13.3
Impact Factor
Theranostics 2019; 9(12):3526-3540. doi:10.7150/thno.32796 This issue Cite
Research Paper
Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma
1. The International Cooperation Laboratory on Signal Transduction, Eastern Hepatobiliary Surgery Hospital, Second Military Medical University, Shanghai 200438, China.
2. Key Laboratory of Computational Biology, CAS-MPG Partner Institute for Computational Biology, Shanghai Institutes for Biological Sciences, University of Chinese Academy of Science, Chinese Academy of Sciences, Shanghai 200031, China
3. Department of Pathology, Eastern Hepatobiliary Surgery Hospital, Second Military Medical University, Changhai Road 225, Shanghai 200438, China
4. Fudan University Shanghai Cancer Center; Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 200032, China
5. Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha, Hunan, China.
6. Department of Oncology, Eastern Hepatobiliary Surgery Hospital, Second Military Medical University, North Moyu Road 700, Shanghai, China
7. The United Innovation of Mengchao Hepatobiliary Technology Key Laboratory of Fujian Province, Mengchao Hepatobiliary Hospital of Fujian Medical University, Fuzhou 350025, China
8. Department of Biliary Surgery I, Eastern Hepatobiliary Surgery Hospital, Second Military Medical University, Changhai Road 225, Shanghai 200438, China
9. School of Public Health, Shanghai JiaoTong University, 200240, China
*These authors contributed equally to this work.
Received 2019-1-4; Accepted 2019-4-17; Published 2019-5-26
Abstract
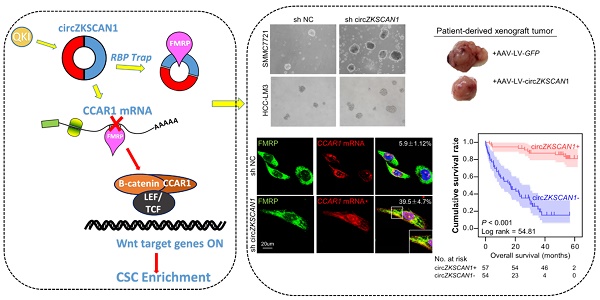
Circular RNA (circRNA) possesses great pre-clinical diagnostic and therapeutic potentials in multiple cancers. It has been reported playing roles in multiple malignant behaviors including proliferation, migration, metastasis and chemoresistance. However, the underlying correlation between circRNAs and cancer stem cells (CSCs) has not been reported yet.
Methods: circZKSCAN1 level was detected in HCC tissue microarrays to clarify its prognostic values. Gain and loss function experiments were applied to investigate the role of circZKSCAN1 in HCC stemness. Bioinformatic analysis was used to predict the possible downstream RNA binding protein and further RNA immunoprecipitation sequencing was carried out to identify the RBP-regulated genes.
Results: The absence of circZKSCAN1 endowed several malignant properties including cancer stemness and tightly correlated with worse overall and recurrence-free survival rate in HCC. Bioinformatics analysis and RNA immunoprecipitation-sequencing (RIP-seq) results revealed that circZKSCAN1 exerted its inhibitive role by competitively binding FMRP, therefore, block the binding between FMRP and β-catenin-binding protein-cell cycle and apoptosis regulator 1 (CCAR1) mRNA, and subsequently restrain the transcriptional activity of Wnt signaling. In addition, RNA-splicing protein Quaking 5 was found downregulated in HCC tissues and responsible for the reduction of circZKSCAN1.
Conclusion: Collectively, this study revealed the mechanisms underlying the regulatory role of circZKSCAN1 in HCC CSCs and identified the newly discovered Qki5-circZKSCAN1-FMRP-CCAR1-Wnt signaling axis as a potentially important therapeutic target for HCC treatment.
Keywords: circular RNA, hepatocellular carcinoma, cancer stem cell, RNA-binding protein
Introduction
Hepatocellular carcinoma (HCC) is one of the most lethal malignancies worldwide, accounting for approximately 700,000 deaths per year [1]. Liver cancer stem cells (CSC) are defined by stemness-related markers including CD133 and epithelial cell adhesion molecule (EpCAM) and are considered to be responsible for the initiation of tumor progression, drug-resistance, metastasis, and recurrence [2]. Numerous long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) were found to be dysregulated in HCC, with pro- or anti-tumor properties. LncTLSNC8 and miRNA-708 acted as tumor suppressors in HCC [3, 4], while lncBRM and miRNA-429 showed tumor-promoting properties [5, 6]. However, few studies have examined the involvement of non-coding RNAs, especially circular RNAs (circRNAs) in the regulation of liver CSCs.
The structure of circRNAs is different as compared with linear mRNAs which have both 5′ and 3′ terminal structures reflecting the start and stop signals for transcription of their corresponding DNA templates. The biogenesis of circRNAs is mostly through the back-splicing mechanism. Transcriptome data for humans, mice, and nematodes suggests the existence of large numbers of circRNAs with unidentified functions [7]. Furthermore, circRNAs are stable, conserved, and resistant to exonucleolytic RNA decay compared with their source mRNA. CircRNAs feature cell-type-specific and developmental-stage-specific expression, indicating their broad participation in various physiological and pathophysiological processes [7, 8].
It has been documented that circRNA-ciRS-7 possess multiple miRNA-targeting sites and act as endogenous competing RNAs or miRNA sponges to regulate transcriptional activity [9]. FOXO3-derived circRNAs prevent tumor growth and metastasis in breast cancer, via performing a 'sponge' function involving eight different miRNAs [10]. A recent study revealed that circMTO1 acted as a tumor suppressor by binding miRNA-9, which upregulated the expression of p21 in HCC [11].
Till now, it is still unclear whether circRNAs could exert their potential activities during tumor progression beyond miRNA or mRNA sponge. RNA-binding proteins (RBPs), play a crucial role in post-transcriptional modification and mRNA translation. However, whether circRNAs could modulate the malignant behavior of tumors through targeting RBPs has not been documented previously.
