13.3
Impact Factor
Theranostics 2018; 8(6):1527-1539. doi:10.7150/thno.22699 This issue Cite
Research Paper
PRL-3 is a potential glioblastoma prognostic marker and promotes glioblastoma progression by enhancing MMP7 through the ERK and JNK pathways
1. Department of Pathophysiology, School of Basic Medicine, Fourth Military Medical University, Xi'an, China, 710032
2. State Key Laboratory of Cancer Biology, Biotechnology Center, School of Pharmacy, Fourth Military Medical University, Xi'an, China, 710032
3. Experimental Teaching Center of Basic Medicine, Fourth Military Medical University, Xi'an, China, 710032
4. Department of Pathology, Tangdu Hospital, The Fourth Military Medical University, Xi'an, China, 710038
5. Department of General Surgery, Tangdu Hospital, The Fourth Military Medical University, Xi'an, China, 710038
6. Department of Neurosurgery, Xijing Hospital, Fourth Military Medical University, Xi'an, China, 710032
7. Department of Health Statistics, School of Public Health, Fourth Military Medical University, Xi'an, China, 710032
*These authors contributed equally to this work.
Received 2017-9-5; Accepted 2017-12-10; Published 2018-2-7
Abstract
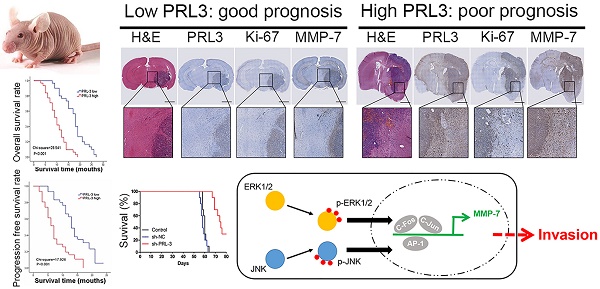
Purpose: Glioblastoma is the most common and aggressive type of primary brain malignancy and is associated with a poor prognosis. Previously, we found that phosphatase of regenerating liver-3 (PRL-3) was significantly up-regulated in glioblastoma as determined by a microarray analysis. However, the function of PRL-3 in glioblastoma remains unknown. We aimed to investigate the clinical relationship between PRL-3 and glioblastoma, and uncover the mechanisms of PRL-3 in the process of glioblastoma.
Methods: PRL-3 expression was evaluated in 61 glioblastoma samples and 4 cell lines by RT-qPCR and immunohistochemistry. Kaplan-Meier analysis was performed to evaluate the prognostic value of PRL-3 for overall survival (OS) and progression-free survival (PFS) for glioblastoma patients. Proliferation was evaluated by Cell Counting Kit-8 (CCK-8) assay and EdU proliferation assay, migration and invasion by wound-closure/Transwell assays, and qRT-PCR/immunoblotting/IHC were used for both in vivo and in vitro investigations.
Result: A high PRL-3 expression level was closely correlated with unfavorable OS and PFS for glioblastoma patients, and was also significantly correlated with Ki-67 expression. Down-regulation of PRL-3 inhibited glioma cell proliferation, invasion and migration through ERK/JNK/matrix metalloproteinase 7 (MMP7) in vitro and in vivo.
Conclusions: PRL-3 expression enhances the invasion and proliferation of glioma cells, highlighting this phosphatase as a novel prognostic candidate and an attractive target for future therapy in glioblastoma.
Keywords: PRL-3, glioblastoma, poor prognosis, prognostic marker
Introduction
Gliomas constitute approximately 80% of all primary malignant brain tumors and result in the loss of more years of life than any other tumor type [1, 2]. Diffuse infiltrative growth is one of the main biological characteristics of glioma [3-5]. The glioblastoma multiforme is known as grade IV glioma, according to the world Health Organization (WHO) classification system [6], which is the most common and aggressive form of glioma. Despite intensive therapeutic efforts, including maximal surgical resection, radiotherapy, and chemotherapy, therapeutic results, as measured by survival time and quality of life, remain unsatisfactory [7, 8]. Genomic alterations, such as co-deletion of 1p/19q, methylation of the O6 methylguanine methyltransferase (MGMT) gene promoter [9], mutations/amplifications of the epidermal growth factor receptor (EGFR) [10], mutations in the isocitrate dehydrogenase 1 (IDH1) gene [11], mutations in PTEN and homozygous deletion of p16INK4a [12] have been confirmed to be closely related to the tumorigenesis of glioblastoma. Therefore, researchers hope that the characterization of molecular alterations that occur in glioblastoma will help improve clinicopathologic determinants of prognosis and choose better therapeutic strategies.
We previously investigated the differences in gene expression profiles between human glioma tissues and normal brain specimens using an Agilent Human A1 array. The phosphatase of regenerating liver-3 (PRL-3, also known as PTP4A3) was one of the most highly up-regulated mRNAs (data not shown). PRL-3, a member of the protein tyrosine phosphatase family (PTPs), was identified as a metastasis-related gene in 2001 [13-16]. The overall folding topology of PRL-3 is similar to that of the dual specificity phosphatases [17]. Substantial data have shown that PRL-3 is overexpressed in various cancer types, including ovarian cancer, myeloma, breast cancer, gastrointestinal cancer, oral squamous cell cancer, hepatocellular carcinoma and intrahepatic cholangiocarcinoma [18, 19]. Furthermore, PRL-3 modulates multiple signaling pathways, including PI3K/AKT, NF-κB, Rho GTPases, MAPK/ERK, and EGFR in various cancer cells [20, 21]. However, to date, the function of PRL-3 in the pathogenesis of glioblastoma remains unknown.
Here, we analyzed the expression of PRL-3 in 61 glioblastoma samples and 8 cell lines using quantitative real-time PCR (qPCR), western blotting, and immunohistochemistry. We also explored the positive rate of Ki-67 and its relationship with PRL-3 expression levels in glioblastoma specimens. Then, we elucidated the impact of high PRL-3 expression on the prognoses of glioblastoma patients, and uncovered that down-regulation of PRL-3 inhibited the progression of glioblastoma through ERK/JNK/MMP7 pathways in vitro and in vivo. According to our research, PRL-3 may be a novel therapeutic target and prognostic marker for patients with glioblastoma.
Materials and Methods
Tissue samples and clinical data
All 61 clinical tumor specimens were collected from patients diagnosed with primary glioblastoma multiforme at the Institute of Neurosurgery, Xijing Hospital. None of the patients received chemotherapy or radiotherapy before surgical tumor resection, and all specimens were obtained during the initial surgery. Patients who died of diseases not directly related to glioblastoma were excluded from this study. Ethics approval was granted by the Ethics Committee of the Fourth Military Medical University. All patients provided signed informed consent prior to study participation. Follow-up data for all eligible patients were obtained every 3 months by telephone and mailed questionnaires. OS was calculated from the date of the initial surgical operation to the date of death. The death of the participants was ascertained by family reports and verified by reviewing public records. Specimens of normal brain tissues were obtained from autopsy material, and to keep these specimens histologically normal, the tissues were acquired within 12 h after death. (Table 1)
Relationship between PRL-3 expression and clinicopathologic variables of patients with glioblastoma (n = 61)
| Clinicopathologic variables | Number | PRL-3 expression | Chi-square value | P value | |
|---|---|---|---|---|---|
| low | high | ||||
| All cases | 61 | 30 | 31 | ||
| Age at diagnosis (years) | 1.428 | 0.300 | |||
| ≤ 60 | 25 | 10 | 15 | ||
| >60 | 36 | 20 | 16 | ||
| Gender | 0.011 | 1.000 | |||
| Male | 37 | 18 | 19 | ||
| Female | 24 | 12 | 12 | ||
| Ki-67 (low/high) | 11.939 | 0.001 | |||
| Low | 29 | 23 | 6 | ||
| High | 32 | 10 | 22 | ||
| Tumor lateralization | 0.262 | 0.184 | |||
| Supratentorial | 22 | 8 | 14 | ||
| Subtentorial | 39 | 22 | 17 | ||
| IDH1 | 1.292 | 0.301 | |||
| Mutation | 9 | 6 | 3 | ||
| Wildtype | 52 | 24 | 28 | ||
Cell lines and cell culture
Human glioma cell lines, U251, U87, LN229, A172, T98G, U118, LN18 and SHG44 were purchased from the China Academia Sinica Cell Repository (Shanghai, China). U251, U87, LN229, A172, T98G, U118, LN18 and SHG44 cells were cultured in DMEM or RPMI 1640 (Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, USA), 50 units/mL penicillin/streptomycin and incubated at 37°C in a humidified incubator with 5% CO 2/95% air.
