13.3
Impact Factor
Theranostics 2011; 1:322-340. doi:10.7150/thno/v01p0322 This volume Cite
Research Paper
Evaluation of 111In-Labeled Cyclic RGD Peptides: Effects of Peptide and Linker Multiplicity on Their Tumor Uptake, Excretion Kinetics and Metabolic Stability
1. School of Health Sciences, Purdue University, IN 47907, USA
2. Medical Isotopes Research Center, Peking University, Beijing 100083, China
Received 2011-6-30; Accepted 2011-7-18; Published 2011-7-25
Abstract
Purpose: The purpose of this study was to demonstrate the valence of cyclic RGD peptides, P-RGD (PEG4-c(RGDfK): PEG4 = 15-amino-4,710,13-tetraoxapentadecanoic acid), P-RGD2 (PEG4-E[c(RGDfK)]2, 2P-RGD4 (E{PEG4-E[c(RGDfK)]2}2, 2P4G-RGD4 (E{PEG4-E[G3-c(RGDfK)]2}2: G3 = Gly-Gly-Gly) and 6P-RGD4 (E{PEG4-E[PEG4-c(RGDfK)]2}2) in binding to integrin αvβ3, and to assess the impact of peptide and linker multiplicity on biodistribution properties, excretion kinetics and metabolic stability of their corresponding 111In radiotracers.
Methods: Five new RGD peptide conjugates (DOTA-P-RGD (DOTA =1,4,7,10-tetraazacyclododecane-1,4,7,10-tetracetic acid), DOTA-P-RGD2, DOTA-2P-RGD4, DOTA-2P4G-RGD4, DOTA-6P-RGD4), and their 111In complexes were prepared. The integrin αvβ3 binding affinity of cyclic RGD conjugates were determined by a competitive displacement assay against 125I-c(RGDyK) bound to U87MG human glioma cells. Biodistribution, planar imaging and metabolism studies were performed in athymic nude mice bearing U87MG human glioma xenografts.
Results: The integrin αvβ3 binding affinity of RGD conjugates follows the order of: DOTA-6P-RGD4 (IC50 = 0.3 ± 0.1 nM) ~ DOTA-2P4G-RGD4 (IC50 = 0.2 ± 0.1 nM) ~ DOTA-2P-RGD4 (IC50 = 0.5 ± 0.1 nM) > DOTA-3P-RGD2 (DOTA-PEG4-E[PEG4-c(RGDfK)]2: IC50 = 1.5 ± 0.2 nM) > DOTA-P-RGD2 (IC50 = 5.0 ± 1.0 nM) >> DOTA-P-RGD (IC50 = 44.3 ± 3.5 nM) ~ c(RGDfK) (IC50 = 49.9 ± 5.5 nM) >> DOTA-6P-RGK4 (IC50 = 437 ± 35 nM). The fact that DOTA-6P-RGK4 had much lower integrin αvβ3 binding affinity than DOTA-6P-RGD4 suggests that the binding of DOTA-6P-RGD4 to integrin αvβ3 is RGD-specific. This conclusion is consistent with the lower tumor uptake for 111In(DOTA-6P-RGK4) than that for 111In(DOTA-6P-RGD4). It was also found that the G3 and PEG4 linkers between RGD motifs have a significant impact on the integrin αvβ3-targeting capability, biodistribution characteristics, excretion kinetics and metabolic stability of 111In-labeled cyclic RGD peptides.
Conclusion: On the basis of their integrin αvβ3 binding affinity and tumor uptake of their corresponding 111In radiotracers, it was conclude that 2P-RGD4, 2P4G-RGD4 and 6P-RGD4 are most likely bivalent in binding to integrin αvβ3, and extra RGD motifs might contribute to the long tumor retention times of 111In(DOTA-2P-RGD4), 111In(DOTA-2P4G-RGD4) and 111In(DOTA-6P-RGD4) than that of 111In(DOTA-3P-RGD3) at 72 h p.i. Among the 111In-labeled cyclic RGD tetramers evaluated in the glioma model, 111In(DOTA-2P4G-RGD4) has very high tumor uptake with the best tumor/kidney and tumor/liver ratios, suggesting that 90Y(DOTA-2P4G-RGD4) and 177Lu(DOTA-2P4G-RGD4) might have the potential for targeted radiotherapy of integrin αvβ3-positive tumors.
Keywords: integrin αvβ3, 111In-labeled cyclic RGD peptides, tumor imaging
Introduction
Over last several years, many multimeric cyclic RGD peptides, such as E[c(RGDfK)]2 (RGD2) and E{E[c(RGDfK)]2}2 (RGD4), have been used to develop the integrin αvβ3-targeted radiotracers for noninvasive imaging of rapidly growing and metastatic tumors [1-14]. Multiple cyclic RGD moieties are utilized to maximize their integrin αvβ3 binding affinity and the radiotracer tumor uptake [9, 15-46]. It was found that the radiolabeled (99mTc, 18F, 64Cu, 68Ga and 111In) cyclic RGD tetramers have better tumor-targeting capability as evidenced by their higher tumor uptake with longer tumor retention time than their corresponding dimeric and monomeric counterparts [20, 22, 29, 32, 34, 35, 44, 46]. It was also found that cyclic RGD tetramers, such as DOTA-RGD4 (DOTA = 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetracetic acid) and DOTA-E{G3-E[G3-c(RGDfK)]2}2 (Figure 1: DOTA-6G-RGD2) are likely bivalent in binding to integrin αvβ3 even though they contain four identical RGD motifs [46]. The key for bivalency is the distance between two RGD motifs [36-38]. For example, RGD2 was monovalent whereas PEG4-E[PEG4-c(RGDfK)]2 (3P-RGD2: PEG4 = 15-amino-4,7,10,13-tetraoxapentadecanoic acid) and G3-E[G3-c(RGDfK)]2 (3G-RGD2: G3 = Gly-Gly-Gly) are bivalent because of the increased distance between two cyclic RGD motifs for simultaneous integrin αvβ3 binding [38, 45].
To further prove the “bivalency concept”, we prepared three new cyclic RGD peptide tetramers (Figure 1: 2P-RGD4, 2P4G-RGD4 and 6P-RGD4). We were particularly interested in these three peptides because they contain different number of G3 and PEG4 linkers between cyclic RGD motifs. The main objective of this study was to demonstrate their valence in binding to integrin αvβ3, and to assess the impact of cyclic RGD peptide and linker multiplicity on biodistribution properties, excretion kinetics and metabolic stability of 111In radiotracers. In this report, we present the synthesis and biological evaluation of RGD tetramer conjugates, DOTA-2P-RGD4, DOTA-2P4G-RGD4 and DOTA-6P-RGD4, and their 111In complexes. A whole-cell competitive displacement assay was used to determine their integrin αvβ3 binding affinities against 125I-c(RGDyK) bound to U87MG human glioma cells. Biodistribution and planar imaging studies performed in the athymic nude mice bearing U87MG glioma xenografts. For comparison purposes, we prepared and evaluated conjugates DOTA-PEG4-c(RGDfK) (DOTA-P-RGD), DOTA-PEG4-E[c(RGDfK)]2 (DOTA-P-RGD2), and their 111In radiotracers using the same in vitro and in vivo assays. In addition, we prepared a “scrambled” peptide conjugate DOTA-E{PEG4-E[PEG4-c(RGKfD)]2}2 (DOTA-6P-RGK4), which has the same amino acid residues as those in DOTA-6P-RGD4 using c(RGKfD) motifs instead of c(RGDfK), and 111In(DOTA-6P-RGK4) to demonstrate the RGD-specificity of 111In radiotracers.
Materials and Methods
Materials and Instruments. Chemicals were purchased from Sigma/Aldrich (St. Louis, MO), and were used without purification. Cyclic peptides, PEG4-c(RGDfK) (P-RGD), PEG4-E[G3-c(RGDfK)]2 (P2G-RGD2), PEG4-E[PEG4-c(RGDfK)]2 (3P-RGD2) and PEG4-E[PEG4-c(RGKfD)]2 (3P-RGK2), were all custom-made by the Peptides International, Inc. (Louisville, KY). DOTA-OSu (1,4,7,10-tetraazacyclododecane-4,7,10-triacetic acid-1-acetate(N-hydroxysuccinamide)) was purchased from Macrocyclics (Dallas, TX). The Boc-protected glutamic acid N-hydroxysuccinamide ester (Boc-E(OSu)2) and PEG4-E[c(RGDfK)]2 (P-RGD2) were prepared according to the methods described in our previous reports [23-26]. Syntheses and biological evaluations of DOTA-3P-RGD2 and 111In(DOTA-3P-RGD2) were reported in our previous communication [45]. ESI (electron-spray ionization) mass spectral data were obtained in positive mode using a Finnigan LCQ classic mass spectrometer, School of Pharmacy, Purdue University. The MALDI (matrix assisted laser desorption ionizations) mass spectral data were collected using an Applied Biosystems Voyager DE PRO mass spectrometer (Framingham, MA), the Department of Chemistry, Purdue University. 111InCl3 was obtained from Perkin-Elmer Life Sciences (North Billerica, MA). The NH4OAc buffer for 111In-labeling studies was passed over a Chelex-100 column (1×15 cm) to minimize the trace metal contaminants.
HPLC Methods. The HPLC method (Method 1) used a LabAlliance HPLC system equipped with a UV/vis detector (λ=254 nm) and Zorbax C18 semi-prep column (9.4 mm x 250 mm, 100 Å pore size; Agilent Technologies, Santa Clara, CA). The flow rate was 2.5 mL/min with the gradient mobile phase going from 90% A (0.1% TFA in water) and 10% (0.1% TFA in acetonitrile) at 0 min to 20% B at 5 min and to 40% B at 20 min. The radio-HPLC method (Method 2) used the LabAlliance HPLC system equipped with a β-ram IN/US detector (Tampa, FL) and Zorbax C18 column (4.6 mm x 250 mm, 300 Å pore size; Agilent Technologies, Santa Clara, CA). The flow rate was 1 mL/min. The mobile phase was isocratic with 90% A (25 mM NH4OAc, pH = 6.8) and 10% B (acetonitrile) at 0 - 5 min, followed by a gradient mobile phase going from 10% B at 5 min to 20% B at 15 min and to 50% B at 20 min.
Cyclic RGD peptide tetramer conjugates: DOTA-6G-RGD4, DOTA-6P-RGD4, DOTA-2P-RGD4 and DOTA-2P4G-RGD4.
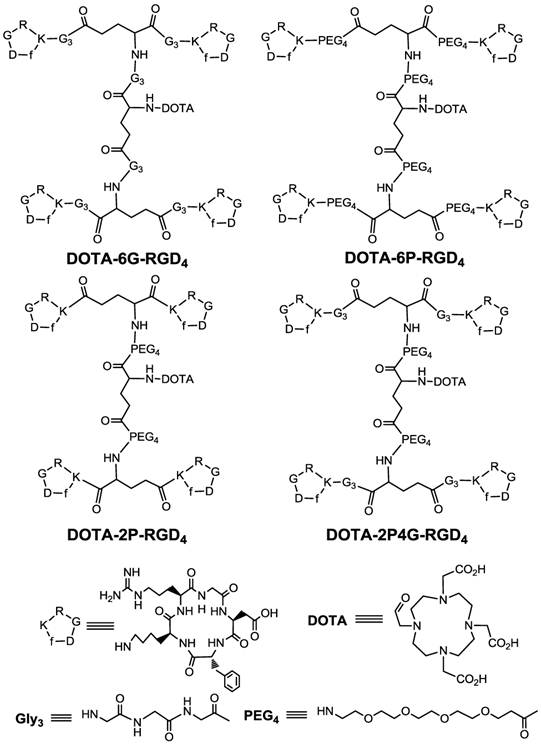
DOTA-PEG4-c(RGDfK) (DOTA-P-RGD). PEG4-c(RGDfK) (7.1 mg, ~8.3 μmol) and DOTA-OSu (5.5 mg, ~11.0 μmol) were dissolved in 1 mL of anhydrous dimethylformamide (DMF). The pH value in the reaction mixture was adjusted to 8.5 - 9.0 using diisopropylethylamine (DIEA). The mixture was then stirred overnight at room temperature. After addition of 3 mL of 100 mM NH4OAc buffer (pH = 7.0), the resulting solution was subjected to HPLC-purification (Method 1). The fraction at ~ 12.5 min was collected. Lyophilization of collected fractions afforded conjugate DOTA-P-RGD. The yield was 4.1 mg (~40%). ESI-MS (positive mode): m/z =1237.58 for [M + H]+ (M =1236.6 calcd. for C54H88N14O19).