Zinc-finger protein with KRAB and SCAN domains 1 (ZKSCAN1) is a member of the zinc-finger protein family, which are classic regulators of gene transcription and closely related to tumorigenesis, tumor progression, and metastasis. ZKSCAN1 expression was shown to be upregulated in esophageal adenocarcinoma might possess pro-tumor properties [12]. Transcriptome sequencing showed exon-exon junction reads connecting the 5′ end of exon 2 and 3′ end of exon 3, suggesting the possible existence of circZKSCAN1 (circBase ID: hsa_circ_0001727) [8]. RNA-seq revealed 372 different gene expression patterns in the circZKSCAN1-knockdown versus control group, while KEGG enrichment analysis suggested that most differentially expressed genes were located in the migration, adhesion, actin cytoskeleton, cytokine interaction, and phosphatidylinositide 3-kinase pathways [13]. However, the mechanisms whereby circZKSCAN1 affects the expression of key genes in specific pathways, and whether its inhibitory role in HCC is related to its regulatory effects on miRNAs and RBPs remains unclear.
In the present study, we demonstrated that circZKSCAN1 suppressed cell stemness in HCC through regulating the function of the RBP fragile X mental retardation protein (FMRP). We also identified the downstream target gene of FMRP as cell cycle and apoptosis regulator 1 (CCAR1), which acts as a co-activator of the Wnt/β-catenin signaling pathway and upregulates cell stemness. The results of this study revealed a novel mechanism whereby circRNAs regulate malignant behavior in cancers, in addition to acting as a miRNA sponge. This information may provide a new angle for further research into the relationship between circRNAs and cancers.
Materials and Methods
Patients and tissue specimens
In total, 112 pairs of HCC and paired para-cancerous tissues were obtained from surgical resections of patients without preoperative treatment at Eastern Hepatobiliary Surgery Hospital (Shanghai, China) from 2009-2010. The procedure of human specimen collection was approved by the Ethics Committee of Eastern Hepatobiliary Surgery Hospital. 10 fresh HCC specimens for RNA-seq were also obtained from patients undergoing hepatectomy in our hospital. The expression of CSC marker, EpCAM in those 10 HCC samples, was evaluated by performing IHC staining with antibody against human EpCAM (Cell Signaling Technology, cat No.2929S)
RIP sequencing and bioinformatics analysis
The RIP procedure was performed with Magna RIP Kit (Catalog No. 17-700, Millipore). HCC cell lines were lysed in RIP lysis buffer and then immunoprecipitated with antibody to RBP of interest with protein A/G magnetic beads. Magnets were used to immobilize magnetic beads bound complexes while washing off the unbound materials. Subsequently, remaining RNA was extracted.
Total FMRP-RIP-RNA from hepatocellular carcinoma cell lines was purified with RNeasy Micro Kit according to manufacturer's instructions (Qiagen, Hilden, Germany).TruSeq Stranded Total RNA with Ribo-Zero kit (Illumina, San Diego, CA) has been used for library preparation following the manufacturer's instructions. Both RNA samples and final libraries were quantified by using the Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA) and quality tested by Agilent 2100 Bioanalyzer RNA Nano assay (Agilent Technologies, Santa Clara, CA). Libraries were then processed with Illumina cBot for cluster generation on the flowcell, following the manufacturer's instructions and sequenced on 150 bp pair-ends mode at the on HiSeq2500 (Illumina, San Diego, CA). The library construction and sequencing were performed at Shanghai Biotechnology Corporation.
Raw sequence files were subjected to quality control analysis using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Sequencing raw reads were preprocessed by filtering out rRNA reads, sequencing adapters, short-fragment reads and other low-quality reads. We used Tophat v2.0.9 to map the cleaned reads to the human reference genome ensemble GRCh38 (hg38) with two mismatches. After genome mapping, Cufflinks v2.1.1 was run with a reference annotation to generate FPKM values for known gene models. Differentially expressed genes were identified using Cuffdiff. The p-value significance threshold in multiple tests was set by the false discovery rate (FDR). The fold-changes were also estimated according to the FPKM in each sample. The differentially expressed genes were selected using the following filter criteria: FDR ≤0.05 and fold-change ≥2 in hepatocellular carcinoma cell lines. Complete RIP-seq datasets can be accessed through GEO via series GSE110048.
Fluorescence in situ hybridization
Fluorescence in situ hybridization was conducted in HCC cell line with CY3-labelled RNA fluorescence probes (GENESEED) against circZKSCAN1 and miR-CCAR1. SMMC7721 cells were grown to 50-75% confluence at the time of fixation. After prehybridization, the cells were hybridized to CY3-labeled probes specific to circZKSCAN1 or CCAR1 at 37 °C overnight. Signals were detected under a microscope (Olympus Corp.). Primers for FISH analysis were Has circ cZKSCAN1-FISH probe: TCTTACAGTCACGAGGAATAGTAAAGAAAC, Has-miR-CCAR1 (Gene ID: 55749, NM_018237.3).
In situ hybridization
In situ RNA hybridization was conducted in formaldehyde-fixed liver tissues with digoxygenin (DIG)-labelled RNA probes (Exiqon) against circZKSCAN1 and sense negative control. Anti-DIGAP (Bosterbio, USA) was used as secondary antibody and staining was developed with NBT/BCIP solution (Bosterbio, USA). circZKSCAN1 probe: TCTTACAGTCACGAGGAATAGTAAAGAAAC.