Patient-derived glioblastoma cells
The patient-derived glioblastoma cells were obtained from patients undergoing surgical treatment at our hospital in accordance with the appropriate Institutional Review Boards, according to previous research [22, 23]. Briefly, within 1 to 3 h after surgical removal, tumor tissues were washed and enzymatically dissociated into single cells; other cells (like red blood cells) were removed by differential centrifugation. To maintain authenticity of the cell lines, frozen stocks were prepared from initial stocks, and every 3 months, a new frozen stock was used for the experiments.
Plasmid construction, Lentivirus packaging and stable cell lines generation
The complete open reading frame (ORF) of human PRL-3 was amplified by PCR using the primer pair 5'-CGGAATTCATGGCTCGGATGAACCGC-3' and 5'-GCTCTAGACTACATAACGCAGCACCGGGT-3' containing EcoRI and XbaI restriction sites within the 5'and 3' termini, respectively. The amplicon was inserted into the pcDNA3.1 vector. The sh-PRL-3 lentivirus was constructed and packaged by GeneCopoeia (Guangzhou, China). To generate the PRL-3 shRNA, three target sequences were desired against human PRL-3 that target different regions of its mRNA. sh-PRL-3 #1: CCAAGTATTTGCACAATATTT; sh-PRL-3 #2: GGTGGAGGTGAGCTACAAACA; sh-PRL-3 #3: GGTGCTGCGTTATGTAGCTCA.
Only sh-PRL-3 #1 presented high efficiency in knocking down PRL-3, which was used for the sh-PRL-3 lentivirus package. For sh Negative Control (sh-NC), the sequence is: GCTTCGCGCCGTAGTCTTA. To generate the stable cell lines, cancer cells were infected by either the sh-PRL-3 or sh-NC viruses at a MOI of 5 in the presence of 8 μg/mL Polybrene (Sigma-Aldrich, USA). On day four after transduction, medium with 5 μg/mL puromycin (Sigma-Aldrich, USA) was used to screen the puromycin-resistant clones. The stable cells were expanded with four weeks of continuous culture. The luciferase reporter plasmid that contains the MMP7 promoter region was purchased from GeneCopoeia (Guangzhou, China).
Quantitative real-time PCR (qPCR) and immunoblot
The total RNA of tissues and cells were extracted with Trizol reagent (Invitrogen, USA) according to the manufacturer's protocol. qPCR and immunoblot were performed as described previously [24]. The primer sequences were as follows:
PRL-3 forward, 5'- GGGACTTCTCAGGTCGTGTC-3';
PRL-3 reverse, 5'-AGCCCCGTACTTCTTCAGGT-3';
β-actin forward: 5'-CCACGAAACTACCTTCAACTCC-3';
β-actin reverse: 5'-TCATACTCCTGCTGCTTGCTGATC-3'.
Antibodies against PRL-3 (1:1000, Abcam, USA); Ki-67 (1:500, Abcam, USA); MMP7 (1:1000, Abcam, USA); p-Akt (Ser473), Akt, GSK-3β, p-GSK-3β (Ser9), Erk1/2, p-Erk1/2 (1:1000, Cell Signaling Technology, USA); and β-actin (1:5000, CWBIO, China) were applied for immunoblotting. All data were performed in triplicate, and the median values were the final outputs.
Immunohistochemistry
Formalin-fixed and paraffin-embedded tissue specimens were cut on a microtome into 4 μm sections. Antibodies against PRL-3 (1:80, Abcam, USA), Ki-67 (1:500, Abcam, USA), MMP7 (1:200, Abcam, USA), and IDH1 (1:100, NewEastBio, USA) were used for immunohistochemical staining. The density of PRL-3 and MMP7 staining in each specimen was scored according to the number of positively stained cells counted in ten high-power fields (200×) and the scores were as follows: 1 for 0-25%, 2 for 25-50%, 3 for 50-75%, and 4 for 75-100%. The intensity of PRL-3 and MMP7 immunostaining were scored as 0, 1, 2, and 3 for no, weak, moderate, and strong immunoreactivity, respectively. The density scores were multiplied by the corresponding immunostaining intensity scores to obtain PRL-3 and MMP7 histological scores. For the assessment of Ki-67 expression, ten high-power fields (200×) from each specimen were chosen randomly, and more than 300 cells were counted to determine the proliferation index, which represented the percentage of the total number of cells that were immunostained. To avoid possible technical errors, cell number counting was performed twice.
Invasion assay
Transwell invasion assays were performed in 6.5 mm Transwell chambers (Corning, USA) with 8 μm pores; the bottom surface of each membrane was pre-coated with 100 μL of Matrigel (BD, USA). Cells (5×104) were seeded in the upper chamber and the lower chamber was filled with 600 μL of culture medium. After 24 h, the cells on the bottom of the membrane were stained with crystal violet and counted under a microscope (100×). Five randomly selected fields were counted for each chamber.
Wound-closure assay
Cells were transfected, as indicated, and the cells (1.5×106 cells) were seeded in 12-well culture plates and cultured until confluent. A straight scratch was generated using a pipette tip to simulate a wound, then the average distance from the edge to the center was measured 24 h later.
EdU(5-ethynyl-2′-deoxyuridine) proliferation assay
Different groups of cells were seeded in 48-well plates at a density of 2×104 cells/well and cultured for 24 h. The assay was performed by EdU kit from RiboBio (Guangzhou, China) according to the manufacturer's instructions. The staining images were acquired using a florescence microscope (OLYMPUS, IX73) and the EdU-positive cells were counted in five random fields.
Cell Counting Kit-8 (CCK-8) proliferation assay
The transfected LN229, SHG44 and U87 cells were seeded into 96-well plates at a density of 5000 cells/well. After culturing for two days, CCK-8 (Dojindo, Japan) reagent was added, and the optical density value of each well was measured using a microplate reader (BioTek, USA) at an absorbance of 450 nm.
Intracranial model of glioma in nude mice
All animal experiments were performed following the Institutional Animal Care and Use Committee (IACUC) guidelines and were approved by the Institutional Animal Ethics Committee of the Fourth Military Medical University. Female BALB/C nude mice aged 4-6 weeks were purchased from the Shanghai Laboratory Animal Center (SLAC, China) and housed within a dedicated SPF facility. Mice were randomly divided into three groups (n=18 per group) and anesthetized (3% chloral hydrate, 10 μL/g weight, i.p.). A small burr hole, about 1.5 mm in diameter, which was located 0.5 mm anterior to bregma, and +2 mm mediolateral from the midline. A total of 5×106 LN229 cells or the cells stably infected with sh-NC or sh-PRL-3 lentivirus were delivered in 10 μL saline at a depth of 2.5 mm, with a Hamilton syringe at a rate of 0.5 µL/min for 20 min, and the syringe was stayed for 5 min before withdrawing the needle. The hole was sealed with bone wax, and the wound was sutured. 50 days after injection, mice were chosen from each group randomly (8 mice per group): three were sacrificed for protein extraction and Western Blot analysis, another five were perfused with saline followed by 4% paraformaldehyde (PFA). Then, the brain was fixed in 4% PFA at 4 °C overnight, paraffin-embedded and cut into 4 μm serial coronal sections, which were stained with H&E and immunohistochemical staining for PRL-3, Ki-67 and MMP7. Other mice in each group were fed for survival curves analysis.
