13.3
Impact Factor
Theranostics 2018; 8(4):874-877. doi:10.7150/thno.23364 This issue Cite
Editorial
LET's sponge: How the lncRNA PFL promotes cardiac fibrosis
1. Goethe-University, Institute for Cardiovascular Physiology, Frankfurt am Main, Germany;
2. German Center of Cardiovascular Research (DZHK), Partner site RheinMain, Frankfurt, Germany
Received 2017-10-17; Accepted 2017-10-26; Published 2018-1-1
Abstract
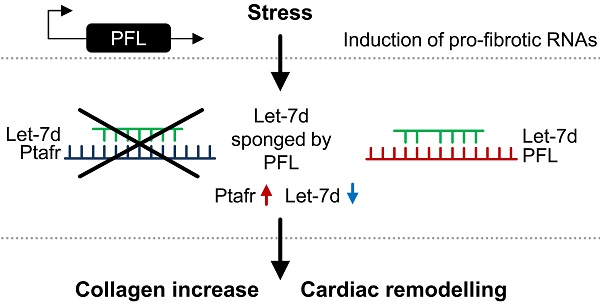
Compared to their protein-coding counterparts, almost nothing is known about the role of long noncoding RNAs (lncRNAs) in cardiac fibrosis. In the current report, Liang and Pan et al. characterized the pro-fibrotic lncRNA PFL in respect to cardiac fibrosis in mice. PFL was upregulated in the hearts of mice after myocardial infarction and in fibrotic cardiac fibroblasts. Moreover, PFL competitively sponged the cardio-protective miRNA let-7d in cardiac fibroblasts. Knockdown of platelet activating factor receptor (PTAFR) was shown to affect the pro-fibrotic collagen production mediated by PFL. PTAFR overexpression also led to collagen production and RNA abundance of PTAFR was also regulated by miRNA let-7d. Therefore, the PFL/PTAFR/let-7d-dependent gene regulatory mechanism proposed by the authors manifests the hypothesis of competing endogenous RNAs to cardiac fibrosis.
Keywords: LncRNA, PFL, miRNA, ceRNA, Cardiac Fibrosis
Heart failure is often a consequence of a stress-triggered cardiac remodelling process. Cardiac fibrosis can occur during pathological cardiac remodelling and is characterized by enhanced fibroblast activity and extracellular matrix production within the myocardium. This affects the structure and function of cardiomyocytes, endothelial cells, and interstitial cells [1].
With technical advances in molecular biology and bioinformatics, numerous new RNA transcripts have been identified; many classes of which have different functions, sizes and structures [2]. This is especially true for non-coding RNAs (ncRNAs) which, by definition, do not encode for proteins. NcRNAs are broadly classified as small (<200 nucleotides, such as microRNAs (miRNA), small interfering RNAs, Piwi-interacting RNAs, small nucleolar RNAs or transfer RNAs) or long (>200 nucleotides) ncRNAs, both of which are highly dynamic and diverse RNA molecules [2,3]. They offer great potential in biomarker research and disease treatment and already several studies exist in which ncRNAs have been shown to improve our understanding of cardiovascular diseases [4,1,5].
Liang and Pan et al. [6] have reported in this issue the pro-fibrotic lncRNA (PFL, also known as NONMMUT022555) to be involved in mouse cardiac fibrosis. Like collagen, fibronectin, α-smooth muscle actin and connective tissue growth factor, PFL was identified as being upregulated in the hearts of mice after myocardial infarction (MI) as well as in cardiac fibroblasts exposed to transforming growth factor-ß1, angiotensin II and 20% serum. Knockdown of PFL reduced collagen deposition, improved cardiac function and decreased fibrotic protein expression in MI mice [6].