DOTA-PEG4-[c(RGDfK)]2 (DOTA-P-RGD2). PEG4-[c(RGDfK)]2 (1.8 mg, 1.1 μmol) and DOTA-OSu (2.1 mg, 4.2 μmol) were dissolved in DMF (1 mL). The pH in the reaction mixture was adjusted to 8.5 - 9.0 using DIEA. The mixture was stirred for 2 h at room temperature. Upon addition of 2 mL NH4OAc buffer (100 mM, pH = 7.0), the product was separated by HPLC (Method 1). Lyophilization of the collected fractions at ~ 16.4 min afforded 0.9 mg of DOTA-P-RGD2 (~41%) with 95% HPLC purity. The yield was ESI-MS (positive mode) for m/z = 1951.5 for [M + H]+, 976.6 for [M + 2H]2+ and 1974.4 for [M + Na]+ (M = 1950.2 calcd. for [C86H134N24O28]+).
Boc-E{PEG4-E[c(RGDfK)]2}2 (Boc-2P-RGD4). To a solution of Boc-E(OSu)2 (1.1 mg, 2.5 μmol) in 1 mL of DMF were added PEG4-[c(RGDfK)]2 (5.8 mg, 3.4 μmol) and DIEA (2 drops). The reaction mixture was stirred for 2 h at room temperature. The reaction was terminated by adding 3 mL NH4OAc buffer (100 mM, pH = 7.0). The product was isolated by HPLC (Method 1). Lyophilization of collected fractions at 18.2 min afforded Boc-2P-RGD4. The yield was 4.5 mg (~79%). ESI-MS (positive mode): m/z = 3344.2 for [M + H]+ and 1671.9 for [M + 2H]2+ (M = 3342.7 calcd. for [C150H229N41O46]+).
E{PEG4-E[c(RGDfK)]2}2 (2P-RGD4). The Boc-2P-RGD4 (4.3 mg, 1.3 μmol) was dissolved in 2 mL of TFA. After standing at room temperature for 5 min, TFA was removed. The residue was dissolved in 2 mL of 0.1 M NH4OAc buffer (100 mM, pH = 7.0). The product was separated by HPLC (Method 1). Lyophilization of collected fractions at 15.7 min afforded 2P-RGD4. The yield was 2.6 mg (~59%). ESI-MS (positive mode): m/z = 3244.0 for [M + H]+ and 1620.5 for [M + 2H]2+ (M = 3242.6 calcd. for [C145H221N41O44]+).
DOTA-E{PEG4-E[c(RGDfK)]2}2 (DOTA-2P-RGD4). DOTA-OSu (3.3 mg, 6.5 μmol) and 2P-RGD4 (2.2 mg, 0.7 μmol) were dissolved in 1 mL of anhydrous DMF. After addition of DIEA (3 drops), the reaction mixture was stirred for 2 h at room temperature. Upon addition of 2 mL NH4OAc buffer (100 mM, pH = 7.0), the product was isolated by HPLC (Method 1). Lyophilization of collected fraction at 16.1 min afforded conjugate DOTA-2P-RGD4 with >95% HPLC purity. The yield was 1.4 mg (~56%). MALDI-MS: m/z = 3626.4 for [M + H]+ (M = 3626.8 calcd. for [C161H247N45O51]+).
Boc-E{PEG4-E[Gly3-c(RGDfK)]2}2 (Boc-2P4G-RGD4). To a solution of Boc-E(OSu)2 (3.8 mg, 8.6 μmol) in DMF (3 mL) were added P2G-RGD2 (20.9 mg, 11.0 μmol) and excess DIEA (3 drops). The mixture was stirred for 3 h at room temperature. The reaction was terminated by addition of 3 mL NH4OAc buffer (100 mM, pH = 7.0). The product was isolated by HPLC (Method 1). Lyophilization of collected fractions at 16.3 min afforded Boc-2P4G-RGD4. The yield was 9.5 mg (~ 43 %). ESI-MS (positive mode): m/z = 4028.0 for [M + H]+ (M = 4027.3 calcd. for [C174H265N53O58]+).
E{PEG4-E[Gly3-c(RGDfK)]2}2 (2P4G-RGD4). The Boc-protected 2P4G-RGD4 (9.0 mg, 2.2 μmol) was dissolved in 2 mL of TFA. After standing at room temperature for 5 min, TFA was removed, and the residue was dissolved in 2 mL of 0.1 M NH4OAc buffer (100 mM, pH = 7.0). The product was separated from the mixture by HPLC (Method 1). Lyophilization of collected fractions at 14.2 min afforded 2P4G-RGD4. The yield was 4.0 mg (~46%). ESI-MS (positive mode): m/z = 3927.9 for [M + H]+ and 1964.6 for [M + 2H]2+ (M = 3927.3 calcd. for [C169H257N53O56]+).
DOTA-E{PEG4-E[Gly3-c(RGDfK)]2}2 (DOTA-2P4G-RGD4). DOTA-OSu (5.8 mg, 11.5 μmol) and 2P4G-RGD4 (3.2 mg, 0.8 μmol) were dissolved in DMF (2 mL). After addition of DIEA (2 drops), the mixture was stirred for 2 h at room temperature. Upon addition of 2 mL NH4OAc buffer (100 mM, pH = 7.0), the product was separated by HPLC (Method 1). Lyophilization of collected fraction at 14.9 min afforded conjugate DOTA-2P4G-RGD4 with >95% HPLC purity. The yield was 2.2 mg (~64%). MALDI-MS: m/z = 4315.4 for [M + H]+ (M = 4313.6 calcd. for [C185H283N57O63]+).
Boc-E{PEG4-E[PEG4-c(RGDfK)]2}2 (Boc-6P-RGD4). To a solution of Boc-E(OSu)2 (2.1 mg, 4.75 μmol) in DMF (2 mL) was added 3P-RGD2 (22 mg, 10.7 μmol). After addition of excess DIEA, the reaction mixture was stirred for 3 h at room temperature. The reaction was terminated by addition of 3 mL NH4OAc buffer (100 mM, pH = 7.0). The product was separated by HPLC (Method 1). Lyophilization of collected fractions at 18.5 min afforded desired product Boc-6P-RGD4. The yield was 15.6 mg (~76%). MALDI-MS: m/z = 4333.8 for [M + H]+ (M = 4331.8 calcd. for [C194H313N45O66]+).
E{PEG4-E[PEG4-c(RGDfK)]2}2 (6P-RGD4). The Boc-protected 6P-RGD4 (15.6 mg, 3.6 μmol) was dissolved in 2 mL of TFA. After standing 5 min at room temperature, TFA was removed, and the residue was dissolved in 2 mL of NH4OAc buffer (100 mM, pH = 7.0). The product was separated from the mixture by HPLC (Method 1). Lyophilization of collected fractions at 17.3 min afforded the desired product 6P-RGD4. The yield was 14.0 mg (~92%). MALDI-MS: m/z = 4233.4 for [M + H]+ (M = 4231.7 calcd. for [C189H305N45O64]+).
DOTA-E{PEG4-E[PEG4-c(RGDfK)]2}2 (DOTA-6P-RGD4). DOTA-OSu (7.7 mg, 15.3 μmol) and 6P-RGD4 (3.8 mg, 0.9 μmol) were dissolved in 1 mL of anhydrous DMF. After addition of DIEA (3 drops), the reaction mixture was stirred for 2 h at room temperature. Upon addition of 2 mL NH4OAc buffer (100 mM, pH = 7.0), the product was separated from the mixture by HPLC (Method 1). Lyophilization of collected fraction at 18.2 min afforded DOTA-6P-RGD4 with >95% purity. The yield was 1.8 mg (~43%). MALDI-MS: m/z = 4622.7 for [M + H]+ (M = 4618.1 calcd. for [C205H331N49O71]+).
Boc-E{PEG4-E[PEG4-c(RGKfD)]2}2 (Boc-6P-RGK4). To a solution of the Boc-E(OSu)2 (5.5 mg, 12.5 μmol) in DMF (2 mL) was added 3P-RGK2 (30 mg, 14.6 μmol). After addition of excess DIEA, the reaction mixture was stirred for 3 h at room temperature. The reaction was terminated by addition of 3 mL NH4OAc buffer (100 mM, pH = 7.0). The product was isolated by HPLC (Method 1). Lyophilization of collected fractions at 18.5 min afforded desired product Boc-6P-RGK4. The yield was 15.6 mg (~49%). MALDI-MS: m/z = 4334.9 for [M + H]+ (M = 4331.8 calcd. for [C194H313N45O66]+).
E{PEG4-E[PEG4-c(RGKfD)]2}2 (6P-RGK4). Boc-6P-RGK4 (15.5 mg, 3.6 μmol) was dissolved in TFA (2 mL). After 5 min at room temperature, TFA was removed, and the residue was dissolved in 2 mL of NH4OAc buffer (100 mM, pH = 7.0). The product was separated from the mixture by HPLC (Method 1). Lyophilization of collected fractions at 15.8 min afforded 6P-RGK4. The yield was 14.0 mg (~93%). MALDI-MS: m/z = 4233.4 for [M + H]+ (M = 4231.7 calcd. for [C189H305N45O64]+).
DOTA-E{PEG4-E[PEG4-c(RGKfD)]2}2 (DOTA-6P-RGK4). DOTA-OSu (5.0 mg, 10.0 μmol) and 6P-RGK4 (3.8 mg, 0.9 μmol) were dissolved in DMF (1 mL). After addition of DIEA (3 drops), the reaction mixture was stirred for 2 h at room temperature. Upon addition of 2 mL NH4OAc buffer (100 mM, pH = 7.0), the product was separated from the mixture by HPLC (Method 1). Lyophilization of the collected fraction at ~18.6 min afforded DOTA-6P-RGK4 with >95% HPLC purity. The yield was 2.8 mg (~68%). MALDI-MS: m/z = 4622.7 for [M + H]+ (M = 4618.1 calcd. for [C205H331N49O71]+).
111In-Labeling. To a clean Eppendorf tube were added 200 μL of the DOTA conjugate solution (2.5 mg/mL in 0.1 M NaOAc buffer, pH = 5.5) and 20 μL of 111InCl3 solution (~500 μCi in 0.05 M HCl). The vial was heated in a water bath at 100 oC for ~ 15 min. After cooling to room temperature, a sample of the resulting solution was analyzed by radio-HPLC (Method 2). The water-octanol partition coefficients of all 111In radiotracers were determined according to literature methods [40, 41]. The log P values were reported as an average of three independent measurements plus the standard deviation.
Dose Preparation. For biodistribution studies, all new 111In radiotracers were purified by HPLC (Method 2). Volatiles in the HPLC mobile phase were removed under vacuum (~5 mmHg) at room temperature. Doses were prepared by dissolving the 111In radiotracer in saline to 70 - 100 μCi/mL. For the blocking experiment, excess RGD2 was dissolved in the dose solution to 7.5 mg/mL. For planar imaging studies, doses were prepared by dissolving the radiotracer (no purification) in saline to ~1 mCi/mL. In all cases, the dose solution was filtered with a 0.20 μm Millex-LG filter before being injected into the animals. Each animal was injected with ~0.1 mL of the dose solution.
In Vitro Whole-Cell Integrin αvβ3 Binding Assay. The integrin binding affinity and specificity of cyclic RGD peptides were assessed using 125I-c(RGDyK) as the integrin-specific radioligand. Briefly, U87MG human glioma cells were grown in Gibco's Dulbecco's medium supplemented with 10% fetal bovine serum (FBS), 100 IU/ml penicillin and 100 µg/ml streptomycin (Invitrogen Co, Carlsbad, CA), at 37 °C in humidified atmosphere containing 5% CO2. Filter multiscreen DV plates were seeded with 105 glioma cells in binding buffer and incubated with 125I-c(RGDyK) in the presence of increasing concentrations of the RGD conjugate. After removing the unbound 125I-c(RGDyK), hydrophilic PVDF filters were collected. The radioactivity was determined using a γ-counter (Packard, Meriden, CT). The IC50 values were calculated by fitting the data by nonlinear regression using GraphPad PrismTM (GraphPad Software, Inc., San Diego, CA). Experiments were carried out twice with triplicate samples. IC50 values are reported as an average plus the standard deviation.
Animal Model. Biodistribution and imaging studies were performed using the athymic nude mice bearing U87MG human glioma xenografts in compliance with NIH animal experiment guidelines (Principles of Laboratory Animal Care, NIH Publication No. 86-23, revised 1985). The animal protocol has been approved by the Purdue University Animal Care and Use Committee. U87MG cells were cultured in the Minimum Essential Medium, Eagle with Earle's Balanced Salt Solution (non-essential amino acids sodium pyruvate) (ATCC, Manassas, VA) in humidified atmosphere of 5% carbon dioxide, and were supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin solution (GIBCO, Grand Island, NY). Female athymic nu/nu mice were purchased from Harlan (Indianapolis, IN) at 4 - 5 weeks of age. Each mouse was implanted subcutaneously with ~5 × 106 U87MG tumor cells into the left and right upper flanks. In this way, one could assess the impact of tumor size on the radiotracer imaging quality in a single tumor-bearing mouse. All procedures were performed in a laminar flow cabinet using aseptic technique. Four weeks after inoculation, the tumor size was 0.1 - 0.4 g, and animals were used for biodistribution and imaging studies.