Xenograft mouse model
Male BALB/C nude mice aged 4-6 weeks were purchased from the Shanghai Laboratory Animal Center (SLAC, China) and housed within a dedicated SPF facility at the Laboratory Animal Center of the Fourth Military Medical University. A total of 1×106 U251 or U87 cells stably infected with sh-NC or sh-PRL-3 virus were injected into the right flanks of the nude mice. The mice were imaged for Fluc activity using a bioluminescence imaging system (IVIS-100 Spectrum, USA) on days 10, 20 and 30 after implantation. Bioluminescence imaging was used to detect subcutaneous tumor growth. Light emission from animal tissue was measured using software provided by the vendor (Xenogen). The data were normalized to the bioluminescence at the initiation of treatment for each animal. In parallel, an equal number of tumor cells were inoculated into the same sites of other athymic mice to record their survival time. A portion of tumor tissues was used to extract proteins, and the remaining tissues were fixed in 4% paraformaldehyde and embedded in paraffin.
Statistical analyses
Statistical analyses were performed using SPSS 19.0 (IBM Corporation, USA) and GraphPad Prism 5.0 (GraphPad Software Inc., CA). The differences among/between sample groups were analyzed by one-way ANOVA or Student's t test. Pearson correlation analysis was used to determine the correlations between PRL-3 and Ki-67 expression. OS and PFS were estimated using the Kaplan-Meier method. The correlations between clinicopathological characteristics and PRL-3 expression were assessed using chi-square tests. Cox's proportional hazards regression model was applied for univariate and multivariate survival analyses. Data are presented as the mean ± standard deviation (SD) from at least three independent experiments and were analyzed with Student's t-test. Statistical significance was assigned at P<0.05 (*), P<0.01 (**) or P<0.001 (***).
Results
PRL-3 is up-regulated in glioblastoma specimens and is associated with poor patient survival
To investigate the potential role of PRL-3 in glioma, the expression of PRL-3 was evaluated by qPCR in 61 specimens from patients with glioblastoma and 10 normal brain specimens. We found that PRL‑3 mRNA expression was significantly higher in glioblastoma tissues than in normal brain specimens (Figure 1A). Western blot analysis of normal (n=7) and glioblastoma (n=7) specimens confirmed that PRL-3 was overexpressed in glioblastoma (Figure 1B).
PRL-3 is up-regulated in glioblastoma specimens and is associated with poor patient prognoses. A. Relative PRL-3 mRNA expression in normal brain specimens (n=10) and glioblastoma specimens (n=61). B. Western blot analysis of normal brain specimens (n=7) and glioblastoma specimens (n=7). C. Representative immunohistochemistry (IHC) staining with PRL-3 and Ki-67 in normal brain specimens and glioblastoma specimens. D. The correlation between Ki-67 expression and IHC PRL-3 score in glioblastoma (r2=0.4991, P<0.01). Pearson's coefficient tests were performed to assess significance. E. Kaplan-Meier analysis was used to evaluate the prognostic value of PRL-3 for overall survival (OS) and progression free survival (PFS) for glioblastoma patients. P values were determined by the log-rank test.
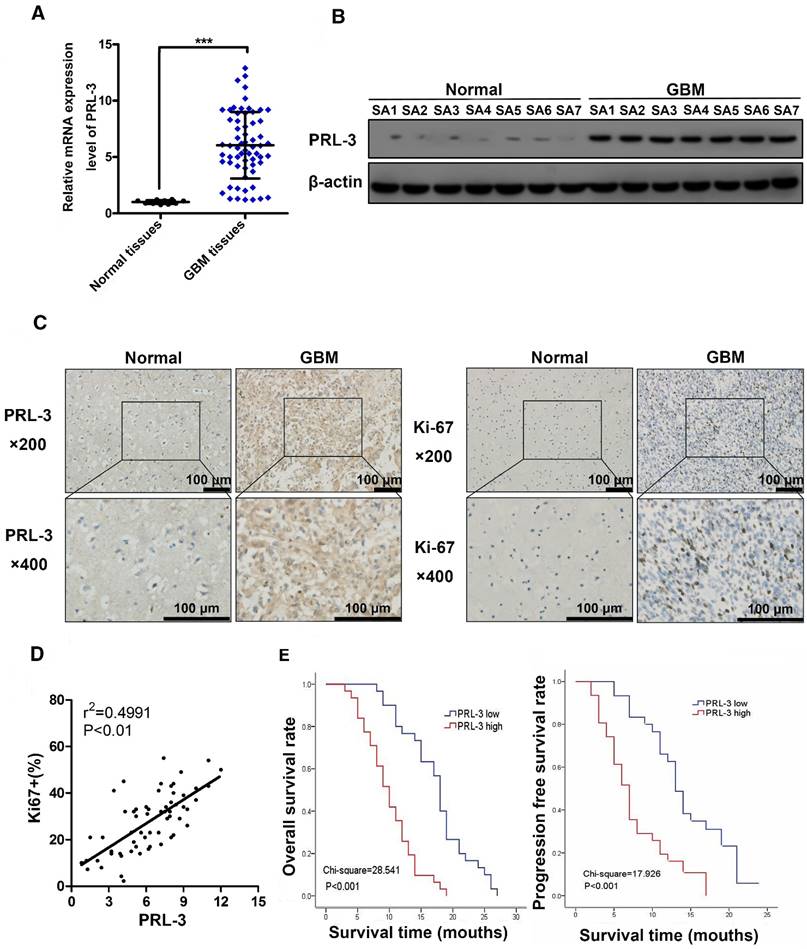
These findings were further supported by immunohistochemical analyses of normal (n=10) and glioblastoma (n=61) specimens (Figure 1C). Among the glioblastoma cases, 31 cases (50.8%) had high PRL-3 expression levels, and others had low or negative PRL-3 expression levels (Table 1). Ki-67 is a validated indicator of tumor cell proliferation activity and is strongly associated with unfavorable prognoses in glioma patients [25-27]. Our result indicated that high Ki-67 labeling index positively correlated with PRL-3 levels (Pearson's coefficient test r2=0.4991, P<0.01, Figure 1D, Table 1). High Ki-67 proliferation index was closely related to shorter overall survival (OS) and progression-free survival (PFS). Patients with glioblastoma were divided into low and high PRL-3 expression groups; the median OS and PFS in the high PRL-3 expression group (n=31) were 10 ± 0.916 and 10 ± 0.666 months, respectively, whereas those in the low PRL-3 expression group were 18 ± 0.537 and 13 ± 0.762 months, respectively (P<0.001, log-rank test, Figure 1E).
The correlation between clinicopathological characteristics and PRL-3 expression in patients with glioblastoma
Our statistical analysis found no significant association of PRL-3 with gender (P=0.300), age (P=1.00), or tumor lateralization (P=0.184), and there was no correlation between the PRL-3 expression level and IDH1 mutation (P=0.301) among the samples (Table 1). The statistical analyses were performed using the chi-square test. Values of P <0.05 were considered significant.
