13.3
Impact Factor
Theranostics 2012; 2(12):1160-1173. doi:10.7150/thno.4492 This issue Cite
Research Paper
Generation of a Focused Poly(amino ether) Library: Polymer-mediated Transgene Delivery and Gold-Nanorod based Theranostic Systems
1. Chemical Engineering;
2. School of Biological and Health Systems Engineering, Arizona State University, Tempe, AZ 85287-6106, USA.
Received 2012-4-19; Accepted 2012-8-14; Published 2012-12-14
Abstract
A focused library of twenty-one cationic poly(amino ethers) was synthesized following ring-opening polymerization of two diglycidyl ethers by different oligoamines. The polymers were screened in parallel for plasmid DNA (pDNA) delivery, and transgene expression efficacies of individual polymers were compared to those of 25 kDa polyethylenimine (PEI), a current standard for polymer-mediated transgene delivery. Seven lead polymers that demonstrated higher transgene expression than PEI in pancreatic and prostate cancer cells lines were identified from the screen. All seven lead polymers showed highest transgene expression at a polymer:pDNA weight ratio of 5:1 in the MIA PaCa-2 pancreatic cancer cell line. Among the conditions studied, transgene expression efficacy correlated with minimal polymer cytotoxicity but not polyplex sizes. In addition, this study indicated that methylene spacing between amine centers in the monomers, amine content, and molecular weight of the polymers are all significant factors and should be considered when designing polymers for transgene delivery. A lead effective polymer was employed for coating gold nanorods, leading to theranostic nanoassemblies that possess combined transgene delivery and optical imaging capabilities, leading to potential theranostic systems.
Keywords: non viral gene delivery, gold nanorods, two photon imaging, combinatorial synthesis, polymer library.
INTRODUCTION
Viruses have been employed for transgene delivery to various cell lines, both in vitro 1-4 and in vivo 5,6, primarily due to high levels of expression observed with these delivery vectors. While the use of viral vectors can result in high transfection efficacies, concerns with high immunogenicity and insertional mutagenesis limit their repeated use in vivo 7. Non-viral vectors, including cationic polymers, have been explored and developed as safer alternatives to viral vectors due to the ease of synthesis, scale up, versatility, and in some cases, biocompatibility and biodegradability 8. In addition, cationic polymers have also been investigated to enhance viral transduction 2,9-11.
Cationic polymers bind negatively charged plasmid DNA (pDNA) forming polymer/pDNA complexes (polyplexes), which can interact with negatively charged cell membranes and other surface proteins. This interaction induces uptake of the polyplex into cell via endocytosis resulting in their delivery to intracellular endosomal compartments. Successful delivery vehicles are then able to escape endosomes, presumably in part due to endosomal buffering by cationic polymers12-14. A fraction of plasmid DNA molecules in the cytoplasm then enter the nucleus where they are transcribed leading to translation in the cytoplasm, and ultimately expression of the transgene. Polyethylenimime (PEI) 14-22, poly amido-amines 23,24, and chitosan 25,26 are among the commonly employed cationic polymers for plasmid DNA delivery. Modification of PEI with moieties such as polyethylene glycol (PEG), resulted in lower cytotoxicity, and in some cases, enhanced transgene expression27-30. However, the low efficacies and high toxicities of conventionally employed polymers, particularly PEI, motivate the discovery of novel polymers with higher efficacies.
We have employed ring-opening polymerization between diglycidyl ethers (diepoxides) and polyamines 31-34 for the formation of combinatorial polymer libraries for transgene delivery35,36. Recently, it was found that a polymer 1,4 C-1,4 Bis, generated in our laboratory from 1,4-cyclohexanedimethanol diglycidyl ether (1,4C) and 1,4-bis(3-aminopropyl) piperazine) (1,4Bis) monomers demonstrated higher transgene expression than 25 kDa PEI at certain polymer:DNA weight ratios 36,37. In the current study, we sought to build on this finding and investigate a focused polymer library built around the monomers of the previously identified 1,4C-1,4Bis polymer. A library of twenty one poly(amino ethers) or PAEs was synthesized and screened for transgene expression efficacy. From these studies, seven new lead polymers that demonstrated higher transgene expression efficacies than PEI were identified, and characterized. It was found that methylene spacing, molecular weight, and amine content correlated with successful polymer-mediated transgene delivery. Additionally, a lead polymer was interfaced with gold nanorods (GNRs) and the cellular uptake of the resulting polymer-gold nanorod assemblies was imaged using two-photon microscopy. Our results demonstrate that both, transgene delivery and two-photon imaging are simultaneously possible with lead polymer-GNR assemblies, which is indicative of their theranostic potential.
EXPERIMENTAL
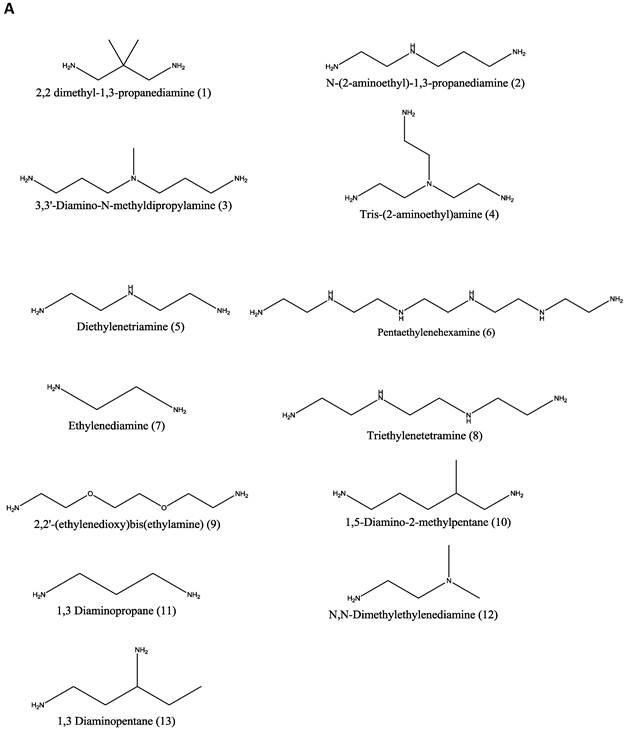
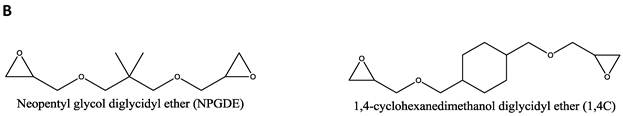
Materials. Thirteen amines (Figure 1A); 2,2 dimethyl-1,3-propanediamine (1), N-(2-aminoethyl)-1,3-propanediamine (2), 3,3'-diamino-N-methyldipropylamine (3), Tris-(2-aminoethyl)amine (4), diethylenetriamine (5), pentaethylenehexamine (6), ethylenediamine (7), triethylenetetramine (8), 2,2'-(ethylenedioxy)bis(ethylamine) (9), 1,5-diamino-2-methylpentane (10), 1,3 diaminopropane (11), N,N-dimethylethylenediamine (12), and 1,3 diaminopentane (13) were obtained from Sigma-Aldrich (St. Louis, MO) and used as received without any further modification. Two diglycidyl ethers (Figure 1B), neopentyl glycol diglycidyl ether (NPGDE) and 1,4-cyclohexanedimethanol diglycidyl ether (1,4C), were also both obtained from Sigma-Aldrich and used as received without any further modification. The control polymer, branched polyethyleneimine (MW ~ 25 kDa, Mn ~ 10kDa) was also obtained from Sigma-Aldrich. Ninhydrin reagent, used for assaying reactive primary and secondary amines, was purchased from Sigma-Aldrich. Luciferase and BCA protein assay kits were purchased from Promega Corporation (Madison, WI) and Thermo Fisher Scientific Inc. (Rockford, IL) respectively. The pGL3 control vector was also purchased from Promega Corporation.