To mechanistically explain the function of PFL, the authors made use of former investigations regarding lncRNAs as competing endogenous RNAs (ceRNA). The ceRNA hypothesis is a proposed explanation for target competition by RNAs with shared miRNA-binding sites (Fig. 1A). This mechanism adds a novel layer of gene regulation [7]. The authors identified two active binding sites for the miRNA let-7d within PFL, which are crucial for the interaction of the lncRNA with the miRNA [6]. As a consequence of the interaction, PFL abundance decreased. Moreover, the expression of genes previously upregulated by PFL, such as the collagens, could be reduced by addition of let-7d (Fig. 1B&C). One has to note that miRNA let-7d was itself reduced up to 28 days post-infarction and its overexpression resulted in a cardioprotective phenotype in cardiac fibrosis and MI [6].
Several studies have already reported the importance of the let-7 family members for cardiac fibrosis. Examples include let-7i, which negatively regulates Angiotensin II-induced cardiac inflammation and fibrosis [8], and let-7c, whose inhibition reduced cardiac fibrosis and improved cardiac function in MI mice [9].
As in the present study, lncRNA-miRNA interactions during cardiac fibrosis have been shown to be very important. LncRNA Gas5 (growth arrest-specific 5) inhibits the activation of cardiac fibroblasts and regulates miR-21 [10]. An opposite effect was shown for lncRNA H19, which increased cardiac fibroblast proliferation [11]. Inhibition of lncRNA Meg3 (maternally expressed gene 3) prevented cardiac fibrosis [12] and lncRNA Wisper (Wisp2 super-enhancer-associated RNA) had a regulatory impact on cardiac fibrosis gene programs and was associated with severe cardiac fibrosis [13]. Furthermore, several ceRNA-mediated mechanisms in cardiac fibrosis or the involvement of let-7 miRNAs with ceRNAs have been shown. The messenger RNA of the proto-oncogene MYCN and lncRNAs H19 and CCAT1 (colon cancer associated transcript 1) sponged let-7 [14-16], and the pro-fibrotic lncRNA myocardial infarction associated transcript (MIAT) sponged miR-21 in cardiac fibroblasts [17]. The present study combines miRNA let-7d, ceRNA PFL and cardiac fibrosis.
How PFL promotes miRNA let-7d abundance. A. A miRNA could have more than one target, thereby opening the possibility that other RNAs compete for miRNA binding. B. Under normal conditions the main target of let-7d is Ptafr, which promotes cell homeostasis. C. Under stress conditions, PFL is upregulated and binds to let-7d, which leads to an upregulation of Ptafr and collagen and increased cardiac remodelling. D. The different aspects of conservation are shown for a typical RNA locus, with the sequence conservation level shown in detail. One should take into account additional sequence elements, potential RNA domains, processing sites and basic conditions, which are required for functional gene expression.
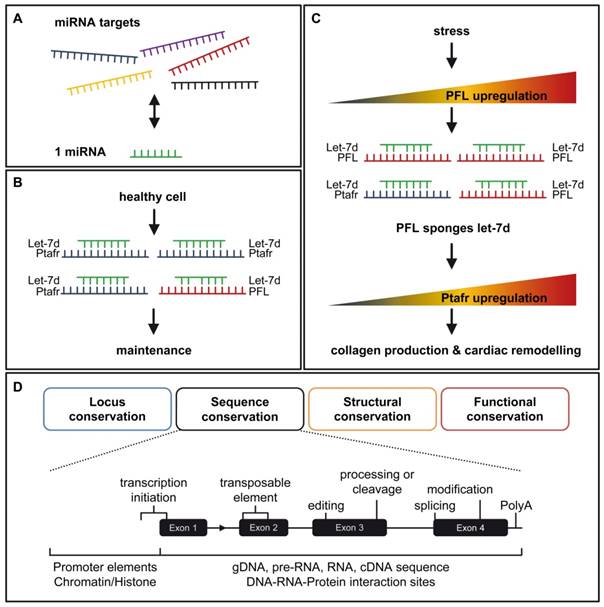
As in the case of the ceRNAs, PFL also required a transcript for competition to miRNA let-7d. Liang and Pan et al. validated two binding sites for let-7d within the platelet-activating factor receptor Ptafr. Both binding sites were conserved from human to mouse and were required for let-7d action. Interestingly, the pro-fibrotic action of PFL was inhibited by Ptafr knockdown [6]. This could qualify Ptafr as a central factor of PFL signaling for the fibrotic process.