Biodistribution Protocol. Twenty-five tumor-bearing mice (20 - 25 g) were randomly divided into five groups. Each tumor-bearing mouse was administered with ~3 μCi of the 111In radiotracer by tail vein injection. Five animals were sacrificed by sodium pentobarbital overdose (~200 mg/kg) at 0.5, 1, 4, 24 and 72 h postinjection (p.i.). Blood samples were withdrawn from the heart of tumor-bearing mice. The tumor and normal organs (brain, eyes, heart, spleen, lungs, liver, kidneys, muscle and intestine) were harvested, washed with saline, dried with absorbent tissue, weighed, and counted on a Perkin Elmer Wizard - 1480 γ-counter (Shelton, CT). The organ uptake was calculated as the percentage of injected dose per gram of organ mass (%ID/g) or the percentage of injected dose per organ (%ID/organ). For the blocking experiment, five tumor-bearing athymic nude mice (20 - 25 g) were used, and each animal was administered with ~3 μCi of 111In(DOTA-6P-RGD4) along with ~350 μg (~14 mg/kg) of RGD2. Such a high dose of RGD2 was used to make sure that the integrin αvβ3 binding was completely blocked. At 1 h p.i., all five tumor-bearing animals were sacrificed for organ biodistribution. Biodistribution data (%ID/g) and target-to-background (T/B) ratios are reported as an average plus the standard deviation based on the results from five tumor-bearing mice (10 tumors) at each time point. Comparison between two radiotracers was made using the two-way ANOVA test (GraphPad Prim 5.0, San Diego, CA). The level of significance was set at p < 0.05.
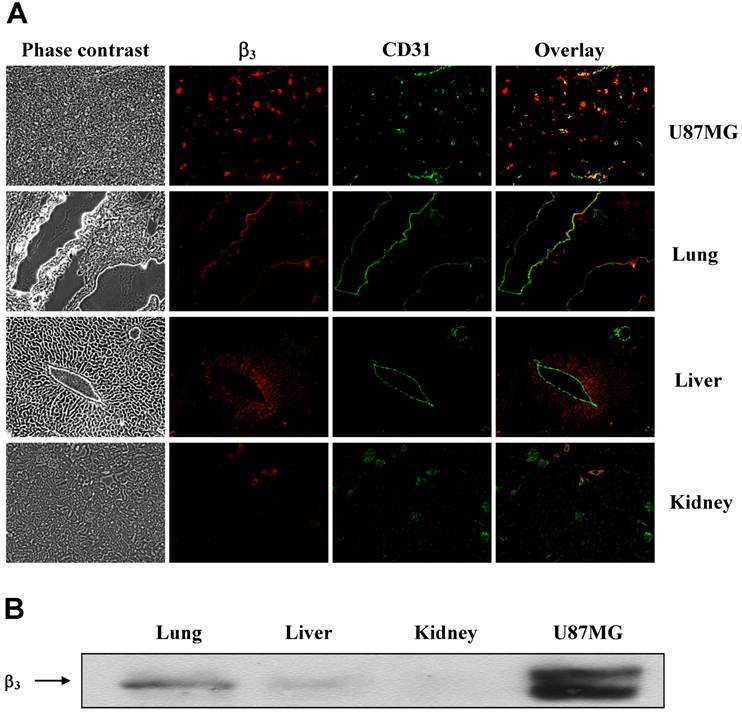
Tissue Immunohistochemistry. The xenografted U87MG tumor and normal organ (lung, liver and kidney) tissues were harvested from the tumor-bearing mice. Tissues were immediately snap-frozen in OCT (optical cutting temperature) solution and cut into 5 μm-thick slices. After thorough drying at room temperature, the slides were fixed with ice-cold acetone for 10 min. The sections were then blocked and incubated with anti-integrin β3 antibody (1:100, Santa Cruz) and CD31 antibody (1:100, BD Biosciences) for 1 h at room temperature. After washing, DyLight 594- and fluorescein isothiocyanate (FITC)-conjugated second antibodies (1:500, Jackson ImmunoResearch Inc., West Grove, PA) were added to link to integrin β3 or CD31 primary antibody, respectively. The fluorescence was visualized with an Olympus BX51 fluorescence microscope (Olympus America Inc., Center Valley, PA). Phase contrast pictures were also obtained as the histological reference to illustrate the texture of the tumor or normal organ tissue and demonstrate the site of fluorescent staining. All the pictures were taken under 100x magnification. The same procedure was used to compare integrin β3 expression levels in the U87MG tumor, kidneys, liver and lungs.
Western Blot. The xenografted U87MG glioma tumor tissue and normal organ tissues (lung, liver and kidney) were excised and homogenized in the RIPA buffer. Protease inhibitors were added freshly into the lysis buffer before experiments. After sonication, lysates were centrifuged at 12,000 g at 4 °C for 15 min. The protein concentration was determined using the Bio-Rad Protein Assay Reagent (Bio-Rad Laboratories Inc., Hercules, CA). The protein lysates (50 μg) were separated by 10% SDS-PAGE and transferred to PVDF membranes for detection of integrin β3. The blots were incubated in blocking solution consisting of 5% milk and 3% BSA in TBS-T for 1 h at room temperature and then were immunoblotted at room temperature for 2 h with the rabbit polyclonal anti-β3 primary antibody (1:1000, Cell Signaling Technology, Danvers, MA). The anti-rabbit secondary antibody was incubated with the blots for another 1 h at room temperature. Detection by enzyme-linked chemiluminescence was performed according to the manufacturer's protocol (Thermo Fisher Scientific, Rockford, IL).
Planar Imaging. The imaging studies were performed on 111In(DOTA-2P-RGD4), 111In(DOTA-2P4G-RGD4) and 111In(DOTA-6P-RGD4) using the athymic nude mice (n = 3) bearing U87MG human glioma xenografts. Each tumor-bearing mouse was administered with ~100 μCi of 111In radiotracer in 0.1 mL saline via tail vein injection. Animals were anesthetized with intraperitoneal injection of ketamine (80 mg/kg) and xylazine (19 mg/kg), and were then placed supine on a single head mini gamma camera (Diagnostic Services Inc., NJ) equipped with a parallel-hole, medium-energy and high-resolution collimator. The whole-body anterior images were acquired at 0.5, 1, 4, 24 and 72 h p.i. and stored digitally in a 128 x 128 matrix. The acquisition count limits were set at 200 K. After completion of imaging, animals were sacrificed by sodium pentobarbital overdose (~200 mg/kg).
Metabolism. Athymic nude mice were used for metabolism studies. Each mouse was administered with ~100 μCi of 111In(DOTA-2P-RGD4), 111In(DOTA-2P4G-RGD4) and 111In(DOTA-6P-RGD4) via tail vein. The urine samples were collected at 30 min and 120 min p.i. by manual void, and were mixed with equal volume of 50% acetonitrile aqueous solution. The mixture was centrifuged at 8,000 rpm for 5 min at room temperature. The supernatant was collected and passed through a 0.20 micron Millex-LG filter unit to remove any precipitate or particles. The filtrate was analyzed by radio-HPLC (Method 2). The feces samples were collected at 120 min p.i. and suspended in a mixture of 20% acetonitrile aqueous solution (2 mL). The resulting mixture was vortexed for 10 min. After centrifuging at 8,000 rpm for 5 min, the supernatant was collected and passed through a 0.20 μm Millex-LG filter unit. The filtrate was analyzed by radio-HPLC (Method 2). The kidney and liver tissues were harvested at 120 min p.i., counted in a Perkin Elmer Wizard - 1480 γ-counter (Shelton, CT) for total radioactivity, and then homogenized. The homogenate was mixed with 2 mL 20% acetonitrile aqueous solution. After centrifuging at 8,000 rpm for 5 min, the supernatant was collected and counted on a γ-counter to determine the percentage of radioactivity recovery. After filtration through a 0.20 μm Millex-LG filter unit to remove foreign particles or precipitate, the filtrate was then analyzed by radio-HPLC (Method 2). The percentage of radioactivity recovery was >95% (by γ-counting) for both urine and feces samples.
Results
Cyclic RGD Conjugates. Syntheses of DOTA-P-RGD and DOTA-P-RGD2 were straightforward by conjugation of DOTA-OSu with PEG4-c(RGDfK) and PEG4-E[c(RGDfK)]2, respectively, in DMF in the presence of excess DIEA. The key step for successful preparation of DOTA-2P-RGD4, DOTA-2P4G-RGD4 and DOTA-6P-RGD4 was the deprotection of Boc-2P-RGD4, Boc-2P4G-RGD4 and Boc-6P-RGD4, respectively. In general, excess TFA must be removed within 10 min of the reaction in order to minimize formation of hydrolyzed products. Conjugation of 2P-RGD4, 2P4G-RGD4 and 6P-RGD4 with DOTA-OSu in anhydrous DMF afforded their corresponding conjugates DOTA-2P-RGD4, DOTA-2P4G-RGD4 and DOTA-6P-RGD4, respectively. DOTA-6P-RGK4 was prepared using the identical procedure to that for DOTA-6P-RGD4. DOTA-6P-RGK4 has the identical amino acids, but their sequence was scrambled to demonstrate the RGD-specificity of DOTA-6P-RGD4. The identities for all new RGD conjugates were confirmed by ESI-MS (positive mode) or MALDI-MS. Their HPLC purity was >95% before being used for 111In-labeling and determination of their integrin αvβ3 binding affinity.
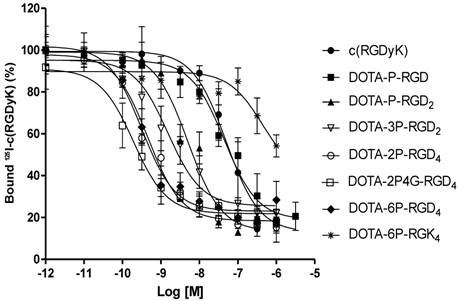
Integrin αvβ3 Binding Affinity. Figure 2 shows the displacement curves of 125I-c(RGDyK) bound to U87MG glioma cells in the presence of cyclic RGD peptides. Their IC50 values were obtained from curve fitting from Figure 2, and were calculated to be 49.9 ± 5.5, 44.3 ± 3.5, 5.0 ± 1.0, 1.5 ± 0.2, 0.5 ± 0.1, 0.2 ± 0.1, 0.3 ± 0.1, and 437 ± 35 nM for c(RGDfK), DOTA-P-RGD, DOTA-P-RGD2, DOTA-3P-RGD2, DOTA-2P-RGD4, DOTA-2P4G-RGD4, DOTA-6P-RGD4 and DOTA-6P-RGK4, respectively. DOTA-6P-RGK4 has much lower binding affinity than DOTA-6P-RGD4 (Figure 2). The IC50 of DOTA-3P-RGD2 was almost identical to that (IC50 = 1.3 ± 0.2 nM) presented in our previous report [45].
Radiochemistry. All 111In radiotracers were prepared by reacting 111InCl3 with the respective DOTA conjugates (DOTA-P-RGD, DOTA-P-RGD2, DOTA-2P-RGD4, DOTA-2P4G-RGD4, DOTA-6P-RGD4 and DOTA-6P-RGK4) in NH4OAc buffer (100 mM, pH = 5.5). Radiolabeling was completed by heating the reaction mixture at 100 oC for ~15 min. The radiochemical purity was >95% with the specific activity of > 40 mCi/μmol for all 111In radiotracers. Their HPLC retention times and log P values were listed in Table 1. We found that all new 111In radiotracers remain stable for more than 72 h in the kit matrix or after HPLC purification in the presence of 3 mM EDTA.