Univariate and multivariate analysis of overall survival in patients with glioblastoma
| Variables | Univariable analysis | Multivariable analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| OR | 95.0% CI for OR | P value | OR | 95.0% CI for OR | P value | |||
| lower | upper | lower | upper | |||||
| Gender | 0.932 | 0.555 | 1.568 | 0.792 | 0.069 | |||
| Age | 1.023 | 0.608 | 1.722 | 0.933 | 0.846 | |||
| Tumor lateralization | 0.937 | 0.550 | 1.597 | 0.811 | 0.062 | |||
| PRL-3 | 4.281 | 2.369 | 7.737 | <0.001 | 3.506 | 1.846 | 6.659 | <0.001 |
| Ki-67 (high vs. low) | 3.686 | 2.047 | 6.640 | <0.001 | 2.086 | 1.112 | 3.911 | 0.022 |
| IDH1 (mutation vs. wildtype) | 0.289 | 0.130 | 0.643 | 0.002 | 0.346 | 0.148 | 0.805 | 0.014 |
OR: odds ratio; CI: confidence interval
*Statistically significant (P < 0.05)
Cox hazard regression analyses of clinicopathological characteristics and PRL-3 expression in patients with glioblastoma
Univariate and multivariate analyses were performed to examine the effect of PRL-3 expression on glioma prognosis (Table 2 and Table 3). As shown in Table 2 and Table 3, both multivariate and univariate analyses indicated that PRL-3 significantly correlated with OS and PFS in glioblastoma patients. Accordingly, we propose that PRL-3 may be a new prognostic biomarker for glioblastoma patients.
Univariate and multivariate analysis of progression-free survival in patients with glioblastoma
| Variables | Univariable analysis | Multivariable analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| OR | 95.0% CI for OR | P value | OR | 95.0% CI for OR | P value | |||
| lower | upper | lower | upper | |||||
| Gender | 0.917 | 0.528 | 1.593 | 0.758 | 0.410 | |||
| Age | 1.040 | 0.598 | 1.808 | 0.890 | 0.470 | |||
| Tumor lateralization | 1.031 | 0.575 | 1.850 | 0.917 | 0.118 | |||
| PRL-3 | 3.267 | 1.788 | 5.969 | <0.001 | 2.592 | 1.373 | 4.893 | 0.03 |
| Ki67 (high vs. low) | 2.909 | 1.617 | 5.232 | <0.001 | 2.229 | 1.199 | 4.146 | 0.011 |
| IDH1 (mutation vs. wildtype) | 0.347 | 0.150 | 0.806 | 0.014 | 0.064 | |||
OR: odds ratio; CI: confidence interval
*Statistically significant (P < 0.05)
PRL-3 is required for tumor growth, migration and invasion
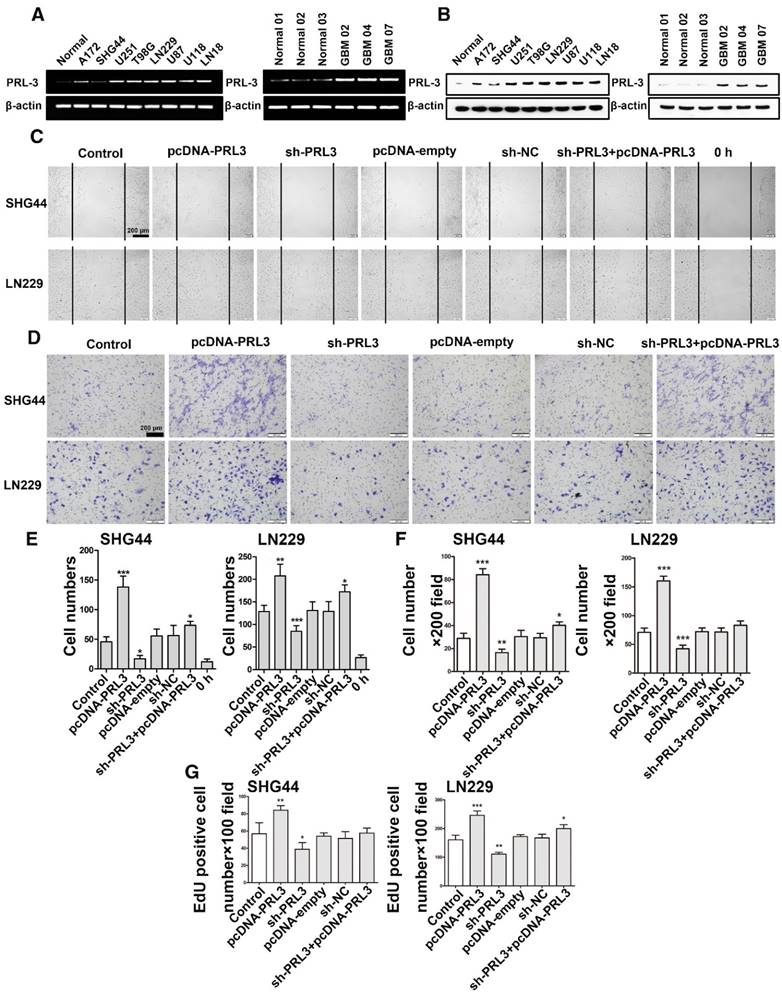
To investigate the biological functions of PRL-3, we detected endogenous PRL-3 expression levels in brain tissue from healthy people and U251, U87, LN229, A172, T98G, U118, LN18 and SHG44 glioma cell lines. Of these cell lines, U87 and SHG44 cells exhibited the highest and lowest PRL-3 expression, respectively (Figure 2A and 2B). In addition, we also detected PRL-3 expression levels in brain tissue from healthy people and three patient-derived glioblastoma cells: PRL-3 was highly expressed in patient-derived glioblastoma cells. We stably knocked down PRL-3 in U87, LN229 and SHG44 cells by using sh-PRL-3 lentivirus (Figure S1A and S1B), and then evaluated the migration ability of these cells using a wound-healing assay. Overexpressed PRL-3 'rescued' the migration ability in stably transfected SHG44, LN229 and U87 cell lines, whereas the sh-PRL-3 group exhibited the opposite effect (Figure 2C, 2E and Figure S1C). Similar results were obtained in Transwell assays, in which the migration and invasion capabilities were significantly increased in PRL-3-overexpressed SHG44, LN229 and U87 cells, and the effects were remarkably weakened by sh-PRL-3 (Figure 2D, 2F and Figure S1D). Furthermore, we determined the effect of PRL-3 on cell proliferation using a CCK-8 assay and an EdU proliferation assay. Compared with the control group and the sh-NC group, the number of EdU-positive cells increased markedly in the pcDNA-PRL-3 group, and inhibition of PRL-3 expression by sh-PRL-3 resulted in the opposite effect (Figure 2G, Figure S1E and Figure S2). The results of the CCK-8 assay were consistent with the EdU assay (Figure S1F). In addition, the migration and invasion capabilities were significantly increased in PRL-3-overexpressed patient-derived glioblastoma cells (GBM02) and the effects were remarkably weakened by sh-PRL-3 (Figure S3). Taken together, these data indicate that PRL-3 increased the degree of malignancy in glioma cells.
PRL-3 promotes diffuse glioma cell proliferation, invasion, and migration. A. Real-time quantitative PCR was performed to detect endogenous PRL-3 mRNA expression in brain tissue from healthy people and U251, U87, LN229 and SHG44 cell lines. B. Western blot analysis of PRL-3 protein expression in brain tissue from healthy people and U251, U87, LN229 and SHG44 cell lines. The results were quantified using densitometry scanning. C. The wound-healing assay was used to investigate the migration capacity of the SHG44 and LN229 cells. D. The effect of PRL-3 on the invasion of SHG44 and LN229 cells were evaluated by the Transwell assay. E and F. The statistical data of C and D. The values are the mean ± SD of three independent experiments. G. EdU proliferation assay of SHG44 and LN229 cells. The values are the mean ± SD of three independent experiments.