Nevertheless, the understanding and interpretation of RNA sequence, function, metabolism, conservation, classification and structure remains challenging due to the extreme fragility of these definitions [18-20]. Recent investigations found that over a thousand long intervening RNAs have conserved functions across mammals, but only several hundred were found beyond mammals. More importantly, major changes to gene architecture are tolerated. This offers the possibility that only short specific parts of the transcript are enough for functional consequences [21]. Assuming these short specific parts are relevant RNA domains, they challenge on the one hand secondary structure conservation for interaction and on the other hand sequence-based alignments with RNAs or DNAs.
Transposable elements, a group of repetitive, mobile sequence elements, are often found in lncRNA exons. It has been proposed that transposable elements are candidates for functional elements of lncRNAs [22]. Among others, MANTIS and ANRIL (antisense noncoding RNA in the INK4 locus) are examples of lncRNAs containing such transposable elements. A small part of MANTIS Exon3 contained a conserved Alu element, which was sufficient for its biological effects [4]. Alu elements were also functionally relevant for ANRIL [23]. The increasingly important class of such small sequence parts justifies the development of novel strategies for a comparative analysis of genes across species and should lead to various strategies for the identification of lncRNA homologues [20] (Fig. 1D). Additionally, the basic conditions for establishing gene expression in the area require extensive research on transcription factor binding motifs, DNA sequence, promoter conservation or RNA processing sites. The impact of RNA modifications is also totally unclear. More than 100 different RNA modifications are known and the number is growing continuously [24]. Initially thought to be important for RNA maturation, it has been shown that RNA modifications are able to change the fate of RNAs. Poly(A)+ RNA N6 methylation (m6A) occurred in response to UV, reflecting an RNA-mediated response to DNA damage by recruiting DNA polymerase K to DNA damage sites [25]. Therefore, it is even more important to add RNA modification conservation to the list of suitable elements used for strategies to find RNA homologues. Together with transposable elements, RNA modifications are also potential disease-relevant elements.
It remains unclear to what extent PFL is modified, and how well its structure and sequence are conserved. However, the finding that lncRNA PFL acts as a ceRNA for miRNA let-7d binding offers an interesting option for therapeutic and biomarker development in the field of cardiac fibrosis.
Acknowledgements
The author thanks Ralf P. Brandes and James A. Oo for critical comments on the manuscript.
Source of Funding
This work was supported by the Goethe-Universität Frankfurt.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Thum T. Noncoding RNAs and myocardial fibrosis. Nat Rev Cardiol. 2014;11:655-663
2. Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77-94
3. Leisegang MS, Schröder K, Brandes RP. Redox Regulation and Noncoding RNAs. Antioxid Redox Signal. 2017
4. Leisegang MS, Fork C, Josipovic I, Richter FM, Preussner J, Hu J, Miller MJ, Epah J, Hofmann P, Günther S, Moll F, Valasarajan C, Heidler J. et al. Long Noncoding RNA MANTIS Facilitates Endothelial Angiogenic Function. Circulation. 2017;136:65-79
5. Uchida S, Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res. 2015;116:737-750
6. Liang H, Pan Z, Zhao X, Liu L, Sun J, Su X, Xu C, Zhou Y, Zhao D, Xu B, Li X, Yang B, Lu Y, Shan H. LncRNA PFL contributes to cardiac fibrosis by acting as a competing endogenous RNA of let-7d. Theranostics. 2018;8(4):1180-1194 doi:10.7150/thno.20846
7. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis. The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-358