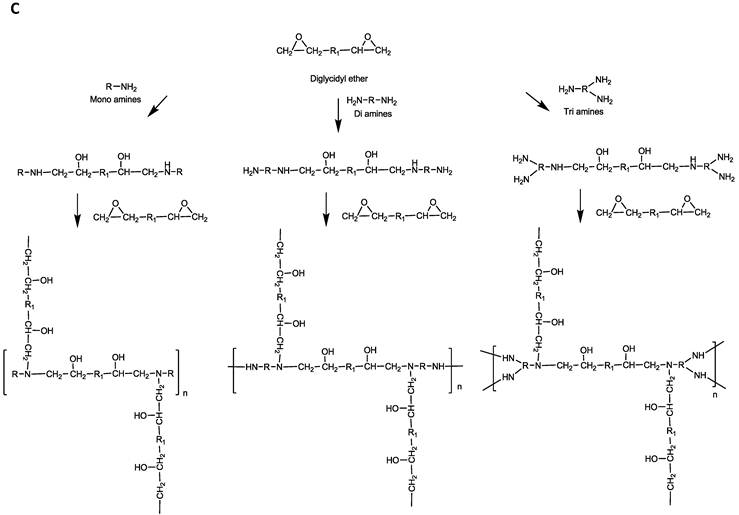
Parallel polymer synthesis. The focused polymer library was generated using a parallel synthesis approach. The library of 26 polymers was synthesized using the two diglycidyl ethers, 1,4C and NPGDE, and the amines listed above (1-13) using ring-opening polymerization (ROP) synthesis 24 resulting in 13 polymers for each diglycidyl ether. Figure 1 shows the structures of all of the monomers used in the synthesis of these polymers, as well as the schematic of the polymerization reaction. The sub-libraries will be referred to henceforth as the NPGDE library and 1,4C library, labeled as such based on the diglycidyl ether used to generate the polymers. Additionally, each polymer was numbered according to the numbering system given to the amines as described in the previous section and shown in Figure 1.
The diglycidyl ether (1,4C or NPGDE) and the amine were mixed in a 1:1 molar ratio in a 4 mL scintillation vial (Wheaton) and incubated at 22°C - 25°C for 16 hours. The volumes for the diglycidyl ethers used were 269.5 µl of 1,4C and 240 µl of NPGDE. For the amines, 138.4, 249.3, 186.1, 173.0, 124.7, 282.4, 77.4, 171.9, 168.6, 155.1, 98.9, 126.1, and 138.0 µl were used of the amines designated as 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13 respectively. After 16 hours, the resulting polymer was collected from the vial and dissolved in 1X PBS (10 mM Na2HPO4, 140 mM NaCl, 27 mM KCl, pH 7.4) to form a polymer solution at a concentration of 10 mg/mL. The pH of the solution was adjusted to 7.4 using 6N hydrochloric acid to compensate for the alkalinity of the cationic polymers. The degree of polymerization was determined by quantifying the amine content using the ninhydrin assay 16. Briefly, a standard curve was generated using glycine with varying amine concentrations (0 μM - 250 μM, total volume of 200 μL for each standard). A 1 mL sample of 5 mg/ml of the resulting polymer was diluted 50X to a total volume of 200 μL. The ninhydrin reagent (100 μL) was added to each of the standards and samples and the absorbance was read using a plate reader (Bio-Tek Synergy 2) at a wavelength of 570 nm to quantify the amine concentration of the polymer. Only those polymers that were soluble in 1X PBS prior to pH adjustment after the 16 hour reaction were employed in transfection studies, in order to maintain the parallel nature of synthesis and screening.
Amine monomers (A) and diglycidyl ether monomers (B) used in the synthesis of the NPGDE and 1,4C libraries. The polymers were numbered according to the amines as labeled above and were named according to the diglycidyl ether used. (C) Schematic of the polymerization reaction.
Cell culture. PC-3 cells (prostate cancer cell line) were obtained from the American Type Culture Collection (Manassas, VA), PC-3 PSMA cells (prostate cancer cell line) were obtained from Dr. Michael Sadelain from the Memorial Sloan-Kettering Cancer center (New York, NY), and MIA PaCa-2 cells (human pancreatic cancer cell line) were obtained as part of a collaboration with Dr. Haiyong Han from the Translational Genomics Research Institute (TGEN, Phoenix, AZ). All cells were cultured in serum-containing media (RPMI 1640, 10% FBS, 1000 units/mL penicillin, and 1000 μg/mL streptomycin) and were grown in an incubator under humidified air containing 5% CO2 at 37°C.
Plasmid DNA. DH5α E.coli cells were transformed with the pGL3 control vector, which encodes for the luciferase protein, under control of the SV40 promoter. The E.coli were cultured in 150 mL of terrific broth (MP Biomedicals, Solon, OH) containing 100 mg/mL ampicillin, overnight in an incubator (200 RPM, 37°C) for 14 hours. Plasmid DNA were isolated according to the QIAGEN plasmid purification maxi kit protocol (QIAGEN Inc., Valencia, CA). DNA concentration and purity was determined by measuring optical density (OD260 and OD280) using the NanoDrop spectrophotometer (ND-1000, NanoDrop Technologies). Only DNA within the acceptable absorbance range of 1.8-2 of the 260 nm:280 nm wavelength ratio were used in the transfection experiments. Plasmid DNA were stored at -20 °C until use.
Polymer-mediated Transgene Expression. PC-3, PC-3 PSMA, and MIA PaCa-2 cells were transfected using DNA/polymer complexes (polyplexes) consisting of pGL3 plasmid DNA and the synthesized polymers. Prior to transfection, cells were seeded at a density of 50,000 cells/well in 500 μl serum-containing medium in a 24-well plate and allowed to attach for 24 hours. Plasmid DNA encoding the luciferase protein, pGL3, was diluted to a concentration of 50 ng/μL using Tris EDTA buffer (10 mM Trizma and 1mM EDTA, Thermo Fisher Scientific, Rockford, IL). Polymer solutions were prepared and diluted with 1X PBS to a concentration of 500 ng/μL and filter-sterilized using a 0.22 μm filter. Polyplexes for transfection were prepared by the drop wise addition of 2000 ng (4 μL) of each polymer on top of 200 ng (4 μL) of pGL3 plasmid DNA to form a polymer:DNA weight ratio 10:1. Complexes were incubated for 20 minutes at room temperature prior to use. Cells were then treated with the polyplexes in serum-free medium (RPMI 1640, 1000 units/mL penicillinand 1000 μg/mL streptomycin) for 6 hours in an incubator under humidified air containing 5% CO2 at 37°C. Subsequently, the medium was replaced with fresh serum-containing medium and allowed to incubate for an additional 48 hours. Luciferase protein expression, expressed in terms of relative luminescence units or RLU, was determined using the luciferase assay kit according to the manufacturer's protocol 48 hours after transfection; luminescence measurements were carried out using a plate reader. The protein content (mg protein) in each well was determined using the BCA Protein Assay Kit. Transgene expression in all cell lines was calculated and normalized with the protein content and expressed as RLU per milligram (mg) protein (RLU/mg protein).
Purification of lead polymers. Screening studies were carried out using as-synthesized polymers. Purification of the lead polymers identified from the screening process was carried out by dialysis using a 1000 Da molecular weight cut off (MWCO) membrane. The polymers were dialyzed against Nanopure water for 48 hours and fresh water was replaced every 12 hours. After 48 hours of purification, the polymer solutions were lyophilized, reconstituted in 1X PBS, and filtered with a 0.22 μm syringe filter before use in transfection experiments.