Results
CircRNA expression spectrum in EpCAMhigh and EpCAMlow HCCs
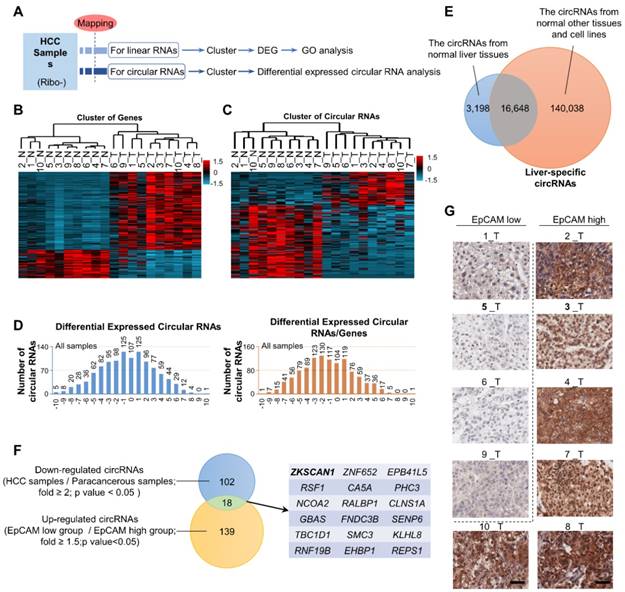
The expression patterns of miRNAs and lncRNAs in HCC have been well studied and their diagnostic and therapeutic potentials have been noted; however, the possible dysregulation of circRNAs in HCC remains unknown. Here, we extracted RNA (except rRNA) from ten HCC samples and paired para-cancerous tissues and performed second-generation sequencing to identify dysregulated RNAs and circRNAs in HCC (Figure 1A and Table S1). Gene-cluster analysis showed that the expression levels of numerous genes were dysregulated, of which most genes were upregulated in HCC samples (Figure 1B and Figure S1A). GO enrichment analysis revealed that these dysregulated genes participated in a wide range of crucial cellular activities (Figure S1B). CircRNA cluster analysis indicated that dysregulated circRNAs were not preferentially up- or downregulated in HCC. However, after circRNA/gene standardization, most circRNAs were shown to be downregulated in HCC (Figure 1C-D). Based on data from CIRCpedia, we identified 3,198 liver-specific circRNAs not expressed in other tissues (Figure 1E and Table S2). After richness standardization of the original linear/circRNA sequencing data (the ratio of circRNAs enrichment to their-corresponding mRNAs enrichment), we identified 120 circRNAs that were significantly downregulated in HCC. Among them, circZKSCAN1 expression was decreased over 2-fold in tumor tissues compared with paired non-tumor tissues (Figure 1F). As EpCAM has been extensively documented as the stem-related gene for CSC expansion in HCC, we then divided HCC tissues into two subgroups according to EpCAM expression level (named as EpCAMhigh and EpCAMlow) basing on RNA-seq results (Figure S1C and Table S1) and immunohistochemical staining scores (Figure 1G). The sequencing data were then reorganized based on the newly defined subgroups (Figure S1D and E), of which 157 circRNAs were found upregulated in EpCAMlow group as compared with EpCAMhigh group. Finally, we identified 18 circRNAs that met the following criteria: 1, target circRNA > 2-fold downregulated in HCC; and 2, target circRNA > 1.5-fold upregulated in EpCAMlow subgroup (Figure 1F), among which circZKSCAN1 was previously shown to play a distinctive role in HCC, though its mechanism is unknown [13]. We, therefore, chose to investigate circZKSCAN1 as a target circRNA.
The exclusive circRNA expression spectrum in hepatocellular carcinoma. (A) Second-generation sequencing workflow. (B) Differentially expressed genes from an RNA-seq of 10 HCC samples and paired para-cancerous tissues. n = 10, red is higher and blue is lower expression. (C) Circular RNAs cluster analysis from an RNA-seq of 10 HCC samples and paired para-cancerous tissues. n = 10, red is higher and blue is lower expression. (D) Differentially expressed Circular RNAs in all cancerous tissue samples(left), as well as Circular RNAs that change independently after richness standardization with the corresponding mRNA(right). (E) Liver-specific circRNAs spectrum. The overlap between circRNAs from normal liver tissues and normal other tissues and cell lines covers 16,648 liver-specific circRNAs. (F) Stemness-related circRNAs from sequencing. The overlap between Down-regulated circRNAs (HCC samples/Para-canceroussamples; fold >= 2;p value < 0.05 ) and Up-regulated circRNAs (EpCAMlow group /EpCAMhigh group; fold >= 1.5;p value<0.05) covers 18 stemness-related circRNAs. (G) Immunohistochemistry staining in formalin-fixed paraffin-embedded (FFPE) sections of 10 HCC sample. Scale bar, 200 μm.
Specific features and prognostic value of circZKSCAN1 in HCC cells
The human ZKSCAN1 gene is localized on human chromosome 17 and is characterized by a 2232 nucleotide-long region containing exons 2 and 3. Deep sequencing of various human cell lines revealed that the 3′ end of exon 3 and 5′ end of exon 2 could be spliced and connected to form a circRNA (Figure 2A). To rule out the insertion of an intron between exons 2 and 3 during circularization, we designed two primers and subjected the PCR products to gel electrophoresis, which confirmed a direct connection between exons 2 and 3 (Figure S2A). Sequencing of the retrieved gel samples confirmed this conclusion (Figure S2B). The expression levels of circZKSCAN1 and linearZKSCAN1 detected in 112 HCC tissues and paired paracancerous tissues were measured by RT-PCR (Figure 2B). Interestingly, there was no change in expression levels of its linear counterpart, suggesting that circZKSCAN1 may function independently of its source gene ZKSCAN1 in HCC. For further confirmation, we established a scoring system based on in situ hybridization staining intensity using a tissue microarray (n = 112; the baseline characteristics of the patients are shown in Table S3-4) with the same results (Figure 2C). We then attempted to investigate the potential role of circZKSCAN1 in HCC CSCs. By using the same 112 pairs of tissues, we identified a negative correlation between EpCAM mRNA and circZKSCAN1 expression (Figure 2D). Relatively, there was no significant difference in expression levels of its linear counterpart (Figure 2E). This negative correlation between EpCAM mRNA and circZKSCAN1 was confirmed using the insitu hybridization and immunohistochemistry scoring system (Figure 2F). As expected, circZKSCAN1 was more resistant to RNase R treatment compared with its linear mRNA (Figure 2G), supporting the existence of natural circZKSCAN1. The half-life of circZKSCAN1 and its linear form were also investigated with the administration of Actinomycin D, which showed that longevity of circZKSCAN1 was much longer than its mRNA (Figure 2H).