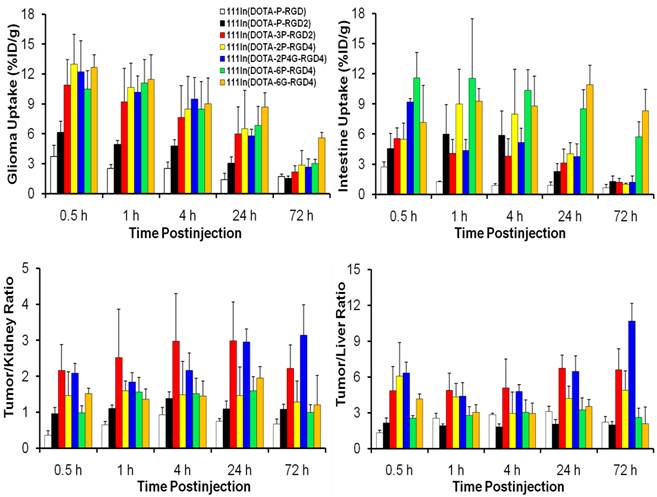
Biodistribution Characteristics. Selected biodistribution data for 111In(L) (L = DOTA-P-RGD, DOTA-P-RGD2, DOTA-2P-RGD4, DOTA-2P4G-RGD4, DOTA-6P-RGD4 and DOTA-6P-RGK4) are listed in Tables 2-6. Figure 3 compares their uptake (%ID/g) in the tumor and intestine, as well as their tumor/kidney and tumor/liver ratios in athymic nude mice (n = 5) bearing U87MG glioma xenografts. The biodistribution data and T/B ratios for 111In(DOTA-3P-RGD3) and 111In(DOTA-6G-RGD4) were obtained from our previous reports [45, 46]. The comparison of their tumor uptake will allow us to determine if the cyclic RGD tetramers (DOTA-2P-RGD4, DOTA-2P4G-RGD4 and DOTA-6P-RGD4) are actually tetravalent in binding to integrin αvβ3.
Competitive inhibition curves of 125I-c(RGDyK) bound to the U87MG human glioma cells by c(RGDyK), DOTA-P-RGD, DOTA-P-RGD2, DOTA-3P-RGD2, DOTA-2P-RGD4, DOTA-2P4G-RGD4, DOTA-6P-RGD4 and DOTA-6P-RGK4. Their IC50 values were calculated to be 49.9 ± 5.5, 44.3 ± 3.5, 5.0 ± 1.0, 1.5 ± 0.2, 0.5 ± 0.1, 0.2 ± 0.1, 0.3 ± 0.1, and 437 ± 35 nM, respectively. The integrin αvβ3 binding affinity follows the order: DOTA-6P-RGD4 ~ DOTA-2P4G-RGD4 ~ DOTA-2P-RGD4 > DOTA-3P-RGD2 > DOTA-P-RGD2 > DOTA-P-RGD >> DOTA-6P-RGK4.
Comparison of the uptake (%ID/g) in tumor and intestine, as well as the tumor/kidney and tumor/liver ratios for 111In(L) (L = DOTA-P-RGD, DOTA-P-RGD2, DOTA-3P-RGD2, DOTA-2P-RGD4, DOTA-2P4G-RGD4, DOTA-6P-RGD4 and DOTA-6G-RGD4) in the athymic nude mice (n = 5) bearing U87MG glioma xenografts. Biodistribution data and T/B ratios for 111In(DOTA-3P-RGD3) and 111In(DOTA-6G-RGD4) were obtained from our previous reports [45, 46].
Radiochemical purity (RCP), HPLC retention time and log P values for 111In-labeled cyclic RGD peptides.
| Radiotracer | RCP (%) | Retention Time (min) | Log P Value |
|---|---|---|---|
| 111In(DOTA-P-RGD) | > 95 | 12.4 | -3.48 ± 0.12 |
| 111In(DOTA-P-RGD2) | > 92 | 16.7 | -3.22 ± 0.09 |
| 111In(DOTA-3P-RGD2) | > 97 | 19.6 | -4.20 ± 0.21 |
| 111In(DOTA-2P-RGD4) | > 97 | 19.7 | -3.87 ± 0.11 |
| 111In(DOTA-2P4G-RGD4) | > 95 | 18.4 | -3.93 ± 0.09 |
| 111In(DOTA-6P-RGD4) | > 95 | 19.9 | -3.68 ± 0.03 |
| 111In(DOTA-6P-RGK4) | > 95 | 19.1 | -3.61 ± 0.07 |
Selected Biodistribution data of 111In(DOTA-P-RGD) in athymic nude mice (n = 5) bearing U87MG human glioma xenografts.
| %ID/gram | 0.5 h | 1 h | 4 h | 24 h | 72 h |
|---|---|---|---|---|---|
| Blood | 7.39 ± 0.78 | 0.63 ± 0.03 | 0.29 ± 0.02 | 0.06 ± 0.00 | 0.02 ± 0.01 |
| Brain | 0.27 ± 0.02 | 0.08 ± 0.02 | 0.04 ± 0.01 | 0.03 ± 0.00 | 0.02 ± 0.00 |
| Eyes | 0.85 ± 0.15 | 0.48 ± 0.20 | 0.24 ± 0.01 | 0.14 ± 0.01 | 0.10 ± 0.01 |
| Heart | 1.83 ± 0.08 | 0.40 ± 0.03 | 0.27 ± 0.03 | 0.14 ± 0.01 | 0.09 ± 0.01 |
| Intestine | 2.72 ± 0.50 | 1.24 ± 0.09 | 0.91 ± 0.16 | 0.91 ± 0.33 | 0.67 ± 0.32 |
| Kidney | 10.94 ± 3.96 | 3.93 ± 0.09 | 2.80 ± 0.29 | 1.92 ± 0.16 | 1.07 ± 0.11 |
| Liver | 2.79 ± 0.02 | 1.00 ± 0.08 | 0.90 ± 0.15 | 0.47 ± 0.07 | 0.30 ± 0.07 |
| Lungs | 4.60 ± 0.39 | 1.01 ± 0.05 | 0.59 ± 0.01 | 0.30 ± 0.06 | 0.16 ± 0.03 |
| Muscle | 1.20 ± 0.35 | 0.38 ± 0.05 | 0.27 ± 0.03 | 0.19 ± 0.04 | 0.13 ± 0.05 |
| Spleen | 2.91 ± 0.51 | 0.83 ± 0.10 | 0.73 ± 0.16 | 0.63 ± 0.11 | 0.37 ± 0.05 |
| U87MG | 3.74 ± 0.55 | 2.54 ± 0.37 | 2.56 ± 0.40 | 1.43 ± 0.09 | 0.74 ± 0.16 |
| Tumor/Blood | 0.52 ± 0.10 | 4.04 ± 0.70 | 8.75 ± 1.74 | 23.72 ± 1.84 | 40.06 ± 16.44 |
| Tumor /Brain | 13.79 ± 1.72 | 36.49 ± 13.29 | 73.08 ± 3.00 | 50.63 ± 5.24 | 37.16 ± 10.39 |
| Tumor /Heart | 2.06 ± 0.32 | 6.36 ± 1.25 | 9.51 ± 1.48 | 9.91 ± 0.73 | 7.62 ± 1.67 |
| Tumor /Kidney | 0.36 ± 0.13 | 0.65 ± 0.10 | 0.93 ± 0.20 | 0.75 ± 0.07 | 0.67 ± 0.15 |
| Tumor /Liver | 1.34 ± 0.20 | 2.55 ± 0.42 | 2.83 ± 0.14 | 3.10 ± 0.47 | 2.22 ± 0.48 |
| Tumor /Lungs | 0.83 ± 0.14 | 2.54 ± 0.45 | 4.36 ± 0.68 | 4.90 ± 1.02 | 4.66 ± 1.02 |
| Tumor /Muscle | 3.46 ± 1.23 | 6.89 ± 1.55 | 9.50 ± 1.31 | 7.90 ± 1.41 | 7.12 ± 2.37 |
Selected Biodistribution data of 111In(DOTA-P-RGD2) in athymic nude mice (n = 5) bearing U87MG human glioma xenografts.
| %ID/gram | 0.5 h | 1 h | 4 h | 24 h | 72 h |
|---|---|---|---|---|---|
| Blood | 1.13 ± 0.13 | 0.51 ± 0.09 | 0.22 ± 0.05 | 0.07 ± 0.02 | 0.03 ± 0.01 |
| Bone | 1.52 ± 0.47 | 1.12 ± 0.14 | 0.70 ± 0.18 | 0.79 ± 0.11 | 0.43 ± 0.06 |
| Brain | 0.11 ± 0.02 | 0.10 ± 0.02 | 0.09 ± 0.05 | 0.06 ± 0.00 | 0.03 ± 0.01 |
| Eyes | 0.94 ± 0.18 | 0.82 ± 0.27 | 0.62 ± 0.22 | 0.39 ± 0.12 | 0.20 ± 0.04 |
| Heart | 1.03 ± 0.26 | 0.69 ± 0.19 | 0.56 ± 0.15 | 0.35 ± 0.05 | 0.14 ± 0.02 |
| Intestine | 4.58 ± 1.49 | 5.99 ± 2.93 | 5.88 ± 2.42 | 2.28 ± 0.78 | 1.30 ± 0.52 |
| Kidney | 6.36 ± 0.80 | 4.45 ± 0.21 | 3.44 ± 0.43 | 2.79 ± 0.33 | 1.44 ± 0.13 |
| Liver | 2.84 ± 0.36 | 2.57 ± 0.42 | 2.60 ± 0.37 | 1.51 ± 0.57 | 0.78 ± 0.34 |
| Lungs | 3.01 ± 0.43 | 1.91 ± 0.25 | 1.52 ± 0.75 | 0.78 ± 0.09 | 0.33 ± 0.04 |
| Muscle | 1.17 ± 0.26 | 0.80 ± 0.24 | 0.70 ± 0.41 | 0.39 ± 0.11 | 0.24 ± 0.10 |
| Spleen | 2.01 ± 0.43 | 1.65 ± 0.13 | 1.75 ± 0.27 | 1.47 ± 0.35 | 0.58 ± 0.09 |
| U87MG | 6.14 ± 1.12 | 4.95 ± 0.39 | 4.79 ± 0.61 | 3.08 ± 0.60 | 1.55 ± 0.21 |
| Tumor/Blood | 5.43 ± 0.99 | 9.80 ± 0.78 | 22.00 ± 2.79 | 41.34 ± 8.03 | 53.22 ± 6.41 |
| Tumor/Kidney | 0.96 ± 0.18 | 1.11 ± 0.09 | 1.39 ± 0.18 | 1.10 ± 0.21 | 1.08 ± 0.14 |
| Tumor/Liver | 2.16 ± 0.39 | 1.92 ± 0.15 | 1.84 ± 0.23 | 2.04 ± 0.40 | 2.00 ± 0.26 |
| Tumor/Lungs | 2.04 ± 0.37 | 2.60 ± 0.21 | 3.15 ± 0.40 | 3.95 ± 0.77 | 4.75 ± 0.63 |
| Tumor/Muscle | 5.22 ± 0.95 | 6.18 ± 0.49 | 6.88 ± 0.87 | 7.90 ± 1.53 | 6.36 ± 0.84 |
Selected biodistribution data of 111In(DOTA-2P-RGD4) in athymic nude mice (n = 5) bearing U87MG human glioma xenografts.
| %ID/gram | 0.5 h | 1 h | 4 h | 24 h | 72 h |
|---|---|---|---|---|---|
| Blood | 1.12 ± 0.08 | 0.31 ± 0.16 | 0.05 ± 0.02 | 0.03 ± 0.00 | 0.02 ± 0.02 |
| Bone | 1.89 ± 0.37 | 1.83 ± 0.70 | 1.46 ± 0.69 | 84 ± 0.34 | 0.34 ± 0.03 |
| Brain | 0.10 ± 0.01 | 0.12 ± 0.04 | 0.07 ± 0.03 | 0.05 ± 0.01 | 0.02 ± 0.01 |
| Eyes | 0.87 ± 0.24 | 1.10 ± 0.22 | 0.78 ± 0.31 | 0.39 ± 0.12 | 0.21 ± 0.12 |
| Heart | 1.12 ± 0.20 | 0.86 ± 0.22 | 0.68 ± 0.31 | 0.31 ± 0.12 | 0.10 ± 0.01 |
| Intestine | 5.50 ± 1.61 | 8.98 ± 3.47 | 7.98 ± 4.47 | 4.04 ± 1.11 | 1.03 ± 0.10 |
| Kidney | 8.89 ± 1.28 | 6.67 ± 1.01 | 5.68 ± 0.66 | 4.38 ± 1.10 | 2.31 ± 0.61 |
| Liver | 2.13 ± 0.74 | 2.39 ± 0. 97 | 2.89 ± 1.36 | 1.56 ± 0.37 | 0.55 ± 0.05 |
| Lungs | 3.19 ± 0.78 | 2.47 ± 0.66 | 1.93 ± 0.77 | 0.96 ± 0.33 | 0.33 ± 0.03 |
| Muscle | 0.93 ± 0.29 | 0.91 ± 0.37 | 0.72 ± 0.23 | 0.39 ± 0.26 | 0.12 ± 0.04 |
| Spleen | 2.36 ± 0.96 | 2.47 ± 0.66 | 1.98 ± 0.97 | 1.22 ± 0.44 | 0.36 ± 0.02 |
| U87MG | 12.98 ± 5.99 | 10.66 ± 2.41 | 8.49 ± 5.28 | 6.52 ± 3.85 | 2.87 ± 1.45 |
| Tumor/Blood | 11.60 ± 5.36 | 34.67 ± 7.84 | 161.22 ± 100.35 | 231.33 ± 136.67 | 192.06 ± 172.1 |
| Tumor /Kidney | 1.46 ± 0.67 | 1.60 ± 0.27 | 1.49 ± 0.93 | 1.46 ± 0.80 | 1.28 ± 0.59 |
| Tumor /Liver | 6.09 ± 2.81 | 4.34 ± 1.12 | 2.93 ± 1.82 | 4.19 ± 1.04 | 4.89 ± 1.62 |
| Tumor /Lungs | 4.06 ± 1.88 | 4.31 ± 0.96 | 4.40 ± 2.74 | 6.76 ± 3.99 | 8.94 ± 5.37 |
| Tumor /Muscle | 13.92 ± 6.43 | 11.76 ± 3.58 | 11.83 ± 7.36 | 16.78 ± 9.92 | 25.87 ± 15.57 |
Selected biodistribution data of 111In(DOTA-2P4G-RGD4) in athymic nude mice (n = 5) bearing U87MG human glioma xenografts.