Molecular weight measurements. Molecular weights (MWs) of polymers were measured by gel permeation chromatography (GPC) system (Waters 1515) with a refractive index detector (Waters 2414) using an ultrahydrogel column (Waters linear) at a flow rate of 1 ml/min at 35°C. Water containing 0.1% trifluoroacetic acid and 40% acetonitrile was used as a mobile phase. Poly (2-vinylpyridine) samples of different molecular weights (MW: 3000; 7,000; 12,000; 35,000; 70,000) were used as standards.
Dose response using lead polymers. Dose response experiments were carried out using the same method as described previously. The lead polymers were identified as those with higher transfection than PEI across all cell lines from each library (NPGDE and 1,4C). These leads were purified according to the dialysis method in the previous section, and used in subsequent treatments of the cells at different polymer:pDNA weight ratios. The weight ratios used in these studies were 1:1, 5:1, 10:1, 20:1, 25:1, and 50:1 corresponding to 200 ng, 1000 ng, 2000 ng, 4000 ng, 5000 ng, and 10,000 ng of the lead polymers respectively. The amount of plasmid DNA was kept constant at 200 ng (4 μL of 50 ng/μL) in all cases.
Polyplex Cytotoxicity. MIA PaCa-2 cells were seeded in 24 well plates at a density of 50,000 cells/well and allowed to attach overnight in a 37oC, 5% CO2 incubator. Polyplexes were formed as previously described. Cells were treated with polyplexes in serum-free media for 6 hours in an incubator under humidified air containing 5% CO2 at 37°C. Subsequently, the medium was replaced with fresh serum-containing medium. Following the 6 hour incubation, cell viability was determined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) cell proliferation assay kit (ATCC). This assay involves the enzymatic conversion of the MTT substrate to purple-colored formazan in metabolically active cells. This activity is widely employed as an indicator of cell viability and proliferation38; loss of metabolic activity was used as an indirect indicator of loss of cell viability upon polyplex treatment. Following addition of the MTT reagent (2 h at 37 °C), cells were treated with a lysis buffer from the kit and kept at room temperature in the dark for 2 h in order to lyse cells and solubilize the MTT product. The absorbance of each well was measured using a plate reader (BioTek Synergy 2) at 570 nm to assay for the blue MTT product. For data analysis, absorbance readouts were normalized to the live (untreated) and dead (5 µL of 30% hydrogen peroxide treated) controls.
Gold Nanorod (GNR) Synthesis. The seed-mediated method 39, using cetyl trimethyl ammonium bromide (CTAB) as the templating surfactant, was used for the synthesis of gold nanorods (CTAB-GNRs). Briefly, a seed solution was prepared by adding 5 ml of 0.2 M CTAB to 5 ml of 0.0005 M auric acid HAuCl4∙3H2O The addition of 0.6 ml of iced water-cooled 0.01 M sodium borohydride was used to reduce the solution. The growth solution was prepared by adding 5 ml of 0.001 M auric acid to 5 ml of 0.2 M CTAB containing 250 µl of 0.004 M silver nitrate. The growth solution was reduced by the addition of 70 µl of 0.0788 M L-ascorbic acid. Seed solution (12 µl) was added to the growth solution and continuously stirred for four hours at 28-30oC in order to allow for the generation of the gold nanorods. Modulating the silver nitrate concentration in the growth solution resulted in the formation of CTAB-GNRs, with absorbance maxima at different wavelengths between 750-900 nm.
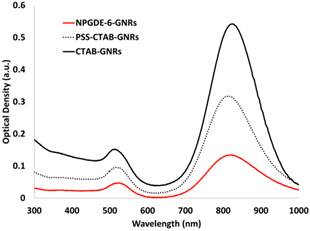
Generation of NPGDE-6-functionalized Gold Nanorod (NPGDE-6-GNR) Assemblies as Potential Theranostic Systems. Dispersions of GNRs with an optical density of 0.5 in 1.5 mL microcentrifuge tubes were centrifuged at 6000 rcf for 10 minutes using a Microfuge 18 centrifuge (Beckman Coulter) in order to remove excess CTAB surfactant. The supernatant was removed and the GNRs were redispersed in 100 µL of a poly(styrene sulfonate) (17kDa) (PSS) solution (10 mg/mL in 0.01X PBS; ~1.5 mM salt concentration). This dispersion was immediately sonicated for 30 minutes to allow for the formation of PSS-coated GNRs (PSS-CTAB-GNRs). Excess PSS was removed by centrifugation at 6000 rcf for 10 minutes. PSS-CTAB-GNRs were then redispersed in 300 µL of nanopure water and 200 µL of NPGDE-6 (10 mg/mL in 0.01X PBS) was added to these dispersions, which were immediately sonicated for 30 min to allow for the formation of the NPGDE-6-GNRs. CTAB-GNRs, PSS-CTAB-GNRs, and NPGDE-6-GNRs were characterized for their UV-Vis absorption spectra from 300-999 nm using a Bio-Tek Synergy 2 plate reader.
Determination of Hydrodynamic Diameters of Polyplexes and NPGDE-6-GNR Assemblies. Polyplexes and GNR assemblies were formed as previously described. The hydrodynamic diameter of polyplexes and different GNR-based assemblies was determined via dynamic light scattering (DLS) using a particle sizer (Corrvus Advanced Optical Instruments). The hydrodynamic diameter was reported in nanometers (nm).
Determination of Zeta Potential of GNR Assemblies. GNR assemblies were synthesized as previously described and zeta potentials of individual assemblies were determined using a Delsa™Nano Submicron Particle Size and Zeta Potential Particle Analyzer (Beckman Coulter). Zeta Potential values are reported in mV.
NPGDE-6-GNR Mediated Transgene Expression. MIA PaCa-2 cells were transfected using DNA-NPGDE-6-GNR nanoassemblies, which consist of pGL3 plasmid DNA loaded on NPGDE-6 functionalized GNRs. Prior to transfection, cells were seeded at a density of 50,000 cells/well in 500 μL serum-containing medium in a 24-well plate and allowed to attach for 24 hours. Sub-toxic amounts of PAE-GNRs loaded with between 10-500 ng of pGL3 plasmid DNA were added to each well in the presence of serum-free media for 6 h. The media was then replaced with serum-containing media for 48 h following which, cells were permeabilized with 150 µL cell lysis buffer (Promega, Madison, WI). The luciferase activity in cell lysates was measured using a luciferase assay kit (Promega, Madison, WI) in a plate reader (BioTek Synergy 2). The relative light unit (RLU) readouts from the assay were normalized to protein amount in milligrams in the respective cell lysates. The protein content was measured using the Pierce BCA Protein Assay Kit (Pierce Biotechnology, Rockford, IL). Transgene expression efficacy was based on luciferase activity in cell lysates, which was expressed as relative light units (RLU) per milligram (mg) of protein or RLU / mg. The transfection experiments were performed at least in triplicate.
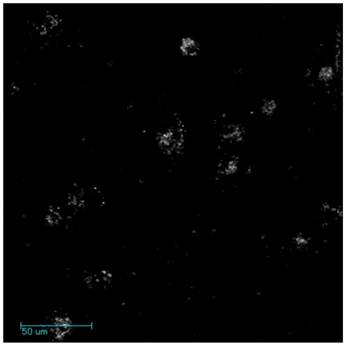
Multi-Photon Imaging of NPGDE-6-GNRs. MIA PaCa-2 cells were seeded at a density of 200,000 cells/well in 2 ml of serum-containing medium in a 6-well plate and allowed 24 hours for attachment. NPGDE-6-GNRs were dispersed in serum-free media at a final optical density of 0.00375 absorbance units. Cells were then treated with the NPGDE-6-GNR assemblies overnight after which serum-free medium was replaced with serum-containing medium. A femtosecond Ti:Sapphire laser (Spectra Physics, Mai Tai) set to a wavelength of 800 nm and 18 mW was used to generate the two photon luminescence of GNRs. Luminescence signals were acquired using an Ultima IV In Vivo Laser Scanning Microscope (Prairie Technologies) with a 63X water immersion lens. The same linear adjustments to brightness and contrast were applied to the entirety of each image using Axiovision processing software to highlight cellular uptake of polymer-coated gold nanorods. All data represented in the original image were presented in the modified image.