Forest plots of overall and progression-free survival in patient subgroups
Specific features of circZKSCAN1 in HCC cells and its significant prognostic values. (A) Schematics of ZKSCAN1 circularization. A circular RNA is formed when the 5' splice site at the end of exon 3 is joined to the 3' splice site at the beginning of exon 2. Repetitive elements in the designated orientations are shown. (B) The relative expression of ZKSCAN1 circRNA and mRNA were determined in fresh RNA samples of HCC tissue and paired non-tumor tissue (distal non-cancerous tissues away from HCC), n=112. (C) The expression level of circZKSCAN1 was measured by in situ hybridization staining using a pair of tissue microarrays (n=112). Score 0 defined as weak expression, score 1+ defined as moderate expression, score 2+ defined as strong expression. The staining intensity criteria was demonstrated on the right. The 100 dots shown in the figure represent the percentage not the exact number of tissues. (D) The correlation of EpCAM mRNA level and circZKSCAN1 expression level using 112 HCC frozen tissues was determined by qPCR. r=-0.3845, P<0.0001. (E) The correlation of EpCAM mRNA level and ZKSCAN1 mRNA expression level using 112 HCC frozen tissues was determined by qPCR. r=0.1495, P=0.1242. (F) The correlation of EpCAM mRNA level and circZKSCAN1 ISH staining intensity. (G) Total RNAs were digested with RNase R followed by qRT-PCRs detection of circZKSCAN1expression. ZKSCAN1 mRNA was detected as the RNase R-sensitive control (n=4, Student t test). (H) qRT-PCR for the abundance of circZKSCAN1 and ZKSCAN1 mRNA in SMMC7721 cells treated with Actinomycin D at the indicated time points. (I)The percent of ZKSCAN1 levels. The expression of ZKSCAN1 circRNA and mRNA were detected by qRT-PCRs in the nuclear and cytoplasm fractions of cells. (J) Immunoblot detection of cell cytoplasm and nucleus circZKSCAN1, GAPDH (cytoplasm control) and Histone3(nuclear control) of indicated cell lines are shown. (K) RNA-FISH assays were conducted to detect circZKSCAN1 expression in SMMC7721 cell using Cy3-labeled antisense probes(circZKSCAN1). Nuclei were stained with 4, 6-diamidino-2-phenylindole (DAPI). Scale bar, 10um. (L) Kaplan-Meier analysis of the correlation between circZKSCAN1 expression levels and overall survival (OS). (M) Univariate or multivariate analysis of HRs for overall survival and relapse-free survival. ***p<0.001; NS, not significant; HR, Hazard ratio; CI, confidence interval.
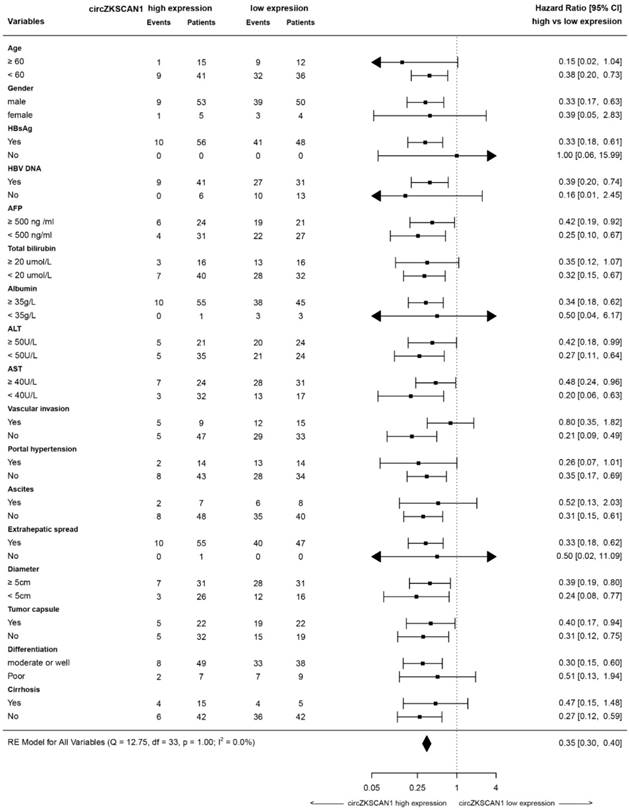
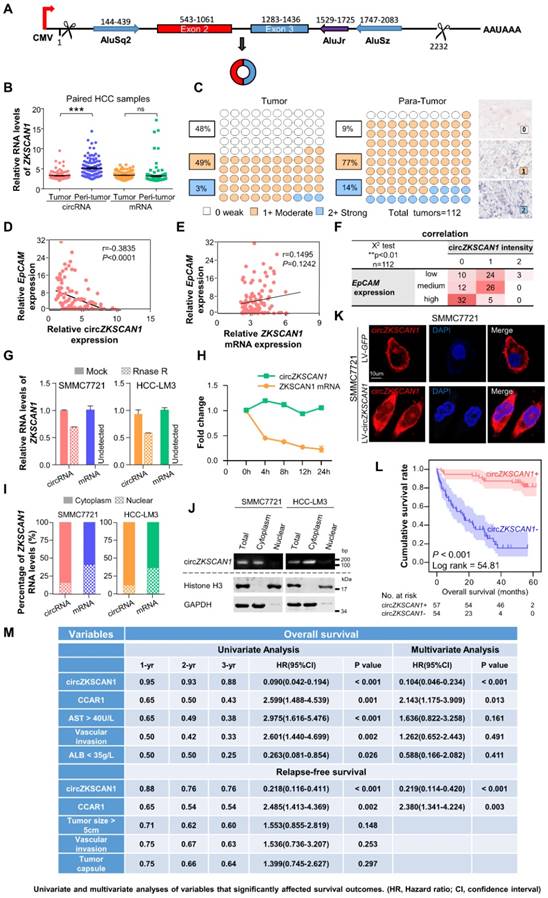
To clarify the function of circZKSCAN1, we identified its subcellular localization using a nucleus-cytoplasm separation technique. CircZKSCAN1 was present preferentially in cytoplasm, while linearZKSCAN1 was distributed in both cytoplasm and nucleus (Figure 2I-K). Based on the patient information for the tissue microarray (n = 112), we analyzed the data by correlation regression analysis and revealed that low circZKSCAN1 expression levels were associated with multiple HCC characteristics (Table S5). Kaplan-Meier survival analysis showed that circZKSCAN1 expression level was positively correlated with overall and recurrence-free survival (Figure 2L and Figure S2C). Univariate and multivariate analysis indicated that circZKSCAN1 expression was an independent and significant factor affecting both overall survival (HR=0.104; 95%CI: 0.046-0.234, p<0.001) and relapse-free survival rate (HR=0.219; 95%CI: 0.114-0.420, p<0.001) of HCC patients(Figure 2M). High circZKSCAN1 expression was a favorable factor for HCC patients despite different characteristics (Table 1).
CircZKSCAN1 suppresses malignant behavior of HCC by modulating cell stemness
We further characterized the role of circZKSCAN1 in regulating stemness in HCC using circZKSCAN1-knockdown and circZKSCAN1-overexpressing HCC cell lines, respectively (SMMC7721 and HCC-LM3), in in vitro and in vivo experiments. The efficiency of lentivirus-mediated knockdown and overexpression were confirmed by PCR (Figure 3A-B). Expression levels of linear ZKSCAN1 were consistently unaffected in the presence or absence of its circular form (Figure S3A).