| %ID/gram | 0.5 h | 1 h | 4 h | 24 h | 72 h |
|---|---|---|---|---|---|
| Blood | 0.84 ± 0.32 | 0.13 ± 0.00 | 0.04 ± 0.00 | 0.02 ± 0.00 | 0.03 ± 0.01 |
| Bone | 2.17 ± 0.42 | 1.76 ± 0.26 | 1.24 ± 0.16 | 0.84 ± 0.10 | 0.42 ± 0.11 |
| Brain | 0.20 ± 0.10 | 0.07 ± 0.00 | 0.06 ± 0.02 | 0.04 ± 0.01 | 0.04 ± 0.02 |
| Eyes | 1.33 ± 0.23 | 0.69 ± 0.07 | 0.66 ± 0.08 | 0.31 ± 0.01 | 0.35 ± 0.24 |
| Heart | 1.79 ± 0.31 | 0.61 ± 0.04 | 0.57 ± 0.06 | 0.26 ± 0..02 | 0.13 ± 0.01 |
| Intestine | 10.36 ± 1.52 | 4.35 ± 1.13 | 5.18 ± 1.39 | 3.78 ± 1.23 | 1.22 ± 0.61 |
| Kidney | 10.72 ± 2.13 | 5.57 ± 0.63 | 4.40 ± 0.63 | 1.98 ± 0.31 | 0.85 ± 0.09 |
| Liver | 3.62 ± 0.85 | 2.36 ± 0.27 | 1.98 ± 0.39 | 0.91 ± 0.08 | 0.25 ± 0.06 |
| Lungs | 4.26 ± 0.78 | 2.38 ± 0.28 | 1.91 ± 0.27 | 1.04 ± 0.04 | 0.30 ± 0.03 |
| Muscle | 1.64 ± 0.42 | 0.98 ± 0.11 | 0.52 ± 0.07 | 0.30 ± 0.16 | 0.27 ± 0.07 |
| Spleen | 2.97 ± 0.61 | 2.02 ± 0.34 | 1.54 ± 0.25 | 1.06 ± 0.19 | 0.54 ± 0.22 |
| U87MG | 11.43 ± 2.92 | 10.18 ± 1.61 | 9.50 ± 2.13 | 5.79 ± 0.67 | 2.66 ± 0.81 |
| Tumor/Blood | 13.58 ± 3.47 | 80.74 ± 10.67 | 218.26 ± 44.57 | 266.19 ± 59.02 | 93.96 ± 22.32 |
| Tumor/Bone | 5.27 ± 1.35 | 5.80 ± 0.65 | 7.87 ± 2.44 | 7.02 ± 1.39 | 6.76 ± 2.74 |
| Tumor /Kidney | 1.07 ± 0.27 | 1.84 ± 0.26 | 2.17 ± 0.48 | 2.96 ± 0.36 | 3.14 ± 0.84 |
| Tumor /Liver | 3.15 ± 0.81 | 4.39 ± 1.13 | 4.79 ± 0.58 | 6.46 ± 1.33 | 10.71 ± 1.45 |
| Tumor /Lungs | 2.68 ± 0.69 | 4.38 ± 1.28 | 5.07 ± 1.52 | 5.59 ± 0.63 | 8.73 ± 1.70 |
| Tumor /Muscle | 6.99 ± 1.79 | 10.43 ± 1.75 | 18.07 ± 2.95 | 24.04 ± 14.24 | 10.16 ± 2.72 |
Selected biodistribution data of 111In(DOTA-6P-RGD4) in athymic nude mice (n = 5) bearing U87MG human glioma xenografts.
| %ID/gram | 0.5 h | 1 h | 4 h | 24 h | 72 h |
|---|---|---|---|---|---|
| Blood | 0.92 ± 0.06 | 0.29 ± 0.14 | 0.20 ± 0.02 | 0.12 ± 0.02 | 0.06 ± 0.00 |
| Bone | 1.27 ± 0.23 | 1.20 ± 0.19 | 089 ± 0.14 | 0.88 ± 0.03 | 0.66 ± 0.01 |
| Brain | 0.15 ± 0.01 | 0.14 ± 0.03 | 0.12 ± 0.01 | 0.08 ± 0.01 | 0.09 ± 0.06 |
| Eyes | 2.78 ± 2.01 | 1.43 ± 0.25 | 1.03 ± 0.14 | 0.63 ± 0.10 | 0.48 ± 0.06 |
| Heart | 0.57 ± 0.15 | 0.78 ± 0.24 | 0.44 ± 0.04 | 0.55 ± 0.06 | 0.37 ± 0.07 |
| Intestine | 11.60 ± 2.52 | 11.53 ± 6.94 | 10.36 ± 2.03 | 8.53 ± 1.88 | 5.74 ± 1.47 |
| Kidney | 10.68 ± 1.79 | 7.21 ± 0.79 | 5.55 ± 0.88 | 4.27 ± 0.22 | 3.10 ± 0.38 |
| Liver | 4.09 ± 0.54 | 4.01 ± 0.41 | 2.82 ± 0.54 | 1.50 ± 0.17 | 1.23 ± 0.27 |
| Lungs | 4.06 ± 0.88 | 3.69 ± 0.92 | 2.18± 0.26 | 1.62 ± 0.21 | 0.66 ± 0.08 |
| Muscle | 1.79 ± 0.11 | 1.04 ± 0.20 | 0.67 ± 0.09 | 0.52 ± 0.05 | 0.23 ± 0.05 |
| Spleen | 2.69 ± 0.63 | 2.12± 0.20 | 2.07 ± 0.37 | 1.67 ± 0.18 | 1.12 ± 0.15 |
| U87MG | 10.48 ± 1.83 | 11.08 ± 2.38 | 8.48 ± 2.75 | 6.86 ± 1.87 | 3.04 ± 0.39 |
| Tumor/Blood | 11.32 ± 1.71 | 46.41 ± 25.60 | 42.10 ± 16.02 | 56.45 ± 10.80 | 52.90 ± 4.95 |
| Tumor /Kidney | 0.99 ± 0.19 | 1.56 ± 0.41 | 1.52 ± 0.43 | 1.60 ± 0.39 | 1.00 ± 0.21 |
| Tumor /Liver | 2.55 ± 0.21 | 2.79 ± 0.73 | 3.04 ± 1.03 | 3.24 ± 1.02 | 2.60 ± 0.80 |
| Tumor /Lungs | 2.61 ± 0.33 | 3.13 ± 1.09 | 3.86 ± 1.09 | 4.68 ± 1.72 | 4.70 ± 0.86 |
| Tumor /Muscle | 5.83 ± 0.80 | 10.90 ± 2.51 | 12.56 ± 3.70 | 13.09 ± 2.85 | 13.47 ± 3.64 |
In the glioma model, 111In(DOTA-6P-RGD4) was excreted mainly via renal route with >75% of the injected radioactivity recovered from urine and <10% of the injected radioactivity from feces. 111In(DOTA-6P-RGD4) had a rapid blood clearance with high tumor/blood ratios (Table 6). It had a high initial tumor uptake (10.5 %ID/g at 0.5 h p.i.) with a slow tumor radioactivity washout (t1/2 ~ 40 h). 111In(DOTA-6P-RGD4) had a moderately low liver uptake with high tumor/liver ratio (Table 6). However, 111In(DOTA-6P-RGD4) had significant uptake in kidneys and intestine. Similar results were also obtained for 111In(DOTA-6G-RGD4) in the same animal model [46]. 111In(DOTA-2P4G-RGD4) was also excreted through the renal route with >80% of the injected radioactivity recovered from urine samples. 111In(DOTA-2P4G-RGD4) had a rapid blood clearance with high tumor/blood ratios (Table 5). The initial tumor uptake of 111In(DOTA-2P4G-RGD4) (11.4 %ID/g at 0.5 h p.i.) was comparable to that of 111In(DOTA-6P-RGD4) (10.5 %ID/g at 0.5 h p.i.) within the experimental error; but its tumor radioactivity washout was slightly faster with t1/2 being ~35 h. 111In(DOTA-2P4G-RGD4) also showed high tumor/muscle ratio (Table 5), and a rapid liver clearance (3.6 and 0.25 %ID/g at 0.5 and 72 h p.i., respectively). As a result, 111In(DOTA-2P4G-RGD4) has the highest tumor/liver ratio at 72 h p.i. (Figure 3). 111In(DOTA-2P4G-RGD4) also had very high initial intestine uptake; but it was much lower (p <0.01) than that of 111In(DOTA-6P-RGD4) at >1 h p.i. The tumor uptake of 111In(DOTA-2P-RGD4) was also high and close to that of 111In(DOTA-6P-RGD4) over the 72 h period (Figure 3). Its intestine uptake was comparable to that of 111In(DOTA-6P-RGD4) within the experimental errors (Figure 3). 111In(DOTA-2P-RGD4) had a rapid clearance from normal organs, such as the blood, kidneys and liver. The tumor/liver and tumor/kidney ratios of 111In(DOTA-2P-RGD4) were well comparable to those obtained for 111In(DOTA-6P-RGD4) and 111In(DOTA-6G-RGD4) (Figure 3).
It seems that 111In(DOTA-2P-RGD4), 111In(DOTA-2P4G-RGD4) and 111In(DOTA-6P-RGD4) might have higher tumor uptake than 111In(DOTA-3P-RGD2) over the first 24 h, but their tumor uptake differences were not significant within the experimental error (p > 0.05). The 72 h tumor uptake follows the general order of 111In(DOTA-6P-RGD4) (3.04 %ID/g) > 111In(DOTA-2P-RGD4) (2.87 %ID/g) ~ 111In(DOTA-2P4G-RGD4) (2.66 %ID/g) > 111In(DOTA-3P-RGD2) (2.18 %ID/g), which is similar to the trend of integrin αvβ3 binding affinity of DOTA-6P-RGD4 (IC50 = 0.3 ± 0.1 nM) ~ DOTA-2P4G-RGD4 (IC50 = 0.2 ± 0.1 nM) ~ DOTA-2P-RGD4 (IC50 = 0.5 ± 0.1 nM) > DOTA-3P-RGD2 (IC50 = 1.5 ± 0.2 nM). Among the 111In radiotracers evaluated in this animal model, 111In(DOTA-6G-RGD4) had the highest uptake in the tumor and intestine (Figure 3) at 72 h p.i. The half-life of 111In(DOTA-6G-RGD4) in the tumor was estimated to be 60 h. In contrast, 111In(DOTA-3P-RGD2) and 111In(DOTA-2P4G-RGD4) had low uptake in the intestine, kidneys and liver over the 72 h period. This was particularly true at > 24 h p.i. As a result, 111In(DOTA-3P-RGD2) and 111In(DOTA-2P4G-RGD4) had the tumor/kidney and tumor/liver ratios that were significantly better (p < 0.05) than those of 111In(DOTA-6G-RGD4) and 111In(DOTA-6P-RGD4) during that period of time (Figure 3).
As expected, 111In(DOTA-P-RGD) had the lowest tumor uptake (3.7 and 0.7 %ID/g at 0.5 and 72 h p.i., respectively) among 111In radiotracers evaluated in this study. 111In(DOTA-P-RGD2) had a relatively high tumor uptake (6.1 at 0.5 h p.i.), but it had a significant washout from the tumor with uptake values being 4.9, 4.8, 3.1 and 1.6 %ID/g at 1, 4, 24 and 72 h p.i., respectively. The tumor uptake follows the trend of 111In(DOTA-3P-RGD2) > 111In(DOTA-P-RGD2) > 111In(DOTA-P-RGD), which is completely consistent with the order of their αvβ3 binding affinity (IC50 value: DOTA-3P-RGD2 < DOTA-P-RGD2 < DOTA-P-RGD).