Statistical Analyses. All experiments were carried out in triplicate unless mentioned otherwise. Data are represented by the mean ± standard deviation. The significance between the control and each experimental test condition was determined using the two-tailed Student's t-test with p-values < 0.05 being considered statistically significant.
RESULTS AND DISCUSSION
Polymer synthesis. The focused polymer library was generated using the ring-opening polymerization between diglycidyl ethers and polyamines as described previously36. The kinetics of polymerization were followed by determining the change in reactive amine content at the beginning and end (16 h) of polymer synthesis; reactive amine content was determined using the ninhydrin assay 16,40. Of the twenty six polymers synthesized, eleven polymers from the NPGDE library (1-11) and ten polymers from the 1,4C library (1-3, 5-6, and 9-13) were soluble in 1X PBS, resulting in a total of 21 polymers that were used for screening transgene expression efficacies. It is possible that modified reaction conditions can be employed in order to overcome the extensive cross-linking that leads to insoluble polymers. However, we did not investigate this further in order to maintain the parallel nature of polymer synthesis.
Screening of NPGDE and 1,4C-based polymers libraries for transgene expression efficacy in MIA PaCa-2, PC-3, and PC3-PSMA cell lines. As-synthesized unpurified polymers were employed in a parallel screen to evaluate transgene expression efficacy in three different human cancer cell lines - MIA-PaCa-2 pancreatic cancer, PC-3, and PC-3 PSMA, both prostate cancer cells. All transfection screening experiments were carried out at a 10:1 polymer:pDNA weight ratio; the pGL3 plasmid, expressing luciferase protein was used in these studies.
Figures 2A and 2B shows the results from screening of the soluble NPGDE polymers and 1,4C polymers respectively. Those polymers that had significantly higher transgene expression compared to PEI (25 kDa, branched) across the three cell lines were identified as leads. Overall, four lead polymers from the NPGDE library and three lead polymers from the 1,4C library were identified from the screening process. From the NPGDE library, polymers based on monomers 3, 6, 10, and 11 corresponding to the amines 3,3'-diamino-N-methyldipropylamine, pentaethylenehexamine, 1,5-diamino-2-methylpentane, and 1,3 diaminopropane, respectively, were the lead candidates. Similarly from the 1,4 C library, polymers based on monomers 3,10, and 11 corresponding to the same amines 3,3'-diamino-N-methyldipropylamine, 1,5-diamino-2-methylpentane, and 1,3 diaminopropane, respectively, were the lead candidates. These seven polymers were employed in further investigations. Table 1 summarizes the results presented in Figures 2A and 2B and compares the efficacy of lead polymers with that of PEI across the three cell lines employed.
Luciferase transgene expression (RLU/mg protein) 48 hours after transfection using NPGDE based polymers (A) and 1,4C based polymers (B) in MIA PaCa-2, PC-3, and PC-3 PSMA cell lines. Only polymers that were soluble in 1X PBS after synthesis were used in this screen which was carried out at a polymer:pDNA weight ratio 10:1 corresponding to 2000 ng (4 μL of 500 ng/μL) of each polymer and 200 ng (4μL of 50 ng/μL) (n = 3 with the exception for 1,4C-3 (n = 2) and 1,4C-10 (n = 2), * = p < 0.05, ** = p < 0.01). The p-values were obtained by comparing data from each polymer to 25 kDa PEI. All negative luminescence values were not considered significant due to lack of physical meaning, and were not included in the calculations.
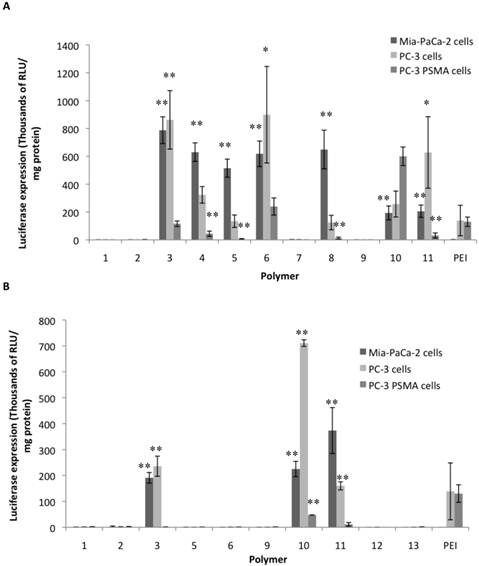
Both, NPGDE and 1,4C-based polymers showed the highest transgene expression in PC-3 cells compared to other cell lines; transgene expression was lowest in the PC3-PSMA cell line as shown in Figure 2. It is likely that the different intracellular trafficking of cargo in these closely related cells results in the observed differences in transgene expression37,41. However in relation with PEI, NPGDE and 1,4C-based polymers demonstrated highest relative efficacies in the MIA-PaCa-2 cell line (a maximum of 700-fold increase over PEI; Table 1), indicating that PEI is not an ideal candidate for transgene delivery to these cells. These polymers were moderately more effective than PEI in PC-3 cells (up to 7-fold increase; Table 1). However, these polymers did not transfect PC3 PSMA cells as well as PEI with the exception of NPGDE-6 and NPGDE-10, which showed 2-fold and 5-fold increase over PEI, respectively. While these results underscore the challenges associated with polymer-mediated transgene delivery in different cell lines, lead polymers from the NPGDE (3,6,10,11) and 1,4C (3,10,11) libraries demonstrated effectiveness across diverse cell lines.
Summary of results from the screening experiments (10:1 polymer:pDNA ratio corresponding to 2000 ng of each polymer and 200 ng of DNA) for identifying lead polymers shown in Figures 2A and 2B, with + and - indicating fold increase and fold decrease, respectively, as compared to 25 kDa PEI.
| Polymer | MIA-PaCa-2 | PC-3 | PC-3 PSMA |
|---|---|---|---|
| NPGDE-3 | + 700 | + 7 | Comparable |
| NPGDE-6 | + 550 | + 6 | + 2 |
| NPDGE-10 | + 170 | + 2 | + 5 |
| NPGDE-11 | + 180 | + 5 | - 4 |
| 1,4C-3 | + 170 | + 2 | - 125 |
| 1,4C-10 | + 200 | + 6 | - 3 |
| 1,4C-11 | + 320 | Comparable | - 11 |
Polymers based on 3,3'-diamino-N-methyldipropylamine (3), 1,5-diamino-2-methylpentane (10), and 1,3 diaminopropane (11) as amine monomers show among the highest levels of transgene (luciferase) expression in MIA PaCa-2 and PC-3 cells. Based on the structures of these three amines, it can be seen that the methylene spacing between amine groups may play a crucial role in transgene delivery. It was determined previously that DNA binding efficiency of polyamines is related to methylene spacing between the amine centers; in general, polyamines with three or four methylene spacing between amines showed higher DNA binding efficacy than those with two methylene spacing 42,43. Based on the results obtained from these experiments, this attribute also seems to enhance transgene expression since all three lead amines (3, 10, and 11) possess between three - five methylene spacing. These polymers also showed higher levels of transgene expression compared to PEI in most cases. This observation is further supported by the structures of other amine monomers including, Tris-(2-aminoethyl)amine (4), diethylenetriamine (5), pentaethylenehexamine (6), and triethylenetetramine (8), all of which have two methylene spacings between amine centers. Polymers based on these amines demonstrated lower levels of transgene expression when compared to the lead amines in most cases. Interestingly, NPGDE-based pentaethylenehexamine (6) polymers demonstrated appreciable transgene expression levels, but those based on 1,4C did not. These results indicate that amine monomers which contain between three and five methylene spacing between amine centers are likely critical in enhancing transgene expression efficacies of NPGDE and 1,4C based polymers, and may play a critical role in other polymeric systems as well. However, more rigorous experimental as well as structure-property relationship modeling approaches are required in order to further establish the validity of these empirical observations.