8. Wang X, Wang H-X, Li Y-L, Zhang C-C, Zhou C-Y, Wang L, Xia Y-L, Du J, Li H-H. MicroRNA Let-7i negatively regulates cardiac inflammation and fibrosis. Hypertension. 2015;66:776-785
9. Tolonen A-M, Magga J, Szabó Z, Viitala P, Gao E, Moilanen A-M, Ohukainen P, Vainio L, Koch WJ, Kerkelä R, Ruskoaho H, Serpi R. Inhibition of Let-7 microRNA attenuates myocardial remodeling and improves cardiac function postinfarction in mice. Pharmacol Res Perspect. 2014;2:e00056
10. Tao H, Zhang J-G, Qin R-H, Dai C, Shi P, Yang J-J, Deng Z-Y, Shi K-H. LncRNA GAS5 controls cardiac fibroblast activation and fibrosis by targeting miR-21 via PTEN/MMP-2 signaling pathway. Toxicology. 2017;386:11-18
11. Tao H, Cao W, Yang J-J, Shi K-H, Zhou X, Liu L-P, Li J. Long noncoding RNA H19 controls DUSP5/ERK1/2 axis in cardiac fibroblast proliferation and fibrosis. Cardiovasc Pathol. 2016;25:381-389
12. Piccoli M-T, Gupta SK, Viereck J, Foinquinos A, Samolovac S, Kramer FL, Garg A, Remke J, Zimmer K, Batkai S, Thum T. Inhibition of the Cardiac Fibroblast-Enriched lncRNA Meg3 Prevents Cardiac Fibrosis and Diastolic Dysfunction. Circ Res. 2017;121:575-583
13. Micheletti R, Plaisance I, Abraham BJ, Sarre A, Ting C-C, Alexanian M, Maric D, Maison D, Nemir M, Young RA, Schroen B, González A, Ounzain S. et al. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci Transl Med. 2017:9
14. Deng L, Yang S-B, Xu F-F, Zhang J-H. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18
15. Kallen AN, Zhou X-B, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi J-S, Zhang H, Min W, Bennett AM, Gregory RI. et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101-112
16. Powers JT, Tsanov KM, Pearson DS, Roels F, Spina CS, Ebright R, Seligson M, Soysa Y de, Cahan P, Theißen J, Tu H-C, Han A, Kurek KC. et al. Multiple mechanisms disrupt the let-7 microRNA family in neuroblastoma. Nature. 2016;535:246-251
17. Qu X, Du Y, Shu Y, Gao M, Sun F, Luo S, Yang T, Zhan L, Yuan Y, Chu W, Pan Z, Wang Z, Yang B. et al. MIAT Is a Pro-fibrotic Long Non-coding RNA Governing Cardiac Fibrosis in Post-infarct Myocardium. Sci Rep. 2017;7:42657
18. Bedoya-Reina OC, Ponting CP. Functional RNA classes. A matter of time? Nat Struct Mol Biol. 2017;24:7-8
19. Ponting CP. Biological function in the twilight zone of sequence conservation. BMC Biol. 2017;15:71
20. Ulitsky I. Evolution to the rescue. Using comparative genomics to understand long non-coding RNAs. Nat Rev Genet. 2016;17:601-614
21. Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015;11:1110-1122
22. Johnson R, Guigó R. The RIDL hypothesis. Transposable elements as functional domains of long noncoding RNAs. RNA. 2014;20:959-976
23. Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, Finstermeier K, Stahringer A, Wilfert W, Beutner F, Gielen S, Schuler G, Gäbel G. et al. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013;9:e1003588
24. Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, Helm M, Bujnicki JM, Grosjean H. MODOMICS. A database of RNA modification pathways-2013 update. Nucleic Acids Res. 2013;41:D262-7
25. Xiang Y, Laurent B, Hsu C-H, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, Ling D, Hsu P-H, Zou L. et al. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543:573-576
Author contact
![]() Corresponding author: Matthias S. Leisegang, PhD; Institut für Kardiovaskuläre Physiologie, Fachbereich Medizin der Goethe-Universität, Theodor-Stern-Kai 7, 60590 Frankfurt am Main, Germany. Tel.: +49-69-6301-6996; Fax.: +49-69-6301-7668; Email: Leiseganguni-frankfurt.de
Corresponding author: Matthias S. Leisegang, PhD; Institut für Kardiovaskuläre Physiologie, Fachbereich Medizin der Goethe-Universität, Theodor-Stern-Kai 7, 60590 Frankfurt am Main, Germany. Tel.: +49-69-6301-6996; Fax.: +49-69-6301-7668; Email: Leiseganguni-frankfurt.de
 Global reach, higher impact
Global reach, higher impact