The sphere-formation ability of HCC cell lines was greatly enhanced by downregulation of circZKSCAN1 expression and markedly reduced after circZKSCAN1 overexpression (Figure 3C-D, Figure S3B-C). Intriguingly, numbers of CSCs increased after the circZKSCAN1 knockdown and showed improved colony-forming ability, while circZKSCAN1 overexpression had the opposite effects in HCC cell lines (Figure 3E-F and Figure S3D-E). Rescue experiments were also performed by transfecting circZKSCAN1 overexpression vector into sh-circZKSCAN1 cell lines (Figure 3C and 3E). As expected, circZKSCAN1 could suppress the expression of EpCAM, SOX2, OCT4 and malignant marker C-MYC in HCC cell lines (Figure 3G and Figure S3F-G). Flow cytometry analysis found a higher frequency of EpCAM+ cells among circZKSCAN1-knockdown cells, and vice versa (Figure 3H). Additionally, the downregulated expression of circZKSCAN1 was observed in sorted EpCAM+ SMMC-7721, HCC-LM3 cells and 10 freshly resected HCC tumor tissues, while its linear mRNA showed no similar trend (Figure 3I-J).
We investigated the role of circZKSCAN1 in vivo using mice injected subcutaneously with modified HCC cell lines. Tumor formation was increased following subcutaneous injection of as few as 10,000 circZKSCAN1-knockdown HCC cells (Figure 3K). Furthermore, tumors in the circZKSCAN1-knockdown group were significantly larger (Figure S3H-I). On the contrary, the circZKSCAN1-overexpressing group showed reduced tumor-forming ability, with a lower incidence of tumors and smaller tumor size (Figure 3K and Figure S3J). We examined the correlation between the number of EpCAM+ cells and circZKSCAN1 expression in xenograft tumors in mice by immunohistochemical staining. The numbers of EpCAM+ cells and Ki-67 positive cells were increased in circZKSCAN1-knockdown and decreased in circZKSCAN1-overexpressing tumors (Figure 3L).
We also determined if circZKSCAN1 expression affected the proliferative ability of HCC. Interestingly, transient silencing of circZKSCAN1 but not linear-ZKSCAN1 facilitated the proliferation of HCC cell lines (Huh-7, HCC-LM3, and SMMC7721) and a normal hepatocyte cell line (QSG7701), indicating a close connection between circZKSCAN1 and malignant behavior in HCC (Figure S3K-N). A similar result was observed in stable cell lines (Figure S3O-P). Silencing circZKSCAN1 also enhanced the metastatic abilities of both normal hepatocytes and HCC cells as shown by cell migration assay (data not shown).
CircZKSCAN1 acts by sponging specific RBPs
CircRNAs mostly act by competing against miRNAs and RBPs to regulate their target genes. We performed bioinformatics analysis and identified 10 RBPs (FMRP, FUS, DGCR8, ELAVL, EIF4A3, PTB, IGF2BP1, IGF2BP2, LIN28A, LIN28B) that may compete against circZKSCAN1 [14]. Knockdown of any single RBP did not affect spheroid formation in HCC control cells, but knockdown of FMRP significantly compromised spheroid-formation ability in circZKSCAN1-deficient HCC cell lines (Figure 4A). Unexpectedly, double knockdown of both circZKSCAN1 and FMRP could ameliorate the enhanced stemness of both SMMC7721 and HCC-LM3 cell lines induced by single knockdown of circZKSCAN1, demonstrating that circZKSCAN1 might suppress stemness by confining the biological functions of FMRP (Figure 4B-C). Flow cytometry analysis showed that double knockdown of circZKSCAN1 and FMRP could reduce the frequency of EpCAM+ cells in sh-circZKSCAN1 cell lines (Figure 4D).
CircZKSCAN1 suppresses malignant behaviors of hepatocellular carcinoma by downregulating cellular stemness. (A)&(B) Expression of circZKSCAN1 was quantified by real-time PCR in SMMC7721/HCC-LM3 after circZKSCAN1 knocked-down or overexpressed using lentivirus, respectively. Data are presented as mean ± SEM (n=4). **P<0.01. (C) &(D) Spheroid formation assay were used to test the sphere formation abilities of the indicated cell lines. Results are shown as mean ± SEM, n=3. *P<0.05, **P<0.01, based on the student's t-test. (E)&(F) In vitro limiting dilution assays were performed to test cancer stem cell frequency of the indicated cell lines. Results are shown as mean ± SEM, n=3. *P<0.05. (G) The EpCAM mRNA expression level of different groups were confirmed by qPCR. Data are presented as mean ± SEM (n=4). *P<0.05, **P<0.01. (H) The EpCAM+ population of different groups was determined by flow cytometry. SMMC7721(left); HCC-LM3(right). Data are presented as mean ± SEM (n=4). *P<0.05, **P<0.01. (I) The expression level of ZKSCAN1 circRNA and mRNA in sorted EpCAM+ and EpCAM- HCC cells. NS, not significant, *P<0.05, **P<0.01. (J) The expression level of ZKSCAN1 circRNA and mRNA in sorted EpCAM+ and EpCAM- primary HCC cells. NS, not significant, ***P<0.001. (K) Nude mice (n=6) were subcutaneously injected with the indicated number of SMMC7721 cells in which circZKSCAN1 was knocked-down or overexpressed using lentivirus before being implanted and the frequency of cancer stem cell was evaluated 2(circZKSCAN1 knock-down group) or 4 months (circZKSCAN1 overexpression group) after injection by a limiting dilution assay. m/n means m out n nice develop a tumor. **p<0.01. (L) H&E and immunohistochemistry staining in formalin-fixed paraffin-embedded (FFPE) sections of SMMC7721 xenograft tumor. Scale bar, 200 μm.