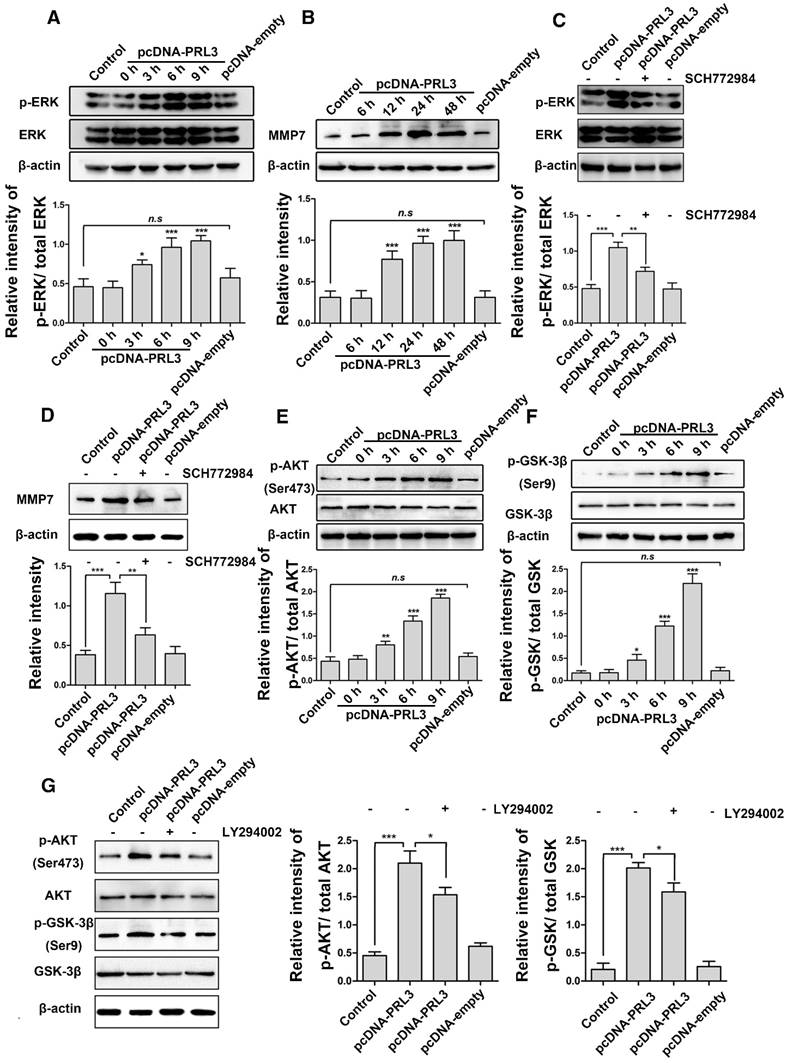
PRL-3 promotes MMP7 expression by activating the ERK pathway in diffuse glioma cells. A. LN229 cells were transfected with pcDNA-PRL-3, and the levels of Erk1/2 and phosphorylated Erk1/2 (p-Erk1/2) were detected by western blotting. B. MMP7 protein expression was increased following Erk1/2 activation. C and D. Both Erk1/2 activation and MMP7 expression were inhibited compared with the group without the inhibitor SCH772984 (lane 2). E and F. Phosphorylated Akt and GSK-3β were analyzed by western blotting as described above. G. The transfected cells pretreated with the PI3K inhibitor LY294002 (lane 3) were analyzed by western blotting to detect levels of Akt, p-Akt, GSK-3β and p-GSK-3β.
PRL-3 promotes MMP7 expression by activeting ERK pathway in diffuse glioblastoma cells
To uncover the molecular mechanism by which PRL-3 regulates glioma cell migration and proliferation, LN229 cells were transfected with pcDNA-PRL-3, and the expression of phosphorylated Erk1/2 protein was determined by western blotting. As shown in Figure 3A, PRL-3 indeed increased the phosphorylation level of Erk1/2. Meanwhile, the expression of MMP7 was also increased accompanied by Erk1/2 activation (Figure 3B). To confirm the role of Erk1/2 in MMP7 expression, we treated LN229 cells (transfected with PRL-3) with the ERK inhibitor SCH772984. As expected, both Erk1/2 activation and MMP7 expression were inhibited compared with the group without inhibitor (Figures 3C and 3D). These data indicate that PRL-3 promotes LN229 cell invasion and migration through activating the Erk1/2/MMP7 pathway.
PI3K/AKT is a major survival pathway in glioblastoma. Next, we explored whether PRL-3 signals through the PI3K/Akt pathway. As shown in Figures 3E and 3F, LN229 cells transfected with pcDNA-PRL-3 exhibited higher levels of phosphorylated Akt at Ser473, which in turn inactivated GSK-3β at Ser9. We determined that the effects of PRL-3 on Akt and GSK-3β were attenuated by the PI3K inhibitor (Figure 3G).
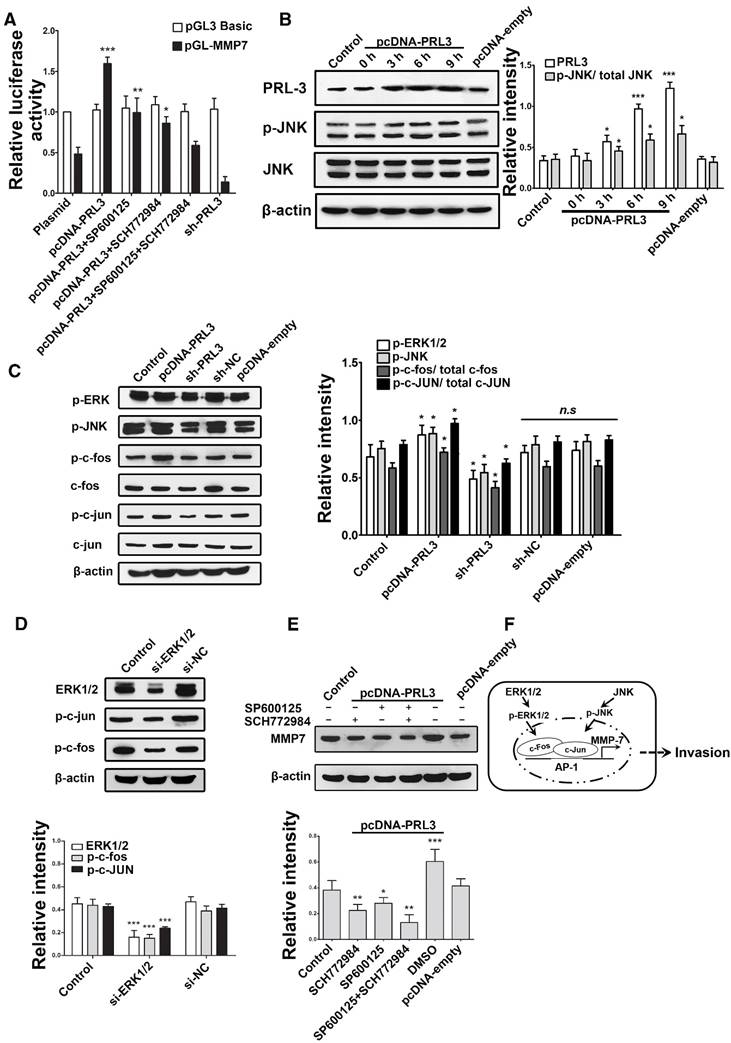
ERK and JNK mediate the effect of PRL-3 by enhancing c-Fos/p-c-Fos and c-Jun/p-c-Jun, which promote MMP7 expression by binding on its promoter region directly
To clarify whether ERK mediates the effect of PRL-3, we used bioinformatics tools to find possible transcription factor binding sites on the MMP7 promoter. The AP-1 (c-Fos and c-Jun) was found to be the potential direct regulator of MMP7, which can also be effected by ERK and JNK. So, we used the luciferase reporter plasmid that contains the MMP7 promoter region and ERK/JNK inhibitor (SCH772984 and SP600125) to investigate which pathway is the direct causation of higher MMP7 expression. The results indicate that both ERK and JNK are responsible for the higher MMP7 expression; ERK seems to contribute more than JNK (Figure 4A), and PRL-3 could also increase the phosphorylation level of JNK (Figure 4B). To further confirm c-Fos and c-Jun were involved in the effect, we detected the expression of p-JNK, c-Fos/p-c-Fos, c-Jun/p-c-Jun and p-ERK after PRL-3 overexpression or knockdown in a U87 model: the results claim that both c-Fos/p-c-Fos and c-Jun/p-c-Jun can be affected by PRL-3 (Figure 4C).