Dose response experiments using lead polymers. Lead polymers were subjected to further experimentation at different polymer:pDNA weight ratios (dose responses) using the same luciferase reporter gene, in order to identify conditions where they demonstrated highest transgene expression efficacies. Based on the results from the screening experiments, it was shown that the lead polymers from the NPGDE (3,6,10,11) and 1,4C (3,10,11) libraries performed most effectively in MIA PaCa-2 cells (Table 1). Thus, these cells were used for subsequent dose response experiments. All lead polymers were subjected to dialysis in order to remove any excess unreacted monomeric amines from the polymer solutions, since these could also bind plasmid DNA along with the polymers. These dialyzed polymers will henceforth be referred to as purified polymers. In order to keep all conditions consistent, PEI was also dialyzed in the same manner; the resulting polymer will also be referred to as purified PEI.
Figure 3 shows the transgene expression efficacies of the lead purified NPGDE polymers (Figure 3A) and 1,4C polymers (Figure 3B) at polymer:pDNA weight ratios of 1:1, 5:1, 10:1, 20:1, 25:1, and 50:1 along with purified PEI in MIA PaCa-2 cells. In most cases, dialysis led to a reduction in transgene expression efficacies. We verified that dialysis results in lower amine content in the polymers (determined using the ninhydrin assay; not shown) presumably due to removal of unreacted monomers or smaller oligomers. This lowering of amine content may also be a potential explanation for the difference in transfection efficacies between the unpurified polymers and the purified polymers both of which were from the same library and same stock.
PEI has been shown to be most effective at a polymer:pDNA weight ratio of 1:1 in several cases 15,37; all purified lead polymers demonstrated transgene expression efficacies that were comparable to that observed with purified PEI at 1:1 polymer: pDNA weight ratio. However, the highest levels of transgene expression occurred at a polymer:pDNA weight ratio of 5:1 for all lead polymers except for NPGDE-3. In particular, the NPGDE-6 polymer demonstrated 200-fold enhancement over that of purified PEI, and was selected as a lead candidate for demonstrating theranostic potential. Interestingly, purified PEI was not as effective as any of the NPGDE and 1,4C-based polymers at any other condition, suggesting that polymers from our library were more effective across a wide range of doses.
Luciferase gene expression (RLU/mg protein) from the dose response transfections using the purified NPGDE polymers (A) and 1,4C polymers (B) at different polymer:DNA weight ratios; 1:1, 5:1, 10:1, 20:1, 25:1, and 50:1 corresponding to 200 ng, 1000 ng, 2000 ng, 4000 ng, 5000 ng, and 10,000 ng of the lead polymers respectively with the plasmid DNA amount kept constant at 200 ng. Experiments were performed in the MIA PaCa-2 cell line (n = 3. * = p < 0.05, ** = p < 0.01). The p-values were obtained by comparing data from each polymer to 25 kDa PEI and apply to all polymers at the specified ratios. All negative luminescence values were not considered significant due to lack of physical meaning, and were not included in the calculations.
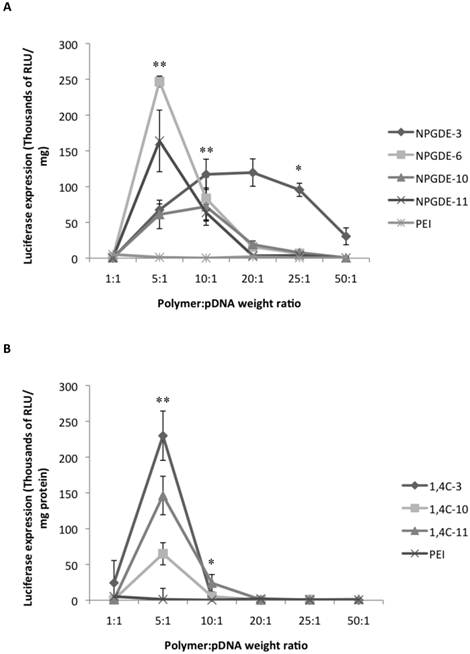
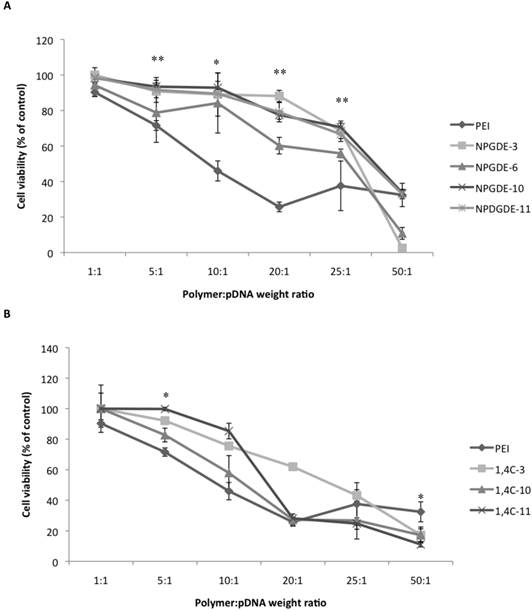
Transfection efficacy decreased at higher polymer:pDNA weight ratios for most polymers. This, in part, could be attributed to cytotoxicity of the polymers beyond weight ratios of 5:1 and 10:1 as seen in Figure 4. It is important to note that all polymers from our library, particularly those based on the NPGDE monomer, demonstrated significantly lower cytotoxicities towards MIA-Pa-Ca-2 cells compared to PEI. Cationic polymers can interact with negatively charged cell membranes, leading to their disruption, which can lead to cell death44-46. Thus, although higher polymer amounts may lead to higher amine content, excess positive charges on the polyplex surface, effective condensation of the DNA and promotion of endosomal escape, this can also lead to increasing toxicity. A balance between amine content and toxicity is therefore critical for designing effective polymers for transgene delivery.
Role of Polymer Molecular Weights
Table 2 shows the molecular weights of the purified lead polymers determined using gel permeation chromatography (GPC). It has been demonstrated previously that transgene expression of PEI increases with the molecular weight of the polymeric species47. However, cytotoxicity of PEI also increases with molecular weight 48, which necessitates an optimization between transgene expression efficacy and cytotoxicity. Most of the NPGDE- and 1,4C-based polymers demonstrated a similar trend at a polymer:pDNA ratio of 5:1, in that, they were minimally toxic to cells which showed >75% viability (Figure 4). While most NPGDE-based polymers demonstrated similar cytotoxicities, the NPGDE-6 polymer demonstrated the highest loss in cell viability (78%), presumably since it had the highest molecular weight among the library polymers. However, the polymer cytotoxicity did not correlate entirely with polymer molecular weight in all cases, as seen in case of 1,4C-based polymers (Table 2 and Figure 4). Despite its comparable molecular weight to that of 25kDa PEI (Table 2), the NPGDE-6 lead polymer demonstrated higher transgene expression efficacies (Figure 3) and lower cytotoxicities (Figure 4) compared to the current standard.
Effect of Polyplex Hydrodynamic Diameter
Table 2 shows the hydrodynamic diameters of the polyplexes at polymer:DNA weight ratios of 5:1, 10:1, and 25:1 using the lead polymers, as determined using dynamic light scattering studies. These ratios were chosen since polymer:DNA weight ratios of 5:1 and 10:1 showed among the highest transgene expression efficacies. A ratio of 25:1 was chosen since transgene expression is low at this ratio. Polyplex size was measured to be between 200-300 nm in most cases. Out of all the lead polymers, only NPGDE-6 formed a polyplex size that was statistically significant when compared to PEI at a polymer:DNA ratio of 5:1 (321 nm). While it is typically shown that transgene expression is most efficient when polyplex sizes are 200 nm or less 7,19,49,50, our results suggest that polyplexes of 300 nm or above (NPGDE-6), also result in transgene expression.