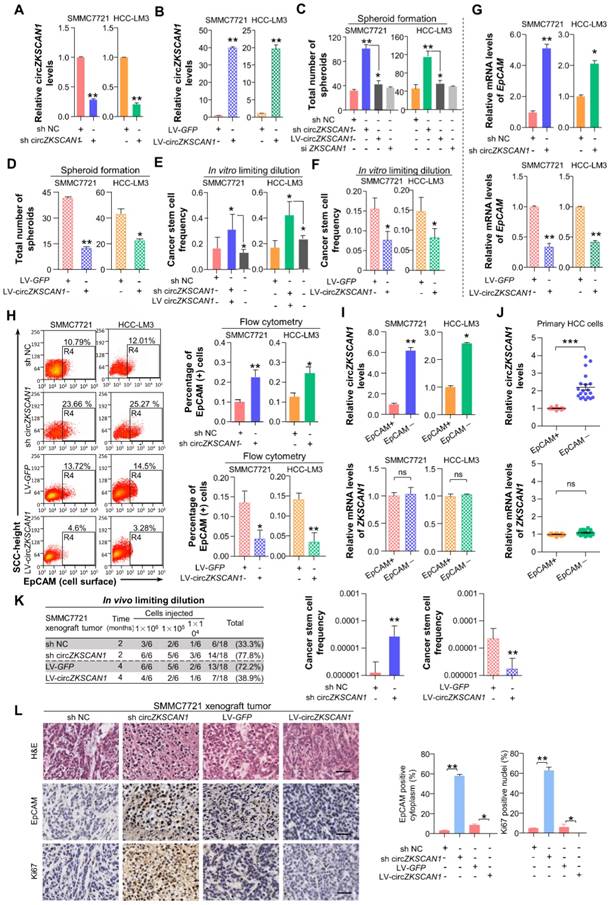
CircZKSCAN1 performs its function through sponging specific RNA-binding protein. (A) Spheroid formation assay result in the context of 10 different RBP knockdown. HCC control cell line (left); HCC lentivirus-mediated circZKSCAN1 deficient cell line (right). Data are presented as mean ± SEM (n=4). *P<0.05. (B) Comparison of the spheroid formation ability between single circZKSCAN1 or FMRP knockdown and double knockdown. Data are presented as mean ± SEM (n=4). *P<0.05. (C) The EpCAM mRNA expression level of different groups were confirmed by qPCR. Data are presented as mean ± SEM (n=4). *P<0.05. (D) The EpCAM+ population of different groups was determined by flow cytometry. SMMC7721(above); HCC-LM3(below). Data are presented as mean ± SEM (n=4). *P<0.05. (E) Graphical abstract of the RNA immunoprecipitation sequencing procedure. (F) Venn diagram of differentially expressed genes in RIP-target genes of FMRP, stemness-related genes, and FMRP CLIP database. (G) RIP-seq results of FMRP binding sites in CCAR1 gene. (H) The correlation of CCAR1 mRNA level and circZKSCAN1 expression level using 112 HCC frozen tissues was determined by qPCR. r==-0.2987, P<0.01. (I) The correlation of CCAR1 mRNA level and ZKSCAN1 mRNA expression level using 112 HCC frozen tissues was determined by qPCR. r=0.1562, P=0.1082. (J)&(K) Whole-cell RNA extract was precipitated with input (positive control), IgG (negative control) and FMRP antibody and the precipitation was then verified by Western Blot. CircZKSCAN1 and CCAR1 level of the precipitations was confirmed by qPCR. The CCAR1 protein level was also detected via WB. (L) The co-localization of FMRP protein and circZKSCAN1 was confirmed by immunofluorescence and fluorescence in situ hybridization. (M) The co-localization of FMRP protein and CCAR1 was confirmed by immunofluorescence and fluorescence in situ hybridization. Sh NC, HCC control cells. Sh circZKSCAN1, HCC lentivirus-mediated circZKSCAN1 knockdown cells. Quantification of the percentage of interaction were marked.
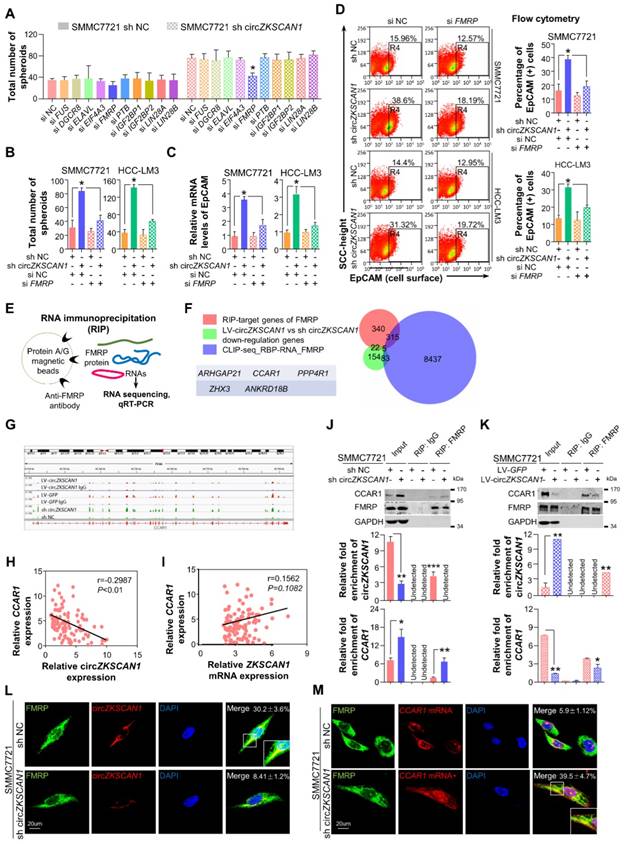
RNA immunoprecipitation (RIP)-sequencing assay was applied to investigate the potential target stemness-related genes of FMRP (Figure 4E). Mapping data revealed that FMRP had 682 potential target genes, and further GO analysis demonstrated that these target genes had broad functions in cellular physiological processes (Figure S4A). After overlapping with a gene signature containing 264 genes negatively co-expressed with circZKSCAN1 and the FMRP CLIP database (8840 genes, http://starbase.sysu.edu.cn/), 5 genes, including CCAR1 [15], ARHGAP21 [16], PPP4R1 [17], ZHX3 [18], and ANKRD18B [19], were identified as the potential targets involving circZKSCAN1-FMRP modulated signaling (Figure 4F and Table S7-9). Further correlation analysis demonstrated that of these 5 genes, only CCAR1 showed negative correlation with circZKSCAN1 rather than linear-ZKSCAN1 (Figure 4G-I). The graphic abstract of the physical binding between FMRP protein and CCAR1 mRNA was shown in Figure S4B. The type-1 KH domain on FMRP protein directly bonds with a 21-nt long sequence on CCAR1 transcript. As shown in Fig, 4j and k, RIP assay showed that circZKSCAN1 deficiency had no effect on the expression level of FMRP, but markedly enhanced the level of CCAR1 interacting with FMRP. Although other four genes might be combined with FMRP, no significant changes of mRNA-FMRP complex were observed in response to circZKSCAN1 expression (Figure S4C-E). We also applied FUS, an RNA-binding protein, as negative controls to further confirm the specific interaction between circZKSCAN1 and FMRP (Figure S4E). Moreover, fluorescence in situ hybridization at a subcellular level confirmed the competitive binding of circZKSCAN1 or CCAR1 mRNA with FMRP protein (Figure 4L-M). After FMRP knockdown, the expression of CCAR1 was significantly inhibited (Figure S4F). Taken together, these data suggest that circZKSCAN1 might serve as CSCs modulator by preventing FMRP-CCAR1-induced signaling.