Similar results were obtained in patient-derived glioblastoma cells (GBM02): PRL-3 may increase the phosphorylation level of Erk1/2 and JNK. At the same time, the expression of MMP7 was also increased, accompanied by Erk1/2 and JNK activation (Figure S3). Then, siRNA was used to interfere with ERK expression and evaluate its effect on p-c-Fos and p-c-Jun; both p-c-Fos and p-c-Jun expression can be affected by ERK (Figure 4D). In addition, both ERK and JNK were responsible for the PRL-3-induced MMP7 expression, with ERK being mainly responsible (Figure 4E). Our data indicate that ERK mediates the effect of PRL-3 by enhancing c-Fos/p-c-Fos and c-Jun/p-c-Jun, which can promote MMP7 expression by binding on its promoter region directly. But, ERK isn't the only mediator: JNK makes a partial contribution to mediating the effect of PRL-3 on MMP7 expression (Figure 4F).
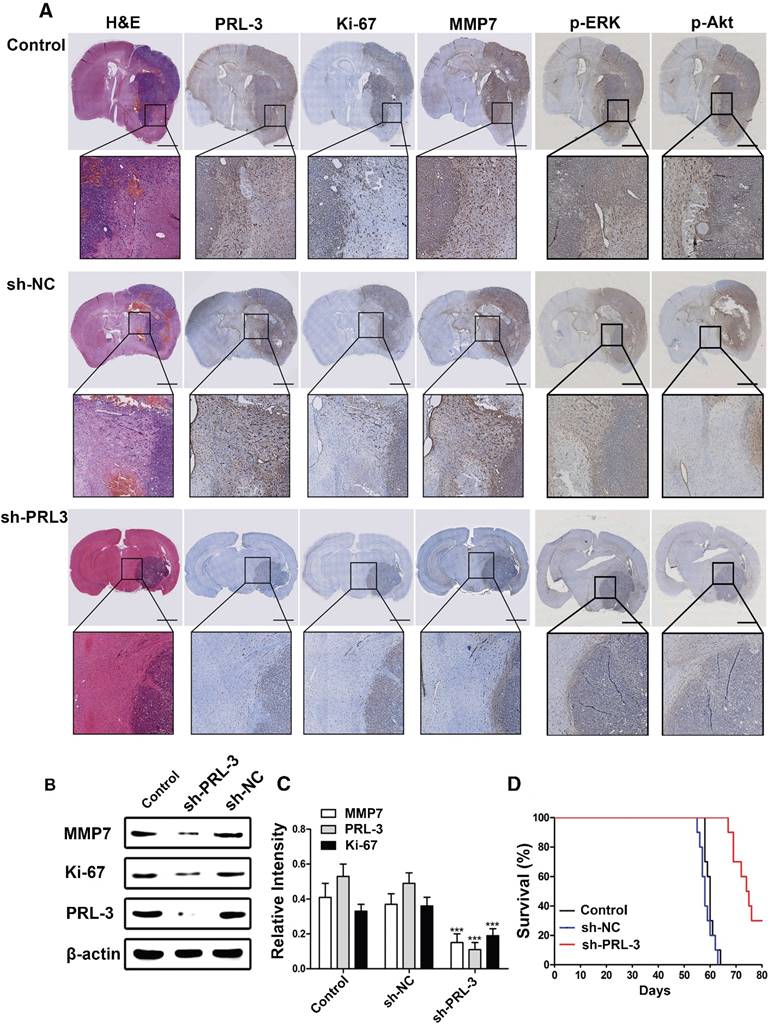
Knockdown of PRL-3 inhibits invasion and prolongs survival time in an orthotopic glioblastoma nude mouse model
To investigate the role of PRL-3 in tumorigenesis, a total of 5×106 LN229 cells or the cells stably infected with sh-NC or sh-PRL-3 lentivirus were injected into the right frontal lobe of nude mice. 50 days after implantation, mice were sacrificed for evaluation. As showed in Figure 5A, the growth and invasion ability of the tumor was significantly inhibited by knockdown of PRL-3, and the IHC and western blot analysis revealed that MMP7 and Ki-67 protein levels decreased due to PRL-3 knockdown (Figure 5B and 5C). Moreover, lower PRL-3 extended the survival time of the sh-PRL-3 group (Figure 5D). To further investigate the role of PRL-3 in tumorigenesis, a total of 5×106 U251 or U87 cells stably transfected with sh-NC or sh-PRL-3 lentivirus were injected into the right flanks of nude mice. In these models, sh-PRL-3 resulted in a significant reduction of the subcutaneous tumor volume compared to the sh-NC and control groups. In vivo bioluminescence imaging analysis of the mice at 10, 20 and 30 days after implantation revealed that the growth and invasion of tumors were significantly inhibited by the decreased expression of PRL-3 (Figure S4A and Figure S5A). PRL-3 knockdown extended the survival time of the sh-PRL-3 group (Figure S4B and Figure S5B). Moreover, western blot analysis revealed that MMP7 and Ki-67 protein levels decreased due to PRL-3 knockdown (Figure S4C, D and Figure S5C, D). Similar results were observed in MMP7-, Ki67- and PRL-3-stained sections (Figure S4E and Figure S5E). These data indicate that PRL-3 promoted tumorigenesis in vivo and in vitro, and targeting PRL-3 is a potential candidate for the clinical treatment of glioblastoma.
ERK and JNK mediate the effect of PRL-3 by enhancing c-Fos/p-c-Fos and c-Jun/p-c-Jun, which promote MMP7 expression by binding on its promoter region directly. A. The luciferase reporter plasmid that contains the MMP7 promoter region was used with ERK/JNK inhibitor (SCH772984 and SP600125) to investigate which pathway is the direct causation of higher MMP7 expression. B. Cells were transfected with pcDNA-PRL-3, and the levels of JNK and phosphorylated JNK (p-JNK) were detected by western blotting. C. Both c-Fos/p-c-Fos and c-Jun/p-c-Jun can be affected by PRL-3. D. Both p-c-Fos and p-c-Jun expression can be affected by ERK. E. Both ERK and JNK are responsible for the PRL-3-induced MMP7 expression. F. Schematic model of PRL-3-induced MMP7 overexpression. *P<0.05, **P<0.01, ***P<0.001 compared with the control group.