Hydrodynamic diameter ± standard deviations for all of the polyplexes formed using purified lead polymers and purified PEI at polymer:pDNA weight ratios of 5:1, 10:1, and 25:1 corresponding to 1000 ng, 2000 ng, and 5000 ng of each polymer respectively. The amount of plasmid DNA was kept constant at 200 ng. Molecular weights of the lead polymers Mn: Number averaged molecular weight, Mw: Weight-averaged molecular weight, PDI: polydispersity index. Values for PEI are those provided by the vendor. * = p < 0.05; the p-values were obtained by comparing data from each polymer to 25 kDa PEI.
| Polymer | Mw | Mn | PDI | 5:1 | 10:1 | 25:1 |
|---|---|---|---|---|---|---|
| NPGDE-3 | 10,798 | 4,240 | 2.5 | 230.3 nm ± 15.1 nm | 292.6 nm ± 37.1 nm | 268.3 nm ± 43.5 nm |
| NPGDE-6 | 28,452 | 5,038 | 5.6 | *320.6 nm ± 32.0 nm | 296.0 nm ± 29.3 nm | 255.0 nm ± 30.8 nm |
| NPGDE-10 | 4,959 | 2,314 | 2.1 | 239.3 nm ± 6.0 nm | 305.3 nm ± 47.5 nm | 266.7 nm ± 39.7 nm |
| NPGDE-11 | 6,289 | 2,900 | 2.2 | 221.0 nm ± 15.7 nm | 284.0 nm ± 40.0 nm | 263.0 nm ± 13.1 nm |
| 1,4,C-3 | 16,898 | 4,101 | 4.1 | 257.3 nm ± 4.0 nm | 271.3 nm ± 28.9 nm | 262.0 nm ± 34.4 nm |
| 1,4C-10 | 10,097 | 2,726 | 3.7 | 281.6 nm ± 30.4 nm | 283.3 nm ± 8.3 nm | 242.0 nm ± 8.9 nm |
| 1,4C-11 | 12,429 | 3,076 | 4.0 | 282 nm ± 20.2 nm | 286.0 nm ± 16.4 nm | 255.3 nm ± 11.1 nm |
| PEI | 25,000 | 10,000 | 2.5 | 250.3 nm ± 15.5 nm | 299.3 nm ± 16.6 nm | 252.0 nm ± 41.6 nm |
Cytotoxicity data obtained from the MTT assay using the purified NPGDE polymers (A) and 1,4C polymers (B) at different polymer:DNA weight ratios; 1:1, 5:1, 10:1, 20:1, 25:1, and 50:1, corresponding to 200 ng, 1000 ng, 2000 ng, 4000 ng, 5000 ng, and 10,000 ng of the lead polymers respectively, in the MIA PaCa-2 cell line (n = 3 * = p < 0.05 and ** = p < 0.01). The amount of plasmid DNA amount was kept constant at 200 ng. The p-values were obtained by comparing data from each polymer to 25 kDa PEI across all polymers at the specified ratios. Note that at a polymer:DNA weight ratio of 5:1, the comparison between NPDGE-6 and PEI cytotoxicities are not statistically significant.
Theranostic Potential of Gold Nanorods coated with the Lead NPGDE-6 Polymer: Simultaneous Transgene Delivery and Imaging
Visualization techniques were explored in order to demonstrate the theranostic potential of the lead cationic polymer for simultaneous imaging and transgene delivery. Previous results indicated that gold nanorod (GNR)-based assemblies exhibited poor stability in phosphate-buffered saline (PBS) and serum-free medium (SFM) compared to DI water and serum-containing media51. Coating GNRs with cationic poly(amino ethers) enhanced their stability in biologically relevant media 52. Additionally, this allows for loading nanoassemblies with plasmid DNA, which can then be delivered to cells, resulting in transgene expression, as previously demonstrated53. Similar strategies were followed for generating lead polymer-GNR assemblies; a layer-by-layer deposition approach was employed wherein positively charged CTAB-GNRs were first coated with negatively charged poly(styrene sulfonate) and then by NPGDE-6 via electrostatic interactions. Successful coating of CTAB-GNRs was confirmed via a shift in the maximum absorption peak of the GNR absorption spectra following deposition of the PSS layer and the subsequent NPGDE-6 layer. As seen in Figure 5, CTAB-GNRs have a maximum absorption at 825 nm, which then decreases to 814 nm following PSS coating and remains at 814 nm following NPGDE-6 coating. This blue shift in the absorption peak can be attributed to a change in the dielectric constant of the gold nanorods surroundings54. Additionally, changes in zeta potential and hydrodynamic diameters further confirm successful coating of the two polymer layers. Table 3 shows the change in zeta potential and hydrodynamic diameter of the GNR assemblies after deposition of each polymer layer. Initially, CTAB-GNRs were found to have a positive zeta potential due to the CTAB layer and a hydrodynamic diameter of approximately 34.5 nm. Following deposition of PSS, it is observed that the zeta potential becomes negative, due to the polyanionic nature of PSS. The hydrodynamic diameter also increase to approximately 46.4 nm, further confirming the PSS coating as indicated by an increase in size due to an additional polymer layer. The zeta potential then reverted to a positive value, following deposition of the cationic NPGDE-6 polymer. The size of the nanoassembly also increased, confirming deposition of the final poly(amino ether) layer. A small decrease in zeta potential was observed upon loading plasmid DNA on NPGDE-6-GNR assemblies, likely due to the negative nature of plasmid DNA; the hydrodynamic diameter of the assemblies also increased upon loading plasmid DNA. A final hydrodynamic diameter of less than 150 nm, and a positive zeta potential following plasmid DNA loading onto the NPGDE-6-GNRs, make this nanoassembly a suitable candidate for simultaneous transgene delivery and imaging.
Zeta potential and hydrodynamic diameter values (mean ± one standard deviation) for GNR assemblies following deposition of PSS and NPGDE-6 layers, and loading with 75 ng of pGL3 plasmid DNA.
| Zeta Potential (mV) | Hydrodynamic Diameter (nm) | |
|---|---|---|
| CTAB-GNRs | 31.1 ± 8.3 | 34.5 ± 2.4 |
| PSS-GNRs | -47.5 ± 14.1 | 46.4 ± 7.1 |
| NPGDE-6-GNRs | 39.3 ± 7.9 | 50.3 ± 4.4 |
| pGL3-NPGDE-6-GNRs | 35.1 ± 3.6 | 117.0 ± 10.4 |
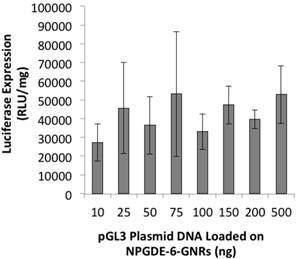
NPGDE-6-GNRs loaded with different amounts of plasmid DNA varying from 10-500 ng were employed for transgene expression studies. Transgene expression of the encoded luciferase gene was observed following treatment of MIA-PaCa-2 cells with different pGL3-NPGDE-6-GNRs (Figure 6). Similar expression of luciferase was observed across all conditions, although the expression values were less than those observed with highest NPGDE-6 polyplex-based luciferase expression (i.e. without the use of GNRs). Thus, GNRs coated with the NPGDE-6 polymer can be employed for transgene delivery and expression.
