannon GJ. RNA interference. Nature. 2002;418:244-51
3. McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature. 2002;418:38-9
4. Shen H, Mittal V, Ferrari M, Chang J. Delivery of gene silencing agents for breast cancer therapy. Breast Cancer Res. 2013;15:205
5. de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6:443-53
6. Kalluri JR, Arbneshi T, Khan SA, Neely A, Candice P, Varisli B. et al. Use of gold nanoparticles in a simple colorimetric and ultrasensitive dynamic light scattering assay: selective detection of arsenic in groundwater. Angew Chem Int Ed Engl. 2009;48:9668-71
7. Shen J, Xu R, Mai J, Kim HC, Guo X, Qin G. et al. High Capacity Nanoporous Silicon Carrier for Systemic Delivery of Gene Silencing Therapeutics. ACS Nano. 2013;7:9867-80
8. Xue HY, Wong HL. Solid lipid-PEI hybrid nanocarrier: an integrated approach to provide extended, targeted, and safer siRNA therapy of prostate cancer in an all-in-one manner. ACS Nano. 2011;5:7034-47
9. Shen H, Sun T, Fang X, Ferrari M. Nanovector delivery of siRNA for cancer therapy. Cancer Gene Ther. 2012;19:367-73
10. Tang F, Li L, Chen D. Mesoporous silica nanoparticles: synthesis, biocompatibility and drug delivery. Adv Mater. 2012;24:1504-34
11. Li Z, Barnes JC, Bosoy A, Stoddart JF, Zink JI. Mesoporous silica nanoparticles in biomedical applications. Chem Soc Rev. 2012;41:2590-605
12. Yu MK, Park J, Jon S. Targeting strategies for multifunctional nanoparticles in cancer imaging and therapy. Theranostics. 2012;2:3-44
13. Cho Y, Borgens RB. The preparation of polypyrrole surfaces in the presence of mesoporous silica nanoparticles and their biomedical applications. Nanotechnology. 2010;21:205102
14. Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D. et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685-96
15. Pun SH, Bellocq NC, Liu A, Jensen G, Machemer T, Quijano E. et al. Cyclodextrin-modified polyethylenimine polymers for gene delivery. Bioconjug Chem. 2004;15:831-40
16. Radu DR, Lai CY, Jeftinija K, Rowe EW, Jeftinija S, Lin VS. A polyamidoamine dendrimer-capped mesoporous silica nanosphere-based gene transfection reagent. J Am Chem Soc. 2004;126:13216-7
17. Liu W, Liu J, Yang X, Wang K, Wang Q, Yang M. et al. pH and ion strength modulated ionic species loading in mesoporous silica nanoparticles. Nanotechnology. 2013;24:415501
18. Dominska M, Dykxhoorn DM. Breaking down the barriers: siRNA delivery and endosome escape. J Cell Sci. 2010;123:1183-9
19. Zintchenko A, Philipp A, Dehshahri A, Wagner E. Simple modifications of branched PEI lead to highly efficient siRNA carriers with low toxicity. Bioconjug Chem. 2008;19:1448-55
20. Thomas M, Klibanov AM. Conjugation to gold nanoparticles enhances polyethylenimine's transfer of plasmid DNA into mammalian cells. Proc Natl Acad Sci U S A. 2003;100:9138-43
21. Yang XZ, Dou S, Wang YC, Long HY, Xiong MH, Mao CQ. et al. Single-step assembly of cationic lipid-polymer hybrid nanoparticles for systemic delivery of siRNA. ACS Nano. 2012;6:4955-65
22. Sonawane ND, Szoka FC Jr, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J Biol Chem. 2003;278:44826-31
23. Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell. 2008;13:472-82
24. Matsumura Y, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986;46:6387-92
25. Jeong J, Mok H, Oh Y-K, Park T. siRNA conjugate delivery systems. Bioconjugate Chem. 2009;20:5-14
Author contact
![]() Corresponding author: cesmzwsysu.edu.cn or hshenorg
Corresponding author: cesmzwsysu.edu.cn or hshenorg
 Global reach, higher impact
Global reach, higher impact