Down-regulation of PRL-3 suppresses tumor growth and MMP7 expression in an orthotopic glioma mouse model. A. A total of 5×106 LN229 cells or the cells stably infected with sh-NC or sh-PRL-3 lentivirus were injected into the right frontal lobe of nude mice. 50 days after implantation, mice were sacrificed for evaluation. Representative H&E and IHC staining of Ki-67, MMP7, PRL-3, p-ERK and p-Akt are shown for coronal sections with indicated administrations. Scale bar=1000 μm. B and C. Total proteins were extracted from the dissected tumor samples. The expressions of PRL-3, Ki-67 and MMP7 were analyzed by western blotting. D. Kaplan-Meier survival analysis was performed to evaluate the overall survival time. P values were determined by the log-rank test (P<0.001). *P<0.05, **P<0.01, ***P<0.001 compared with the control group.
Discussion
Glioblastoma is one of the most aggressive human tumors. Although multimodal treatment strategies involving surgical resection, radiation, and chemotherapy have been used, the prognosis of glioblastoma patients remains very poor [6, 28]. Significant efforts have been made to identify prognostic molecular biomarkers that could provide knowledge regarding glioblastoma formation and progression, such as 1p/19q co-deletion, the methylation of MGMT promoter, EGFR amplification and IDH1 mutations [9-12, 28].
PRL-3 is a protein tyrosine phosphatase that was previously identified to be a metastasis-related gene [15]. High PRL-3 expression promotes gastric cancer proliferation and migration and has a negative impact on prognosis [29-31]. Through interaction with repressor/activator protein 1 (RAP1), PRL-3 activates NF-κB signaling and downstream cascade signal transduction [32]. Recently, PRL-3 has been demonstrated to be a predictive molecular marker of prognosis after curative surgery in certain malignant tumor types. Kong et al. [3] confirmed PRL-3 expression in gliomas by in situ hybridization and immunohistochemistry. However, the clinical relevance of PRL-3 and its mechanism in glioma and glioblastoma remain unknown.
In the present study, we used relatively large numbers of clinical tissue samples to evaluate PRL-3 expression levels and the clinical significance in glioblastoma. We detected PRL-3 mRNA and protein expression levels in 61 glioblastoma multiforme tissue samples by qPCR and IHC, respectively. Ki-67 is necessary for DNA synthesis and an independent prognostic molecular factor in glioblastoma [11, 25, 27]. Subsequently, we analyzed Ki-67 expression in our glioblastoma samples and the Pearson correlation analysis revealed that the Ki-67 indexes were positively correlated with PRL-3 expression. These results imply that aberrantly high expression levels of PRL-3 are likely associated with adverse prognoses. We also downloaded a data file (Glioblastoma Multiforme (TCGA) 604 samples/591 patients) from cBioPortal [33, 34], which contains U133 microarray results. We chose pathological data containing survival time and divided the specimens into 2 groups (PRL-3 high expression and low expression) based on 50% quantile. After that, we performed a survival analysis and log-rank test for comparation, and the result (data not shown) indicates that patients with higher PRL-3 expression had a poor prognosis. This is in accordance with our finding in this manuscript. To further investigate the relationship between PRL-3 and the poor prognosis of glioblastoma, we examined the OS and PFS rates using Kaplan-Meier survival analyses. Higher PRL-3 expression levels were associated with unfavorable prognoses, but there was no correlation between the PRL-3 expression level and IDH1 mutation for OS and PFS, according to the chi-square test result. In addition, univariate and multivariate analyses indicated that PRL-3 expression was a new prognostic factor for OS and PFS. Taken together, the above results confirm that PRL-3 might serve as a novel prognostic factor and a potential therapeutic target for patients with glioblastoma. To clarify the biological behavior of PRL-3 in glioblastoma, we detected PRL-3 expression in 8 glioma cell lines, and constructed a PRL-3 overexpression plasmid and lentiviral vector to stably knock down PRL-3, with which we conducted stable transduction of LN229, U87 and SHG44 cell lines, and pcDNA-PRL-3 was applied to 'rescue' the function of PRL-3. Our results demonstrated that PRL-3 could promote the proliferation, invasion and migration of LN229, U87, SHG44 and patient-derived glioblastoma cells. Hoverer, there was no previous research on the mechanisms of PRL-3 in cell invasion, migration and proliferation in glioblastoma cells until now. Jiang et al. [35] demonstrated that the up-regulation of PRL-3 induced the activation of the PI3K/Akt pathway and then enhanced survival and resistance to stress-induced apoptosis in colon cancer cells. A recent study confirmed that PRL-3 promotes gastric cancer migration and invasion through a NF-κB-HIF-1α-miR-210 axis [36]. The latest research showed that the loss of PRL-3 alters ERK and p38 signaling in triple-negative breast cancer cells [37]. Zhan et al. [38] found that the knockdown of PRL-3 weakened MMP9 expression in endometrial stromal cells. In this study, we provided evidence that PRL-3 could activate PI3K, Akt, and Erk1/2, whereas GSK-3β was inhibited. We further detected the MMPs, which are downstream target proteins of ERK and JNK. Increased MMP7 expression was accompanied by the phosphorylation of Erk1/2 and activated JNK. In vivo, consistent with the above results, the inhibition of PRL-3 expression suppressed tumor growth and prolonged the survival time of orthotopic glioblastoma and subcutaneous tumors in nude mice.
In summary, high PRL-3 expression is associated with a poor prognosis in glioblastoma patients. The down-regulation of PRL-3 inhibits the proliferation, invasion and migration of glioblastoma cells in vitro and in vivo. More importantly, our findings demonstrate that PRL-3 can serve as a valuable prognostic marker and is a promising therapeutic target for glioblastoma multiforme.
Abbreviations
PRL-3: phosphatase of regenerating liver-3; OS: overall survival; PFS: progression-free survival; CCK-8: Cell Counting Kit-8; MMP7: matrix metalloproteinase 7; MGMT: methylation of the O6 methylguanine methyltransferase; IDH1: isocitrate dehydrogenase 1; qRT-PCR: quantitative real-time PCR; RAP1: repressor/activator protein 1.
Acknowledgements
The authors thank Pro. Jing Ye in Department of Pathology of our Hospital, for help in IDH1 mutation analysis of tissue samples, and Pro. Minggao Zhao in the Department of Pharmacology of our university, for help in intracerebral infusion of nude mice. In addition, we appreciate the services of Nature Publishing Group Language Editing for manuscript polishing.