FMRP modulates cellular stemness in HCC through its target gene CCAR1, which assists in the activation of the Wnt/β-catenin pathway
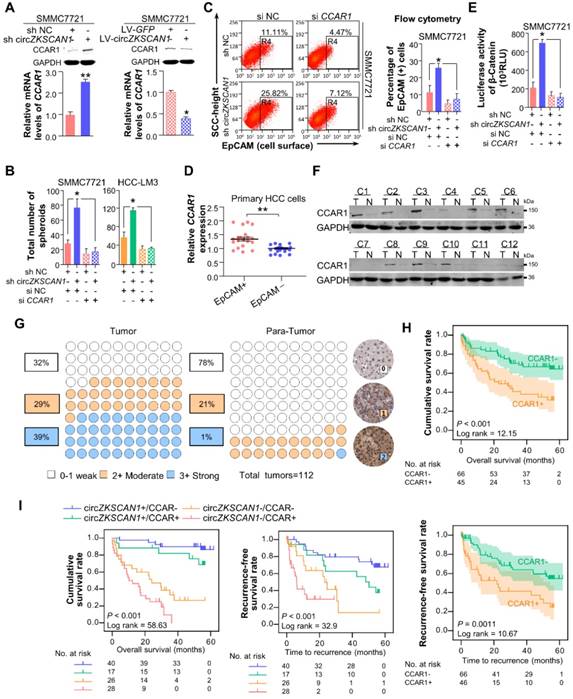
As shown in Figure 5A, the negative correlation between circZKSCAN1 and CCAR1 was observed in HCC cell lines in the presence or absence of circZKSCAN at both protein and mRNA levels. The loss of CCAR1 (Figure S5A) could counteract the enhancement of stemness induced by circZKSCAN1 deficiency, suggesting that CCAR1 and circZKSCAN1 had opposite effects on stemness (Figure 5B). Interestingly, knockdown of circZKSCAN1 boosted the expression of CCAR1, while changes in the expression of CCAR1 had no effect on circZKSCAN1, suggesting that the circZKSCAN1-FMRP-CCAR1 axis might not involve positive or negative feedback (Figure S5B). Moreover, flow cytometry indicated that the percentage of EpCAM+ cells were reduced in CCAR1-knockdown cells regardless the presence of circZKSCAN1 (Figure 5C), consistent with the findings that CCAR1 level was increased in sorted EpCAM+ tumor cells from 10 freshly resected HCC tissues (Figure 5D). CCAR1 was previously reported as a co-activator dependent on β-catenin function in the Wnt signaling pathway [20]. As expected, luciferase analysis showed that knockdown of CCAR1 ameliorated the transcriptional activity of Wnt signaling (Figure 5E). As shown in Figure 5F, CCAR1 expression was markedly upregulated in HCC as compared with para-cancerous tissues. Immunohistochemical staining using a tissue microarray consisting of paired cancerous and para-cancerous tissues from 112 patients showed the significant enrichment of CCAR1 in HCC tissues (39% in tumor versus 1% in para-tumor with strong staining, respectively) (Figure 5G) and their tight correlation with worse outcome clinically (Figure 5H). Multivariate analysis also identified CCAR1 as an independent significant factor in OS (overall survival) and RFS (recurrence-free survival) in HCC patients (Figure 2I). More, the combination of circZKSCAN1 and CCAR1 showed significant prognostic value for predicting overall and recurrence-free survival in HCC patients, with improved predictive power compared with either parameter alone (Figure 5I).
Overexpression of circZKSCAN1 directly inhibits HCC growth in vivo which was regulated by QKI
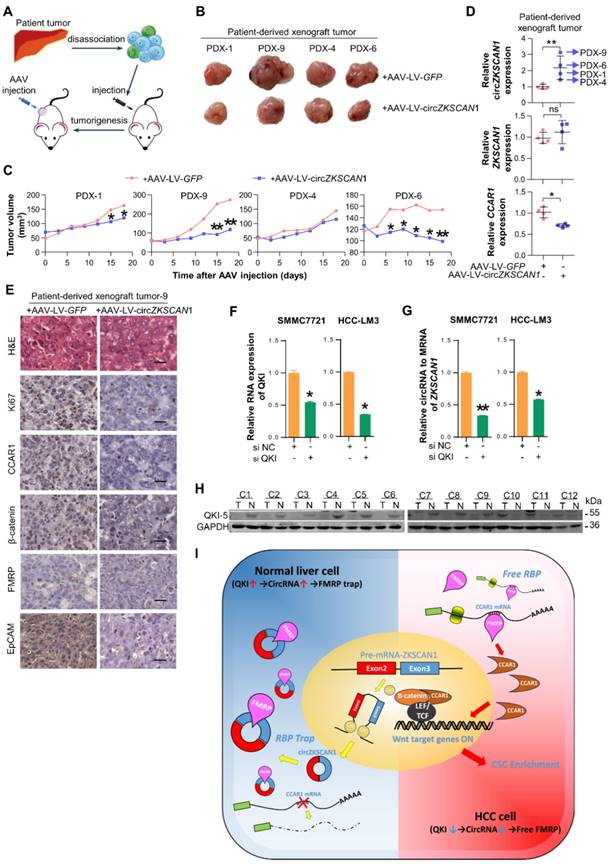
To explore its therapeutic potential, we constructed four PDX (patient-derived xenograft) mouse models. The mice were injected with Adeno-associated virus that introducing the expression of circZKSCAN1 (AAV-LV-circZKSCAN1) in tumor mass (Figure 6A), and the tumors were harvested 18 days after injection. CircZKSCAN1 overexpression significantly reduced the growth rate of the tumor xenografts compared with the control group (Figure 6B-C), coupled with markedly downregulated CCAR1 (Figure 6D). It should be noted that no obvious shrinkage of tumor might be due to the relatively lower expression level of circZKSCAN1 in PDX-4 in comparison with other models (Figure 6D). Furthermore, we detected the expression change of Ki-67/CCAR1/FMRP/β-catenin/EpCAM in PDX-9 which exhibited the greatest inhibition induced by circZKSCAN1. Consistent with our previous findings, except FMRP which showed no expression change after circZKSCAN1 overexpression, the expression of all other proteins was downregulated (Figure 6E). For rescue experiments, we constructed xenograft tumor models with sh-circZKSCAN1 HCC cells and injected circZKSCAN1 overexpression AAV into tumor mass (Figure S6A). The expression of CCAR1 and β-catenin was also detected (Figure S6B).