Integrin αvβ3 Specificity. Figure 4A compares the 60-min organ uptake of 111In(DOTA-6P-RGD4) in the absence/presence of excess RGD2 (~350 μg/mouse). Co-injection of excess RGD2 significantly blocked the tumor uptake of 111In(DOTA-6P-RGD4) (0.7 %ID/g with RGD2 vs. 11.1 %ID/g without RGD2 at 1 h p.i.). The normal organ uptake of 111In(DOTA-6P-RGD4) was also blocked by the presence of excess RGD2. For example, the uptake of 111In(DOTA-6P-RGD4) in the eyes, heart, intestine, liver, lungs and spleen was 1.4, 0.8, 11.5, 4.0, 3.7 and 2.1 %ID/g, respectively, without excess RGD2, while its uptake values in the same organs were 0.1, 0.4, 0.5, 0.7, 1.2, and 0.5 %ID/g, respectively, with excess RGD2. Similar properties were also observed for other radiolabeled (99mTc, 64Cu and 111In) cyclic RGD dimers [18, 25, 36-38, 43, 45] and tetramers [20, 31, 46].
A: Comparison of the 60-min biodistribution data of 111In(DOTA-6P-RGD4) in the athymic nude mice (n = 5) bearing U87MG human glioma xenografts in the absence/presence of excess RGD2 to demonstrate its integrin αvβ3-specificity. B: Comparison of the 60-min biodistribution data of 111In(DOTA-6P-RGD4) and 111In(DOTA-6P-RGK4) in the athymic nude mice (n = 5) bearing U87MG glioma xenografts to demonstrate the RGD-specificity.
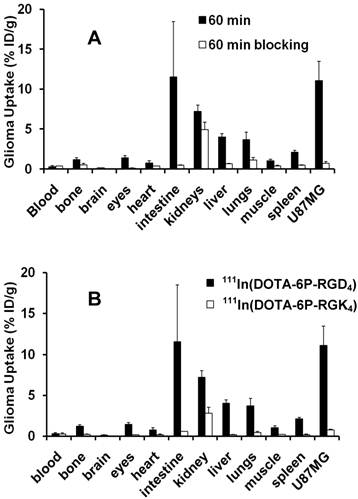
RGD Specificity. Figure 4B compares the 60-min uptake of 111In(DOTA-6P-RGD4) and 111In(DOTA-6P-RGK4) in the U87MG glioma and normal organs. As expected, 111In(DOTA-6P-RGK4) had much lower (p < 0.01) tumor uptake (0.8 %ID/g) than 111In(DOTA-6P-RGD4) (11.1 %ID/g) due to the low integrin αvβ3 binding affinity of DOTA-6P-RGK4. 111In(DOTA-6P-RGK4) also had significantly lower uptake than 111In(DOTA-6P-RGD4) in the normal organs, such as intestine, kidneys, liver, lungs and spleen (Figure 4B). Apparently, replacing c(RGDfK) motifs with c(RGKfD) moieties resulting in a dramatic uptake decrease in the tumor and integrin αvβ3-positive organs, particularly intestine, liver, lungs and spleen.
Integrin αvβ3 Expression in Normal Organs. To further understand the localization mechanism of 111In radiotracers, the tumor, lung, liver and kidney tissues were harvested, and were then double-stained for the CD31 and integrin β3 expression. As shown in Figure 5A, U87MG glioma had a high integrin β3 expression level on both tumor cells and tumor neovasculature. Most of the blood vessels expressing integrin β3 were seen from the overlay of positive CD31 and β3 positive staining. Both the liver and lungs had much lower integrin β3 expression levels than the U87MG tumor. The integrin β3 expression in kidneys was very low, which seems consistent with the results from blocking experiment (Figure 4A). These results were further confirmed by the Western blot data (Figure 5B).
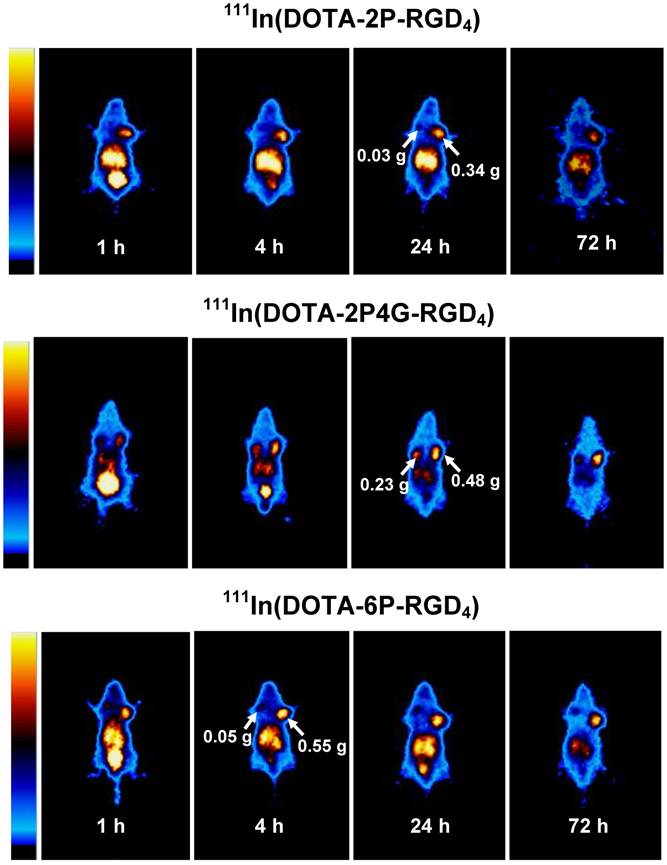
Imaging Study. Figure 6 illustrates whole-body images of the glioma-bearing mice administered with 111In(DOTA-2P-RGD4), 111In(DOTA-2P4G-RGD4) and 111In(DOTA-6P-RGD4 at 1, 4, 24 and 72 h p.i. The glioma tumors could be clearly visualized with excellent T/B contrast as early as 1 h after injection of 111In(DOTA-2P-RGD4), 111In(DOTA-2P4G-RGD4) and 111In(DOTA-6P-RGD4). All three radiotracers had a long tumor retention time with t1/2 > 30 h, which was completely consistent with the results obtained from ex-vivo biodistribution studies (Tables 2-4).
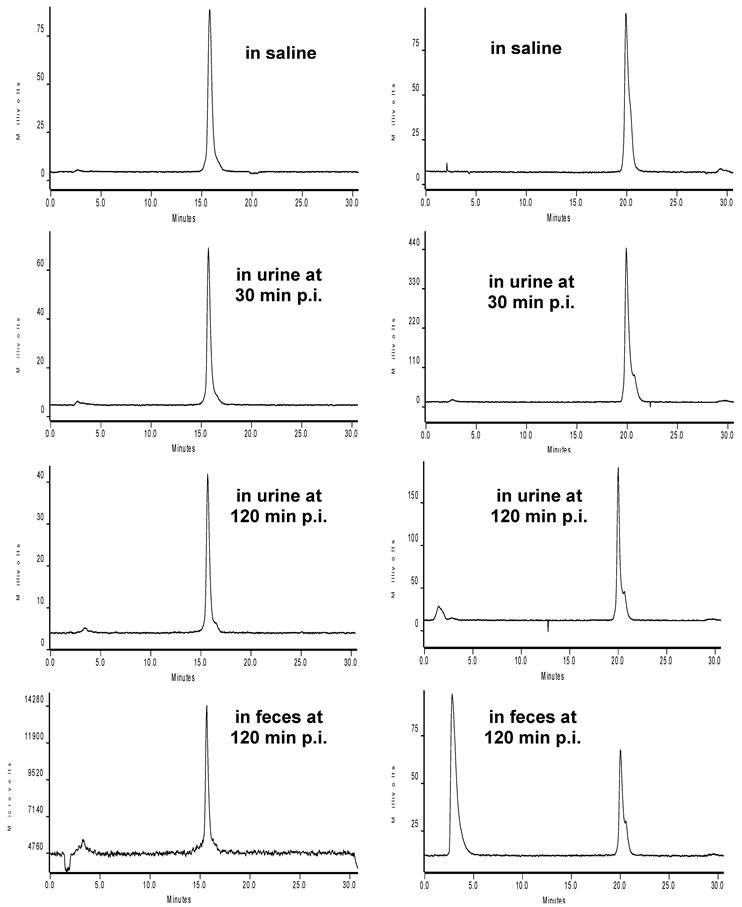
Metabolic Stability. Figure 7 shows typical radio-HPLC chromatograms of 111In(DOTA-2P-RGD4) and 111In(DOTA-6P-RGD4) in saline before injection, in urine at 30 min and 120 min p.i., and in feces at 120 min p.i. The radioactivity recovery from the urine and feces samples was >95% in both cases. Attempts to extract radioactivity from the liver and kidneys were not successful due to very limited radioactivity accumulation in both organs. There was little metabolism detected in the urine or feces samples over the 2 h study period for 111In(DOTA-2P-RGD4). 111In(DOTA-2P4G-RGD4) had the same metabolic stability as 111In(DOTA-2P-RGD4). 111In(DOTA-6P-RGD4) was able to maintain its integrity during its excretion via the renal route. However, there was a significant metabolism during its excretion via the hepatobiliary route. As a matter of fact, only ~45% of 111In(DOTA-6P-RGD4) remained intact in the feces sample. The metabolic instability was also reported for 111In(DOTA-6G-RGD4) [46].
A: Representative microscopic fluorescence images of the organ tissues (U87MG glioma, lung, liver and kidney) obtained from the U87MG tumor-bearing mice. For β3 staining (red), frozen tissue slices were incubated with a β3 primary antibody followed by a DyLight 594-conjugated secondary antibody. For CD31 staining (green), slices were incubated with a CD31 primary antibody followed by a FITC-conjugated secondary antibody (×100). Yellow color indicates the overlay of integrin β3 and CD31. Phase contrast pictures were shown as histological reference. B: Western blot results showing expression of integrin β3 in U87MG glioma, lungs, liver and kidneys obtained from the U87MG tumor-bearing mice.
The whole-body planar images of the tumor-bearing mice (bearing U87MG human glioma xenografts) administered with ~100 μCi of 111In(DOTA-3P-RGD2) (top), 111In(DOTA-2P-RGD4) (middle) and 111In(DOTA-6P-RGD4) (bottom) to illustrate the long tumor retention times of 111In-labeled cyclic RGD tetramers. Arrows indicate the presence of tumors.
Representative radio-HPLC chromatograms of 111In(DOTA-2P-RGD4) (left) and 111In(DOTA-6P-RGD4) (right) in saline before injection, in urine at 30 min and 120 min p.i., and in feces at 120 min p.i. Each mouse was administered with ~100 μCi radiotracer. 111In(DOTA-2P4G-RGD4) has the same metabolic stability in the same animal model.
Discussion
In this study, we successfully prepared 111In complexes of DOTA-P-RGD, DOTA-P-RGD2, DOTA-2P-RGD4, DOTA-2P4G-RGD4 and DOTA-6P-RGD4. We found that their integrin αvβ3 binding affinity follows the order: DOTA-6P-RGD4 ~ DOTA-2P4G-RGD4 ~ DOTA-2P-RGD4 > DOTA-P-RGD2 > DOTA-P-RGD (Figure 2). Previously, we showed that DOTA-3P-RGD2 binds to the integrin αvβ3 on U87MG glioma cells in a bivalent fashion due to the increased distance (38 bonds excluding lysine residues) between two cyclic RGD motifs [38, 45]. This study further confirms this observation. For example, the integrin αvβ3 binding affinity of DOTA-3P-RGD2 (IC50 = 1.5 ± 0.2 nM) is significantly higher (p < 0.01) than that of both DOTA-P-RGD (IC50 = 44.3 ± 3.5 nM) and DOTA-P-RGD2 (IC50 = 5.0 ± 1.0 nM). The bivalency is further supported by the higher tumor uptake of 111In(DOTA-3P-RGD2) than 111In(DOTA-P-RGD2) and 111In(DOTA-P-RGD) over the 72 h study period (Figure 3).
The integrin αvβ3 binding affinities of DOTA-2P-RGD4, DOTA-2P4G-RGD4 and DOTA-6P-RGD4 are only marginally higher than that of DOTA-3P-RGD2 (Figure 2), suggesting that they might share the same bivalency in binding to the integrin αvβ3. This conclusion is completely consistent with the similar tumor uptake of 111In(DOTA-3P-RGD2), 111In(DOTA-2P-RGD4), 111In(DOTA-2P4G-RGD4) and 111In(DOTA-6P-RGD4) over the first 24 h (Figure 3). If DOTA-2P-RGD4, DOTA-2P4G-RGD4 and DOTA-6P-RGD4 were tetravalent, they would have had much better integrin αvβ3 binding affinity than DOTA-3P-RGD2 while 111In(DOTA-2P-RGD4), 111In(DOTA-2P4G-RGD4) and 111In(DOTA-6P-RGD4) would have had significantly higher initial tumor uptake than that of 111In(DOTA-3P-RGD2).