Funding
This work was supported by Natural Science Foundation of China. [NO. 81172395, 30801364, 81273279 and 81373201]
Supplementary Material
Supplementary figures.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Schwartzbaum JA, Fisher JL, Aldape KD. et al. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol. 2006;2:494-503
2. Shan S, Hui G, Hou F. et al. Expression of metastasis-associated protein 3 in human brain glioma related to tumor prognosis. Neurol Sci. 2015;36:1799-804
3. Kong L, Li Q, Wang L. et al. The value and correlation between PRL-3 expression and matrix metalloproteinase activity and expression in human gliomas. Neuropathology. 2007;27:516-21
4. Barbano R, Palumbo O, Pasculli B. et al. A miRNA signature for defining aggressive phenotype and prognosis in gliomas. PLoS One. 2014;9:e108950
5. Huang J, DeWees TA, Badiyan SN. et al. Clinical and dosimetric predictors of acute severe lymphopenia during radiation therapy and concurrent temozolomide for high-grade glioma. Int J Radiat Oncol Biol Phys. 2015;92:1000-7
6. Louis DN, Perry A, Reifenberger G. et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803-20
7. Al-Aidaroos AQ, Zeng Q. PRL-3 phosphatase and cancer metastasis. J Cell Biochem. 2010;111:1087-98
8. Pryczynicz A, Guzińska-Ustymowicz K, Chang XJ. et al. PTP4A3 (PRL-3) expression correlate with lymphati metastases in gastric cancer. Folia Histochem Cytobiol. 2010;48:632-6
9. Hegi ME, Diserens AC, Gorlia T. et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997-1003
10. Camara-Quintana JQ, Nitta RT, Li G. Pathology: commonly monitored glioblastoma markers: EFGR, EGFRvIII, PTEN, and MGMT. Neurosurg Clin N Am. 2012;23:237-46
11. Zeng A, Hu Q, Liu Y. et al. IDH1/2 mutation status combined with Ki-67 labeling index defines distinct prognostic groups in glioma. Oncotarget. 2015;6:30232-88
12. Verhaak RG, Hoadley KA, Purdom E. et al. An integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98-110
13. Ustaalioglu BB, Bilici A, Barisik NO. et al. Clinical importance of phosphatase of regenerating liver-3 expression in breast cancer. Clin Transl Oncol. 2012;14:911-22
14. Scott LM, Lawrence HR, Sebti SM. et al. Targeting protein tyrosine phosphatases for anticancer drug discovery. Curr Pharm Des. 2010;16:1843-62
15. Saha S, Bardelli A, Buckhaults P. et al. A phosphatase associated with metastasis of colorectal cancer. Science. 2001;294:1343-6
16. Navis AC, van den Eijnden M, Schepens JT. et al. Protein tyrosine phosphatases in glioma biology. Acta Neuropathol. 2010;119:157-75
17. Jeong KW, Kang DI, Lee E. et al. Structure and backbone dynamics of vanadate-bound PRL-3: comparison of 15N nuclear magnetic resonance relaxation profiles of free and vanadate-bound PRL-3. Biochemistry. 2014;53:4814-25
18. Ming J, Jiang Y, Jiang G. et al. Phosphatase of regenerating liver-3 induces angiogenesis by increasing extracellular signal-regulated kinase phosphorylation in endometrial adenocarcinoma. Pathobiology. 2014;81:1-7
19. Campbell AM, Zhang ZY. Phosphatase of regenerating liver: a novel target for cancer therapy. Expert Opin Ther Targets. 2014;18:555-69
20. Bessette DC, Qiu D, Pallen CJ. PRL PTPs: mediators and markers of cancer progression. Cancer Metastasis Rev. 2008;27:231-52
21. Al-Aidaroos AQ, Yuen HF, Guo K. et al. Metastasis-associated PRL-3 induces EGFR activation and addiction in cancer cells. J Clin Invest. 2013;123:3459-71
22. Lee J, Kotliarova S, Kotliarov Y. et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391-403
23. Fael Al-Mayhani TM, Ball SL, Zhao JW. et al. An efficient method for derivation and propagation of glioblastoma cell lines that conserves the molecular profile of their original tumours. J Neurosci Methods. 2009;176:192-9
24. Mu N, Gu J, Huang T. et al. A novel NF-κB/YY1/microRNA-10a regulatory circuit in fibroblast-like synoviocytes regulates inflammation in rheumatoid arthritis. Sci Rep. 2016;6:20059
25. Raghavan R, Steart PV, Weller RO. Cell proliferation patterns in the diagnosis of astrocytomas, anaplastic astrocytomas and glioblastoma multiforme: a Ki-67 study. Neuropathol Appl Neurobiol. 1990;16:123-33
26. Colman H, Giannini C, Huang L. et al. Assessment and prognostic significance of mitotic index using the mitosis marker phospho-histone H3 in low and intermediate-grade infiltrating astrocytomas. Am J Surg Pathol. 2006;30:657-64
27. Sallinen PK, Haapasalo HK, Visakorpi T. et al. Prognostication of astrocytoma patient survival by Ki-67 (MIB-1), PCNA, and S-phase fraction using archival paraffin-embedded samples. J Pathol. 1994;174:275-82
28. Jansen M, Yip S, Louis DN. Molecular pathology in adult gliomas: diagnostic, prognostic, and predictive markers. Lancet Neurol. 2010;9:717-26
29. Dai N, Lu AP, Shou CC. et al. Expression of phosphatase regenerating liver 3 is an independent prognostic indicator for gastric cancer. World J Gastroenterol. 2009;15:1499-505
30. Wang Z, He YL, Cai SR. et al. Expression and prognostic impact of PRL-3 in lymph node metastasis of gastric cancer: its molecular mechanism was investigated using artificial microRNA interference. Int J Cancer. 2008;123:1439-47
31. Wang H, Vardy LA, Tan CP. et al. PCBP1 suppresses the translation of metastasis-associated PRL-3 phosphatase. Cancer Cell. 2010;18:52-62
32. Teo H, Ghosh S, Luesch H. et al. Telomere-independent Rap1 is an IKK adaptor and regulates NF-kappaB-dependent gene expression. Nat Cell Biol. 2010;12:758-67
33. Cerami E, Gao J, Dogrusoz U. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-4
34. Gao J, Aksoy BA, Dogrusoz U. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1
35. Jiang Y, Liu XQ, Rajput A. et al. Phosphatase PRL-3 is a direct regulatory target of TGFbeta in colon cancer metastasis. Cancer Res. 2011;71:234-44
36. Zhang C, Tian W, Meng L. et al. PRL-3 promotes gastric cancer migration and invasion through a NF-κB-HIF-1α-miR-210 axis. J Mol Med (Berl). 2016;94:401-15
37. den Hollander P, Rawls K, Tsimelzon A. et al. Phosphatase PTP4A3 promotes triple-negative breast cancer growth and predicts poor patient survival. Cancer Res. 2016;76:1942-53
38. Zhan H, Ma J, Ruan F. et al. Elevated phosphatase of regenerating liver 3 (PRL-3) promotes cytoskeleton reorganization, cell migration and invasion in endometrial stromal cells from endometrioma. Hum Reprod. 2016;31:723-33
Author contact
![]() Corresponding authors: Qingdong Guo, Department of Neurosurgery, Xijing Hospital, Fourth Military Medical University, #127 West Changle Road, Xi'an, China; Email: guoqd1991cn; Tel: +86 (0) 29-84773488; Fax: +86 (0) 29-84773488. Wei Zhang, State Key Laboratory of Cancer Biology, Biotechnology Center, School of Pharmacy, Fourth Military Medical University, #169 West Changle Road, Xi'an, China; Email: zhangw90edu.cn; Tel: +86 (0) 29-84774775; Fax: +86 (0) 29 84774775. Lei Shang, Department of Health Statistics, School of Public Health, Fourth Military Medical University, Xi'an, China; Email: shangleiedu.cn; Tel: +86 (0) 29-84773490; Fax: +86 (0) 29 84773490.
Corresponding authors: Qingdong Guo, Department of Neurosurgery, Xijing Hospital, Fourth Military Medical University, #127 West Changle Road, Xi'an, China; Email: guoqd1991cn; Tel: +86 (0) 29-84773488; Fax: +86 (0) 29-84773488. Wei Zhang, State Key Laboratory of Cancer Biology, Biotechnology Center, School of Pharmacy, Fourth Military Medical University, #169 West Changle Road, Xi'an, China; Email: zhangw90edu.cn; Tel: +86 (0) 29-84774775; Fax: +86 (0) 29 84774775. Lei Shang, Department of Health Statistics, School of Public Health, Fourth Military Medical University, Xi'an, China; Email: shangleiedu.cn; Tel: +86 (0) 29-84773490; Fax: +86 (0) 29 84773490.
 Global reach, higher impact
Global reach, higher impact