GNRs demonstrate highly efficient two-photon induced luminescence both in vitro and in vivo55. Furthermore, due to the facile control over their longitudinal plasmonic resonance in the near infrared (NIR) region, excitation wavelengths can be tuned to this region which can result in minimal tissue scattering and phototoxicity56,57. Thus, the synthesis of NPGDE-6-GNRs allows for a nanoscale assembly that can interact with cells, deliver transgene payload and be visualized using two-photon imaging. Figure 6 shows the GNR luminescence signal following NIR excitation at 800 nm. NPGDE-6-GNR assemblies were seen to enter cells which was indicative of cellular uptake of these delivery vehicles. Taken together, lead polymers from the screening could be interfaced with gold nanorods resulting in the formation of nanoscale assemblies that can be used for simultaneous non-viral transgene delivery and optical-based imaging.
CONCLUSIONS
We have generated and evaluated a new, focused library of poly(amino ethers) for transgene delivery. The library was generated by reacting polyamines with two diglycidyl ethers, NPGDE and 1,4C. Parallel screening of the polymer library for delivery of pGL3 plasmid DNA to three cancer cell lines resulted in the identification of four polymers from the NPGDE library and three polymers from the 1,4 C library, that showed significantly higher transgene expression compared to PEI at a 10:1 polymer:pDNA weight ratio. Based on the structures of the amine monomers, it was observed that methylene spacing between amines may play a key role in determining efficacy of transgene expression. Evaluation of different doses indicated that transgene expression was highest at a polymer:plasmid DNA ratio of 5:1. In general, poly(amino ether) leads identified in this investigation demonstrated higher efficacies and lower cytotoxicities than PEI across a wide range of doses. We also demonstrated that a lead polymer could be interfaced with gold nanorods resulting in the formation of nanoscale assemblies that can be employed for plasmid DNA delivery as well as for two-photon imaging of cells. These results indicate a route towards discovery and development of theranostic nanoassemblies which can be employed for simultaneous transgene delivery and imaging.
UV-vis absorption spectra for CTAB-GNRs, PSS-GNRs, and NPGDE-6-GNRs. Spectra were scaled for better comparison.
Luciferase transgene expression (RLU/mg protein) from NPGDE-6-GNRs loaded with different amounts of pGL3 plasmid DNA in the MIA PaCa-2 cell line (n = 3).
Representative microscopy images of two photon-induced luminescence of NPGDE-6-GNR assemblies in MIA PaCa-2 cells.
ACKNOWLEGEMENTS
The authors gratefully acknowledge financial support of the National Science Foundation (Grant CBET-082198), National Institutes of Health (Grant 1R01GM093229-01A1 and 5R21CA143505-02) and the Arizona Biomedical Research Commission (ABRC; Grant 1009). JR was partially supported by a fellowship from the Science Foundation of Arizona. The authors thank Dr. Yanqing Tian, Dr. Roger Johnson, and Professor Deirdre R. Meldrum, Center for Biosignatures Discovery Automation (CBDA), Biodesign Institute, Arizona State University for access to the GPC instrument for molecular weight determination.
COMPETING INTERESTS
The authors have declared that no competing interest exists.
References
1. Simeone DM, Cascarelli A, Logsdon CD. Adenoviral-mediated gene transfer of a constitutively active retinoblastoma gene inhibits human pancreatic tumor cell proliferation. Surgery. 1997;122:428-33
2. Kasman LM, Barua S, Lu P, Rege K, Voelkel-Johnson C. Polymer-Enhanced Adenoviral Transduction of CAR-Negative Bladder Cancer Cells. Molecular Pharmaceutics. 2009;6:1612-9
3. Park Y, Kang E, Kwon OJ. et al. Ionically crosslinked Ad/chitosan nanocomplexes processed by electrospinning for targeted cancer gene therapy. Journal of Controlled Release. 2010;148:75-82
4. Varga CM, Tedford NC, Thomas M, Klibanov AM, Griffith LG, Lauffenburger DA. Quantitative comparison of polyethylenimine formulations and adenoviral vectors in terms of intracellular gene delivery processes. Gene Therapy. 2005;12:1023-32
5. Jounaidi Y, Waxman DJ. Use of replication-conditional adenovirus as a helper system to enhance delivery of P450 prodrug-activation genes for cancer therapy. Cancer Research. 2004;64:292-303
6. Jounaidi Y, Chen CS, Veal GJ, Waxman DJ. Enhanced antitumor activity of P450 prodrug-based gene therapy using the low K-m cyclophosphamide 4-hydroxylase P4502B11. Molecular Cancer Therapeutics. 2006;5:541-55
7. Merdan T, Kopecek J, Kissel T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Advanced Drug Delivery Reviews. 2002;54:715-58
8. Elsabahy M, Nazarali A, Foldvari M. Non-Viral Nucleic Acid Delivery: Key Challenges and Future Directions. Current Drug Delivery. 2011;8:235-44
9. Han JF, Zhao D, Zhong ZR, Zhang ZR, Gong T, Sun X. Combination of adenovirus and cross-linked low molecular weight PEI improves efficiency of gene transduction. Nanotechnology. 2010;21:10
10. Fasbender A, Zabner J, Chillon M. et al. Complexes of adenovirus with polycationic polymers and cationic lipids increase the efficiency of gene transfer in vitro and in vivo. Journal of Biological Chemistry. 1997;272:6479-89
11. MeunierDurmort C, Grimal H, Sachs LM, Demeneix BA, Forest C. Adenovirus enhancement of polyethylenimine-mediated transfer of regulated genes in differentiated cells. Gene Therapy. 1997;4:808-14
12. Berthold PR, Shiraishi T, Nielsen PE. Cellular Delivery and Antisense Effects of Peptide Nucleic Acid Conjugated to Polyethyleneimine via Disulfide Linkers. Bioconjugate Chemistry. 2010;21:1933-8
13. Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. Journal of Gene Medicine. 2005;7:657-63
14. Boussif O, Lezoualch F, Zanta MA. et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in-vivo - polyethylenimine. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:7297-301
15. Cherng JY, Hung WC, Kao HC. Blending of Polyethylenimine with a Cationic Polyurethane Greatly Enhances Both DNA Delivery Efficacy and Reduces the Overall Cytotoxicity. Current Pharmaceutical Biotechnology. 2011;12:839-46
16. Gabrielson NP, Pack DW. Efficient polyethylenimine-mediated gene delivery proceeds via a caveolar pathway in HeLa cells. Journal of Controlled Release. 2009;136:54-61
17. Kang HC, Kang HJ, Bae YH. A reducible polycationic gene vector derived from thiolated low molecular weight branched polyethyleneimine linked by 2-iminothiolane. Biomaterials. 2011;32:1193-203
18. Mishra D, Kang HC, Bae YH. Reconstitutable charged polymeric (PLGA)(2)-b-PEI micelles for gene therapeutics delivery. Biomaterials. 2011;32:3845-54
19. Wang YH, Zheng M, Meng FH, Zhang J, Peng R, Zhong ZY. Branched Polyethylenimine Derivatives with Reductively Cleavable Periphery for Safe and Efficient In Vitro Gene Transfer. Biomacromolecules. 2011;12:1032-40
20. Lee M. Apoptosis induced by polyethylenimine/DNA complex in polymer mediated gene delivery. Bulletin of the Korean Chemical Society. 2007;28:95-8
21. Coll JL, Chollet P, Brambilla E, Desplanques D, Behr JP, Favrot M. In vivo delivery to tumors of DNA complexed with linear polyethylenimine. Human Gene Therapy. 1999;10:1659-66