Two factors (bivalency and the “locally enriched” RGD concentration) contribute to the higher αvβ3 binding affinity of multimeric RGD peptides than that of their monomeric counterpart, and the higher tumor uptake of radiolabeled RGD dimers and tetramers than their monomeric analogs [15, 40-46]. The concentration factor exists in all dimers and tetramers. Because of the short distance between two RGD motifs in RGD2, it is unlikely for DOTA-P-RGD2 to be bivalent [15]. However, once the first one is bonded, the “local concentration” of second RGD motif will be dramatically increased in the vicinity of neighboring integrin αvβ3 sites. This may explain why DOTA-P-RGD2 has the better integrin αvβ3 binding affinity (Figure 2) than DOTA-P-RGD, and 111In(DOTA-P-RGD2) has higher tumor uptake (Figure 3) than 111In(DOTA-P-RGD). Since IC50 values follow the order of DOTA-3P-RGD2 > DOTA-P-RGD2 > DOTA-P-RGD (Figure 2), we believe that both bivalency and concentration factors contribute to the higher tumor uptake of 111In(DOTA-3P-RGD2) than 111In(DOTA-P-RGD) (Figure 3).
DOTA-3P-RGD2 has two cyclic RGD motifs with only one possibility to achieve the bivalency. In DOTA-2P-RGD4, DOTA-2P4G-RGD4 and DOTA-6P-RGD4, however, any two of the four RGD motifs can achieve bivalency due to the longer distance between them. Even though DOTA-2P-RGD4, DOTA-2P4G-RGD4 and DOTA-6P-RGD4 may not be tetravalent, the two extra RGD motifs might contribute to their better integrin αvβ3 binding affinity than that of DOTA-3P-RGD2 (Figure 2), and the slightly higher 72-h tumor uptake (Figure 3) for 111In(DOTA-6P-RGD4) than that for 111In(DOTA-3P-RGD2). Higher tumor uptake was also observed for 111In(DOTA-6G-RGD4) than that of 111In(DOTA-3P-RGD3) at 72 h p.i. in the same tumor-bearing animal model [46].
It is very important to note that the ability of a multimeric cyclic RGD peptide to achieve bivalency depends largely on the integrin αvβ3 density on tumor cells and tumor neovasculature. If the integrin αvβ3 density is high, the distance between two neighboring integrin αvβ3 sites will be short, which makes it easier for a multimeric RGD peptide to achieve bivalency. If the integrin αvβ3 density is low, the distance between two neighboring integrin αvβ3 sites will be long, and it might be more difficult for the same multimeric RGD peptide to achieve simultaneous integrin αvβ3 binding. In that situation, the “concentration factor” will be the dominant force contributing to the better integrin αvβ3 binding affinity of a multimeric cyclic RGD peptide than that of its monomeric counterpart, and to the higher tumor uptake of the radiolabeled cyclic RGD multimers as compared to their monomeric analogs.
The integrin αvβ3-specificity of 111In(DOTA-6P-RGD4) was demonstrated by using RGD2 as the blocking agent. The uptake blockage of 111In(DOTA-6P-RGD4) in the tumor (Figure 4A) suggests that its glioma uptake is indeed mainly integrin αvβ3-mediated. The partial blockage of the uptake for 111In(DOTA-6P-RGD4) in eyes, intestine, kidneys, lungs, liver and spleen indicates that the accumulation of 111In(DOTA-6P-RGD4) in these organs is partially integrin αvβ3-mediated. This conclusion is supported by the tissue-staining studies (Figure 5A). The low integrin β3 expression in kidneys (Figure 5B) suggests that the radiotracer kidney uptake is caused mainly by tubular re-absorption instead of specific integrin αvβ3 binding. The RGD-specificity of 111In(DOTA-6P-RGD4) was demonstrated by the extremely low integrin αvβ3 binding affinity of DOTA-6P-RGK4 (Figure 2: IC50 = 437 ± 35 nM), and the significantly lower tumor uptake (Figure 4B) of 111In(DOTA-6P-RGK4) as compared to that of 111In(DOTA-6P-RGD4). In addition, 111In(DOTA-6P-RGK4) also has low uptake in the intestine, kidneys, liver, lungs and spleen, suggesting that the high uptake of 111In(DOTA-6P-RGD4) in these organs is also RGD-specific.
Non-invasive imaging of integrin αvβ3 is highly desirable for patient selection before anti-angiogenic treatment and for more effective monitoring of therapeutic efficacy in the integrin αvβ3-positive cancer patients [47-49]. For the tumors to be detectable by SPECT (single photon emission tomography) or PET (positron emission tomography), they must have sufficient accumulation of radioactivity. In this study, we found that small tumors (0.03 - 0.05 g) were clearly visualized (Figure 6) with good contrast in the tumor-bearing mice administered with 111In(DOTA-2P-RGD4), 111In(DOTA-2P4G-RGD4) and 111In(DOTA-6P-RGD4). The tumor detection limit was 3 - 4 mm in diameter using a modified clinical γ-camera. We believe that the tumor detection limit might be < 3 mm using the higher resolution SPECT cameras or SPECT/CT systems with 111In(DOTA-3P-RGD2), 111In(DOTA-2P-RGD4), 111In(DOTA-2P4G-RGD4) and 111In(DOTA-6P-RGD4) as radiotracers. From this view point, all these 111In radiotracers are useful for the early detection of integrin αvβ3-positive tumors.
It is well-established that multimerization of cyclic RGD peptides can significantly improve their integrin αvβ3-targeting capability [9, 15-41]. However this often leads to more accumulation of the radiolabeled RGD multimers in the integrin αvβ3-positive organs, as evidenced by the higher uptake of 111In(DOTA-2P-RGD4) and 111In(DOTA-6P-RGD4) than 111In(DOTA-3P-RGD2) in the intestine, liver and kidneys. As a result, 111In(DOTA-2P-RGD4) and 111In(DOTA-6P-RGD4) have the tumor/liver and tumor/kidney ratios significantly (p < 0.01) lower than those of 111In(DOTA-3P-RGD2) (Figure 3). On the basis of tumor-to-background ratios, we believe that 111In(DOTA-3P-RGD2) is a better diagnostic radiotracer than 111In(DOTA-2P-RGD4) and 111In(DOTA-6P-RGD4). At this moment, we are not clear why 111In(DOTA-2P4G-RGD4) has lower (p < 0.05) intestine uptake than 111In(DOTA-6P-RGD4) and 111In(DOTA-6G-RGD4) (Figure 3) even though they share the same four cyclic RGD motifs. However, it is clear that the G3 and PEG4 linkers between cyclic RGD motifs have a significant impact on biodistribution characteristics of 111In-labeled cyclic RGD tetramers.
For diagnostic radiotracers, the contrast between the targeted organ and surrounding tissues is the first priority. For therapeutic radiotracers, however, maximal delivery of tumoricidal radiation dose becomes critically important. The radiotracer with longer retention time will deliver higher radiation dose to tumors during the same period of time. From this viewpoint, DOTA-6G-RGD4 remains the best for the integrin αvβ3-targeted therapeutic 90Y and 177Lu radiotracers since 111In(DOTA-6G-RGD4) has the longest retention time (t1/2 ~ 60 h) among four 111In-labeled RGD tetramers (Figure 3). However, one cannot totally ignore its high intestine uptake and low tumor/liver ratios over the 72 h period. Considering the radiotracer uptake in normal organs (particularly intestine and liver), we believe that DOTA-3P-RGD2 and DOTA-2P4G-RGD4 have significant advantages over DOTA-6P-RGD4 and DOTA-6G-RGD4 because the intestine and liver uptake of 111In(DOTA-3P-RGD2) and 111In(DOTA-2P4G-RGD4) is significantly lower than that of 111In(DOTA-6P-RGD4) and 111In(DOTA-6P-RGD4) (Figure 3). It must be noted that there is always a subtle balance between maximizing the radiation dose to tumor cells and minimizing the radiation burden to the surrounding normal tissues or organs.
It is interesting to point out that 111In(DOTA-2P-RGD4) is stable during its renal and hepatobiliary excretion over the 2 h study period (Figure 7). In contrast, 111In(DOTA-6P-RGD4) has a significant metabolism during its hepatobiliary excretion while it remains relatively unchanged during its renal excretion (Figure 7). 111In(DOTA-6G-RGD4) has similar metabolic instability except that only 25% of it remains intact in the feces samples in the same animal model [46]. Although it remains unclear as for what causes this metabolic instability, one thing is clear that the linkers between RGD motifs have a significant impact on the metabolic stability of 111In-labeled cyclic RGD tetramers.
Conclusion
In summary, we evaluated five new radiotracers 111In(L) (L = DOTA-P-RGD, DOTA-P-RGD2, DOTA-2P-RGD4, DOTA-2P4G-RGD4 and DOTA-6P-RGD4) to explore the effect of peptide and PEG4 linker multiplicity on their tumor uptake and T/B ratios. We found that the addition of two PEG4 linkers between two RGD motifs significantly improves the integrin αvβ3-targeting capability of cyclic RGD dimers as evidenced by their higher integrin αvβ3-binding affinity and the high tumor uptake and better T/B ratios of their radiotracers. We also found that DOTA-2P-RGD4, DOTA-2P4G-RGD4 and DOTA-6P-RGD4 are not tetravalent in binding to integrin αvβ3 because increasing the RGD multiplicity from dimers to tetramers didn't significantly improve the radiotracer's initial tumor uptake. However, the extra RGD motifs in DOTA-2P-RGD4, DOTA-2P4G-RGD4 and DOTA-6P-RGD4 might contribute to slightly higher tumor uptake of 111In(DOTA-2P-RGD4), 111In(DOTA-2P4G-RGD4) and 111In(DOTA-6P-RGD4) than that of 111In(DOTA-3P-RGD2) at 72 h p.i. Among the 111In-labeled cyclic RGD tetramers evaluated in this animal model, 111In(DOTA-2P4G-RGD4) has the best tumor/kidney and tumor/liver ratios. The combination of its high tumor uptake, long retention time and low uptake in normal organs suggests that 90Y(DOTA-2P4G-RGD4) and 177Lu(DOTA-2P4G-RGD4) may have the potential for radiotherapy of integrin αvβ3-positive tumors [50]. Evaluations of their therapeutic efficacy are still in progress, and the result from these studies will be communicated elsewhere.
Abbreviations
c(RGDfK): cyclo(Lys-Arg-Gly-Asp-D-Phe); c(RGDyK): cyclo(Lys-Arg-Gly-Asp-D-Tyr);
E[c(RGDfK)]2 (RGD2): Glu[cyclo(Lys-Arg-Gly-Asp-D-Phe)-cyclo(Lys-Arg-Gly-Asp-D-Phe)];
DOTA: 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetracetic acid;
PEG4: 15 amino-4,710,13-tetraoxapentadecanoic acid;
PEG4-E[G3-c(RGDfK)]2: PEG4-Glu{cyclo[Lys(Gly-Gly-Gly)-Arg-Gly-Asp-D-Phe]}-cyclo[Lys(Gly-Gly-Gly)-Arg-Gly-Asp-D-Phe]};
PEG4-E[PEG4-c(RGDfK)]2: PEG4-Glu{cyclo[Lys(PEG4)-Arg-Gly-Asp-D-Phe]-cyclo[Lys(PEG4)-Arg-Gly-Asp-D-Phe]};
PEG4-E[PEG4-c(RGKfD)]2: PEG4-Glu{cyclo[Arg-Gly-Lys(PEG4)-D-Phe-Asp]-cyclo[Arg-Gly-Lys(PEG4)-D-Phe-Asp]};
E{E[c(RGDfK)]2}2 (RGD4): Glu{Glu[cyclo(Lys-Arg-Gly-Asp-D-Phe)]-cyclo(Lys-Arg-Gly-Asp-D-Phe)}-{Glu[cyclo(Lys-Arg-Gly-Asp-D-Phe)]-cyclo(Lys-Arg-Gly-Asp-D-Phe)};
E{PEG4-E[c(RGDfK)]2}2: Glu{PEG4-Glu[cyclo(Lys-Arg-Gly-Asp-D-Phe)]-cyclo(Lys-Arg-Gly-Asp-D-Phe)}-{PEG4-Glu[cyclo(Lys-Arg-Gly-Asp-D-Phe)]-cyclo(Lys-Arg-Gly-Asp-D-Phe)};
E{PEG4-E[G3-c(RGDfK)]2}2: Glu{PEG4-Glu[cyclo(Lys(Gly-Gly-Gly)-Arg-Gly-Asp-D-Phe)]-cyclo(Lys(Gly-Gly-Gly)-Arg-Gly-Asp-D-Phe)}-{PEG4-Glu[cyclo(Lys(Gly-Gly-Gly)-Arg-Gly-Asp-D-Phe)]-cyclo(Lys(Gly-Gly-Gly)-Arg-Gly-Asp-D-Phe)};
E{PEG4-E[PEG4-c(RGDfK)]2}2: Glu{PEG4-Glu[cyclo(Lys(PEG4)-Arg-Gly-Asp-D-Phe)]-cyclo(Lys(PEG4)-Arg-Gly-Asp-D-Phe)}-{PEG4-Glu[cyclo(Lys(PEG4)-Arg-Gly-Asp-D-Phe)]-cyclo(Lys(PEG4)-Arg-Gly-Asp-D-Phe)};
E{PEG4-E[PEG4-c(RGKfD)]2}2: Glu{PEG4-Glu[cyclo(Arg-Gly-Lys(PEG4)-D-Phe-Asp)]-cyclo(Arg-Gly-Lys(PEG4)-D-Phe-Asp)}-{PEG4-Glu[cyclo(Arg-Gly-Lys(PEG4)-D-Phe-Asp)]-cyclo(Arg-Gly-Lys(PEG4)-D-Phe-Asp)}.