22. Chumakova OV, Liopo AV, Andreev VG. et al. Composition of PLGA and PEI/DNA nanoparticles improves ultrasound-mediated gene delivery in solid tumors in vivo. Cancer Letters. 2008;261:215-25
23. Emilitri E, Ranucci E, Ferruti P. New poly(amidoamine)s containing disulfide linkages in their main chain. Journal of Polymer Science Part a-Polymer Chemistry. 2005;43:1404-16
24. Guo ST, Huang YY, Wei T. et al. Amphiphilic and biodegradable methoxy polyethylene glycol-block-(polycaprolactone-graft-poly(2-(dimethylamino)ethyl methacrylate)) as an effective gene carrier. Biomaterials. 2011;32:879-89
25. Jean M, Alameh M, Buschmann MD, Merzouki A. Effective and safe gene-based delivery of GLP-1 using chitosan/plasmid-DNA therapeutic nanocomplexes in an animal model of type 2 diabetes. Gene Therapy. 2011;18:807-16
26. Sarkar K, Srivastava R, Chatterji U, Kundu PP. Evaluation of Chitosan and Their Self-Assembled Nanoparticles with pDNA for the Application in Gene Therapy. Journal of Applied Polymer Science. 2011;121:2239-49
27. Zhang ZF, Yang CH, Duan YJ. et al. Poly(ethylene glycol) analogs grafted with low molecular weight poly(ethylene imine) as non-viral gene vectors. Acta Biomaterialia. 2010;6:2650-7
28. Chen XA, Zhang LJ, He ZJ. et al. Plasmid-encapsulated polyethylene glycol-grafted polyethylenimine nanoparticles for gene delivery into rat mesenchymal stem cells. International Journal of Nanomedicine. 2011;6:843-53
29. Peng J, Zou F, Liu L. et al. Preparation and characterization of PEG-PEI/Fe3O4 nano-magnetic fluid by co-precipitation method. Transactions of Nonferrous Metals Society of China. 2008;18:393-8
30. Zhuang BX, Zhang YD, Peng J. et al. Effect of Recombinant Plasmid pEGFP-AFP-hTNF on Liver Cancer Cells (HepG2 Cells) in vitro when Delivered by PEG-PEI/Fe3O4 Nanomagnetic Fluid. Journal of the Formosan Medical Association. 2011;110:326-35
31. Ghaemy M, Barghamadi M, Behmadi H. Cure kinetics of epoxy resin and aromatic diamines. Journal of Applied Polymer Science. 2004;94:1049-56
32. Rosu D, Mititelu A, Cascaval CN. Cure kinetics of a liquid-crystalline epoxy resin studied by non-isothermal data. Polymer Testing. 2004;23:209-15
33. Klee JE, Hagele K, Przybylski M. Metal-template synthesis of cyclic epoxide-amine oligomers: Uncrosslinked epoxide-amine addition polymers, 47. Journal of Polymer Science Part a-Polymer Chemistry. 2003;41:2047-52
34. Theis J, Ritter H. Formation of epoxide-amine oligo-adducts as OH-functionalized initiators for the ring-opening polymerization of epsilon-caprolactone. Beilstein Journal of Organic Chemistry. 2010;6:938-44
35. Barua S, Ramos J, Potta T. et al. Discovery of cationic polymers for non-viral gene delivery using combinatorial approaches. Comb Chem High Throughput Screen. 2011;14:908-24
36. Barua S, Joshi A, Banerjee A. et al. Parallel synthesis and screening of polymers for nonviral gene delivery. Mol Pharm. 2009;6:86-97
37. Barua S, Rege K. The influence of mediators of intracellular trafficking on transgene expression efficacy of polymer-plasmid DNA complexes. Biomaterials. 2010;31:5894-902
38. Hayon T, Dvilansky A, Shpilberg O, Nathan I. Appraisal of the MTT-based assay as a useful tool for predicting drug chemosensitivity in leukemia. Leuk Lymphoma. 2003;44:1957-62
39. Nikoobakht B, El-Sayed MA. Preparation and growth mechanism of gold nanorods (NRs) using seed-mediated growth method. Chemistry of Materials. 2003;15:1957-62
40. Friedman M. Applications of the ninhydrin reaction for analysis of amino acids, peptides, and proteins to agricultural and biomedical sciences. Journal of Agricultural and Food Chemistry. 2004;52:385-406
41. Barua S, Rege K. Cancer-cell-phenotype-dependent differential intracellular trafficking of unconjugated quantum dots. Small. 2009;5:370-6
42. Rege K, Ladiwala A, Hu SH, Breneman CM, Dordick JS, Cramer SM. Investigation of DNA-binding properties of an aminoglycoside-polyamine library using quantitative structure-activity relationship (QSAR) models. Journal of Chemical Information and Modeling. 2005;45:1854-63
43. van Dam L, Korolev N, Nordenskiold L. Polyamine-nucleic acid interactions and the effects on structure in oriented DNA fibers. Nucleic Acids Research. 2002;30:419-28
44. Choksakulnimitr S, Masuda S, Tokuda H, Takakura Y, Hashida M. In-vitro cytotoxicity of macromolecules in different cell - culture systems. Journal of Controlled Release. 1995;34:233-41
45. Lv HT, Zhang SB, Wang B, Cui SH, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. Journal of Controlled Release. 2006;114:100-9
46. Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: Implications for gene transfer/therapy. Molecular Therapy. 2005;11:990-5
47. Godbey WT, Wu KK, Mikos AG. Size matters: Molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. Journal of Biomedical Materials Research. 1999;45:268-75
48. Brunot C, Ponsonnet L, Lagneau C, Farge P, Picart C, Grosgogeat B. Cytotoxicity of polyethyleneimine (PEI), precursor base layer of polyelectrolyte multilayer films. Biomaterials. 2007;28:632-40
49. Akinc A, Lynn DM, Anderson DG, Langer R. Parallel synthesis and biophysical characterization of a degradable polymer library for gene delivery. Journal of the American Chemical Society. 2003;125:5316-23
50. Lai TC, Bae Y, Yoshida T, Kataoka K, Kwon GS. pH-Sensitive Multi-PEGylated Block Copolymer as a Bioresponsive pDNA Delivery Vector. Pharmaceutical Research. 2010;27:2260-73
51. Huang H-C, Barua S, Kay DB, Rege K. Simultaneous Enhancement of Photothermal Stability and Gene Delivery Efficacy of Gold Nanorods using Polyelectrolytes. ACS Nano. 2009;3:2941-52
52. Huang HC, Rege K, Heys JJ. Spatiotemporal temperature distribution and cancer cell death in response to extracellular hyperthermia induced by gold nanorods. ACS Nano. 2010;4:2892-900
53. Ramos J, Rege K. Transgene delivery using poly(amino ether)-gold nanorod assemblies. Biotechnol Bioeng. 2012;109:1336-46
54. Sivapalan ST, Vella JH, Yang TK. et al. Plasmonic Enhancement of the Two Photon Absorption Cross Section of an Organic Chromophore Using Polyelectrolyte-Coated Gold Nanorods. Langmuir. 2012;28:9147-54
55. Durr NJ, Larson T, Smith DK, Korgel BA, Sokolov K, Ben-Yakar A. Two-photon luminescence imaging of cancer cells using molecularly targeted gold nanorods. Nano Letters. 2007;7:941-5
56. Li J-L, Gu M. Surface plasmonic gold nanorods for enhanced two-photon microscopic imaging and apoptosis induction of cancer cells. Biomaterials. 2010;31:9492-8
57. Wang HF, Huff TB, Zweifel DA. et al. In vitro and in vivo two-photon luminescence imaging of single gold nanorods. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15752-6
Author contact
![]() Corresponding author: Kaushal Rege, Chemical Engineering, Arizona State University, 501 E. Tyler Mall, ECG 303, Tempe, AZ 85287-6106, Email: kregeedu, Phone: 480-727-8616 Fax: 480-727-9321
Corresponding author: Kaushal Rege, Chemical Engineering, Arizona State University, 501 E. Tyler Mall, ECG 303, Tempe, AZ 85287-6106, Email: kregeedu, Phone: 480-727-8616 Fax: 480-727-9321
 Global reach, higher impact
Global reach, higher impact