Acknowledgements
This work is supported, in part, by Purdue University and the research grant: R01 CA115883 from the National Cancer Institute (NCI).
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Weber WA, Haubner R, Vabuliene E. et al. Tumor angiogenesis targeting using imaging agents. Q J Nucl Med. 2001;45:179-82
2. Liu S, Edwards DS. Fundamentals of receptor-based diagnostic metalloradiopharmaceuticals. Top Curr Chem. 2002;222:259-78
3. Van de Wiele C, Oltenfreiter R, De Winter O. et al. Tumor angiogenesis pathways: related clinical issues and implications for nuclear medicine imaging. Eur J Nucl Med. 2002;29:699-709
4. Liu S, Robinson SP, Edwards DS. Integrin αvβ3 directed radiopharmaceuticals for tumor imaging. Drugs Fut. 2003;28:551-64
5. Haubner R, Wester HJ. Radiolabeled tracers for imaging of tumor angiogenesis and evaluation of anti-angiogenic therapies. Curr Pharm Des. 2004;10:1439-55
6. Liu S, Robinson SP, Edwards DS. Radiolabeled integrin αvβ3 antagonists as radiopharmaceuticals for tumor radiotherapy. Top Curr Chem. 2005;252:193-216
7. Chen X. Multimodality imaging of tumor integrin αvβ3 expression. Mini-Rev Med Chem. 2006;6:227-34
8. Meyer A, Auernheimer J, Modlinger A. et al. Targeting RGD recognizing integrins: drug development, biomaterial research, tumor imaging and targeting. Curr Pharm Des. 2006;12:2723-47
9. Liu S. Radiolabeled multimeric cyclic RGD peptides as integrin αvβ3-targeted radiotracers for tumor imaging. Mol. Pharm. 2006;3:472-87
10. Cai W, Chen X. Multimodality molecular imaging of tumor angiogenesis. J Nucl Med. 2008;49(Suppl 2):113S-128S
11. Haubner R, Wester HJ, Senekowitsch-Schmidtke R. et al. RGD-peptides for tumor targeting: biological evaluation of radioiodinated analogs and introduction of a novel glycosylated peptide with improved biokinetics. J Lab Compd Radiopharm. 1997;40:383-5
12. Haubner R, Bruchertseifer F, Bock M. et al. Synthesis and biological evaluation of 99mTc-labeled cyclic RGD peptide for imaging integrin αvβ3 expression. Nuklearmedizin. 2004;43:26-32
13. Haubner R, Wester HJ, Reuning U. et al. Radiolabeled αvβ3 integrin antagonists: a new class of tracers for tumor imaging. J Nucl Med. 1999;40:1061-71
14. Haubner R, Wester HJ, Weber WA. et al. Noninvasive imaging of αvβ3 integrin expression using 18F-labeled RGD-containing glycopeptide and positron emission tomography. Cancer Res. 2001;61:1781-5
15. Liu S. Radiolabeled cyclic RGD peptides as integrin αvβ3-targeted radiotracers: maximizing binding affinity via bivalency. Bioconj Chem. 2009;20:2199-213
16. Thumshirn G, Hersel U, Goodman SL. et al. Multimeric cyclic RGD peptides as potential tools for tumor targeting: solid-phase peptide synthesis and chemoselective oxime ligation. Chem Eur J. 2003;9:2717-25
17. Poethko T, Schottelius M, Thumshirn G. et al. Chemoselective pre-conjugate radiohalogenation of unprotected mono- and multimeric peptides via oxime formation. Radiochimica Acta. 2004;92:317-27
18. Chen X, Liu S, Hou Y. et al. MicroPET imaging of breast cancer αv-integrin expression with 64Cu-labeled dimeric RGD peptides. Mol. Imaging Biol. 2004;6:350-9
19. Chen X, Tohme M, Park R. et al. MicroPET imaging of breast cancer αv-integrin expression with 18F-labeled dimeric RGD peptide. Mol. Imaging. 2004;3:96-104
20. Wu Y, Zhang X, Xiong Z. et al. MicroPET imaging of glioma integrin αvβ3 expression using 64Cu-labeled tetrameric RGD peptide. J Nucl Med. 2005;46:1707-18
21. Zhang X, Xiong Z, Wu Y. et al. Quantitative PET imaging of tumor integrin αvβ3 expression with 18F-FRGD2. J Nucl Med. 2006;47:113-21
22. Wu Z, Li Z, Chen K. et al. Micro-PET of tumor integrin αvβ3 expression using 18F-labeled PEGylated tetrameric RGD peptide (18F-FPRGD4). J Nucl Med. 2007;48:1536-44
23. Liu S, Edwards DS, Ziegler MC. et al. 99mTc-labeling of a hydrazinonictotinamide-conjugated vitronectin receptor antagonist useful for imaging tumor. Bioconj Chem. 2001;12:624-9
24. Liu S, Hsieh WY, Kim YS. et al. Effect of coligands on biodistribution characteristics of ternary ligand 99mTc complexes of a HYNIC-conjugated cyclic RGDfK dimer. Bioconj Chem. 2005;16:1580-8
25. Jia B, Shi J, Yang Z. et al. 99mTc-labeled cyclic RGDfK dimer: initial evaluation for SPECT imaging of glioma integrin αvβ3 expression. Bioconj Chem. 2006;17:1069-76
26. Liu S, He ZJ, Hsieh WY. et al. Impact of PKM linkers on biodistribution characteristics of the 99mTc-labeled cyclic RGDfK dimer. Bioconj Chem. 2006;17:1499-507
27. Janssen ML, Oyen WJG, Dijkgraaf I. et al. Tumor targeting with radiolabeled αvβ3 integrin binding peptides in a nude mouse model. Cancer Res. 2002;62:6146-51
28. Janssen M, Oyen WJG, Massuger LFAG. et al. Comparison of a monomeric and dimeric radiolabeled RGD-peptide for tumor targeting. Cancer Biother Radiopharm. 2002;17:641-6
29. Dijkgraaf I, Kruijtzer JAW, Liu S. et al. Improved targeting of the αvβ3 integrin by multimerization of RGD peptides. Eur J Nucl Med Mol Imaging. 2007;34:267-73
30. Dijkgraaf I, Liu S, Kruijtzer JAW. et al. Effect of linker variation on the in vitro and in vivo characteristics of an 111In-labeled RGD Peptide. Nucl Med Biol. 2007;34:29-35
31. Liu S, Hsieh WY, Jiang Y. et al. Evaluation of a 99mTc-labeled cyclic RGD tetramer for non-invasive imaging integrin αvβ3-positive breast cancer. Bioconj Chem. 2007;18:438-46
32. Liu S, Kim YS, Hsieh WY. et al. Coligand effects on solution stability, biodistribution and metabolism of 99mTc-labeled cyclic RGDfK tetramer. Nucl Med Biol. 2008;35:111-21
33. Jia B, Liu Z, Shi J. et al. Linker effects on biological properties of 111In-labeled DTPA conjugates of a cyclic RGDfK dimer. Bioconj Chem. 2008;19:201-10
34. Wang JJ, Kim YS, He ZJ. et al. 99mTc-labeling of HYNIC-conjugated cyclic RGDfK dimer and tetramer using EDDA as coligand. Bioconj Chem. 2008;19:634-42
35. Li ZB, Chen K, Chen X. 68Ga-labeled multimeric RGD peptides for microPET imaging of integrin αvβ3 expression. Eur J Nucl Med Mol Imaging. 2008;35:1100-8
36. Wang L, Shi J, Kim YS. et al. Improving tumor-targeting capability and pharmacokinetics of 99mTc-labeled cyclic RGD dimers with PEG4 linkers. Mol Pharm. 2009;6:231-45
37. Shi J, Wang L, Kim YS. et al. Improving tumor uptake and excretion kinetics of 99mTc-labeled cyclic Arginine-Glycine-Aspartic (RGD) dimers with triglycine linkers. J Med Chem. 2008;51:7980-90
38. Shi J, Kim YS, Zhai S. et al. Improving tumor uptake and pharmacokinetics of 64Cu-labeled cyclic RGD peptide dimers with Gly3 and PEG4 Linkers. Bioconj. Chem. 2009;20:750-9
39. Liu Z, Liu S, Wang F. et al. Non-invasive imaging of tumor integrin expression using 18F-labeled RGD dimer peptide with PEG4 linkers. Eur J Nucl Med Mol Imaging. 2009;36:1296-307
40. Shi J, Kim YS, Chakraborty S. et al. 2-Mercaptoacetylglycylglycyl (MAG2) as a bifunctional chelator for 99mTc-labeling of cyclic RGD dimers: effects of technetium chelate on tumor uptake and pharmacokinetics. Bioconj Chem. 2009;20:1559-68
41. Shi J, Wang L, Kim YS. et al. 99mTcO(MAG2-3G3-dimer): a new integrin αvβ3-targeted SPECT radiotracer with high tumor uptake and favorable pharmacokinetics. Eur J Nucl Med Mol Imaging. 2009;36:1874-84
42. Liu Z, Niu G, Shi J. et al. 68Ga-labeled cyclic RGD dimers with Gly3 and PEG4 linkers: promising agents for tumor integrin αvβ3 PET imaging. Eur J Nucl Med Mol Imaging. 2009;36:947-57
43. Liu Z, Jia B, Shi J. et al. Tumor uptake of the RGD dimeric probe 99mTc-G3-2P4-RGD2 is correlated with integrin αvβ3 expressed on both tumor cells and neovasculature. Bioconj Chem. 2010;21:548-55
44. Dijkgraaf I, Yim CB, Franssen GM. et al. PET imaging of αvβ3 integrin expression in tumours with 68Ga-labeled mono-, di- and tetrameric RGD peptides. Eur J Nucl Med Mol Imaging. 2011;38:128-37
45. Shi J, Kim YS, Chakraborty S. et al. Impact of bifunctional chelators on biological properties of 111In-labeled cyclic peptide RGD dimers. Amino Acids. 2010 [Epub ahead of print]
46. Chakraborty S, Shi J, Kim YS. et al. Evaluation of 111In-labeled cyclic RGD peptides: tetrameric not tetravalent. Bioconj Chem. 2010;21:969-78
47. Cai W, Rao J, Gambhir SS. et al. How molecular imaging is speeding up antiangiogenic drug development? Mol Cancer Ther. 2006;5:2624-33
48. Niu G, Chen X. Has molecular and cellular imaging enhanced drug discovery and drug development? Drugs R D. 2008;9:351-68
49. Cai W, Niu G, Chen X. Imaging of integrins as biomarkers for tumor angiogenesis. Curr Pharm Des. 2008;14:2943-73
50. Liu Z, Wang F, Chen X. Integrin targeted delivery of radiotherapeutics. Theranostics. 2011;1:201-10
Author contact
![]() Corresponding author: School of Health Sciences, Purdue University, 550 Stadium Mall Drive, West Lafayette, IN 47907. Phone: 765-494-0236; Fax 765-496-1377; Email: liu100edu
Corresponding author: School of Health Sciences, Purdue University, 550 Stadium Mall Drive, West Lafayette, IN 47907. Phone: 765-494-0236; Fax 765-496-1377; Email: liu100edu
 Global reach, higher impact
Global reach, higher impact