The Quaking (QKI) protein family was previously reported to play an important role in RNA circularization, and QKI5 was shown to regulate circZKSCAN1 formation[21]. We knocked down QKI expression using targeted siRNA (Figure 6E) and found QKI knockdown significantly reduced the expression of circZKSCAN1 (Figure 6F). Besides, the knockdown of QKI significantly decreased the expression of CCAR1 and showed no effect on FMRP (Figure S6C). To further clarify the relationship between QKI and circZKSCAN1, we investigated the QKI level in our animal studies which showed that the expression change in circZKSCAN1 had no effect on QKI expression (Figure S6D-E). Together with the downregulated expression of QKI protein in HCC tissue (Figure 6G), we concluded that abnormal downregulation of QKI in HCC tissues might contribute to the reduced expression of circZKSCAN1.
FMRP modulates cellular stemeness in HCC through its target gene, CCAR1 which assists in the activation of Wnt/β-catenin pathway. (A) The expression level of CCAR1 protein and mRNA in stable constructed circZKSCAN1 knockdown or overexpression HCC cell line were determined by Western Blot or qPCR. Data are presented as mean ± SEM (n=4). *P<0.05; **P<0.01. (B) Comparison of the spheroid formation ability between single circZKSCAN1 or CCAR1 knockdown and double knockdown. Data are presented as mean ± SEM (n=4). *P<0.05. (C) The EpCAM+ population of different groups was determined by flow cytometry. Data are presented as mean ± SEM (n=4). *P<0.05. (D) The expression level of CCAR1 in sorted EpCAM+ and EpCAM- primary HCC cells. P<0.001. (E) The transcriptional activity of β-catenin downstream targets was measured by luciferase reporter gene assay. Data are presented as mean ± SEM (n=4). *P<0.05. (F) The expression level of CCAR1 protein was measured in 12 HCC tissues and paired para-cancerous tissues. (G) The expression level of CCAR1 was measured by immunohistochemistry staining using a pair of tissue microarrays (n=112). Score 0-1 defined as weak expression, score 2 defined as moderate expression, score 3 defined as strong expression. The staining intensity criteria were demonstrated on the right. The 100 dots shown in the figure represent the percentage not the exact number of tissues. (H) Kaplan-Meier analysis of the correlation between CCAR1 expression levels and overall survival (OS) or recurrence free survival (RFS) (I) The prognostic value of the combination of circZKSCAN1 and CCAR1 was compared with individual parameter.
The overexpression of circZKSCAN1 directly inhibits HCC growth in vivo and the regulation of circZKSCAN1. (A) Graphical abstract of the intratumoral injection. (B) Harvested tumor mass from 4 PDX models were presented. Control group (above), CircZKSCAN1 overexpression group (below). (C) The volume of PDX tumors were measured every 3 days. Growth curves were presented. (D) The expression level of circZKSCAN1, ZKSCAN1 mRNA and CCAR1 mRNA of the PDX tumors (n=4) were determined by qPCR. The different degrees of significance were indicated as follows in the graphs: *P<0.05; **P<0.01. (E) H&E and immunohistochemistry staining in formalin-fixed paraffin-embedded (FFPE) sections of PDX-9 with or without circZKSCAN1 overexpression. Scale bar, 200 μm. (F) The knockdown efficiency of small interfering RNA targeting pan-QKI was determined by qPCR. Data are presented as mean ± SEM (n=4). The different degrees of significance were indicated as follows in the graphs: *P<0.05. (G) The expression level of standardized circZKSCAN1 in the context of pan-QKI deficiency was determined by qPCR. Data are presented as mean ± SEM (n=4). *P<0.05; **P<0.01. (H) The expression level of QKI-5 protein in 12 pairs of tumor and para-tumor tissues was measured by Western Blot. The internal control band of GAPDH is the same with figure 5e since they are generated from the same membrane. (I) Graphical abstract of the whole QKI5-circZKSCAN1-FMRP-CCAR1 axis.
Discussion
CircRNAs were first discovered in RNA viruses [22] but received little attention because of their low levels in cells. However, second-generation sequencing techniques and bioinformatics have revealed that some circRNAs are much more abundant than the mRNAs from which they originate. CircRNAs are now widely recognized as a novel subset of competing for endogenous RNAs that regulate target genes by affecting the functions of miRNAs and RBPs. The roles of circRNAs have been studied in various cancers, including liver [11], lung [23], breast [24], bladder [25], and gastrointestinal cancer[26], and have been shown to facilitate proliferation, metastasis, and chemotherapeutic resistance [23, 27]. However, to the best of our knowledge, few studies have reported on the possible effects of circRNAs on CSCs. CSCs are responsible for tumor initiation, progression, metastasis, chemoresistance, and relapse [28]. The results of the current study provide the first evidence to show that circZKSCAN1 suppresses cell stemness in HCC by competing against the RBP, FMRP.
We identified possible circRNAs with regulatory functions in HCC stem cells using paired tumor and para-cancerous tissues from 10 HCC patients, separated into EpCAMhigh (n = 6) and EpCAMlow (n = 4) groups based on EpCAM expression level. We then acquired the complete expression profile by next-generation sequencing of all samples and successfully filtered out 157 circRNAs significantly enriched in the EpCAMlow group using real-time PCR and siRNAs targeting specific circRNAs. After comparing the expression levels of these circRNAs and their corresponding mRNAs in cancerous and para-cancerous tissues, we selected circZKSCAN1, which was expressed independently of its mRNA, as a circRNA with a potentially unique role in HCC. Further experiments confirmed that circZKSCAN1 inhibited cell stemness, proliferation, and metastasis in HCC. In the previous published study[13], the authors discovered that circZKSCAN1 regulated several malignant behaviors in HCC cell lines such as proliferation and metastasis, however, the exact molecular mechanism remains unclear since the authors only reported the dysregulated genes after circZKSCAN1 knockdown. In our study, we looked from a different angle and discovered that circZKSCAN1 was a potential stemness regulator in HCC and uncovered the possible regulating pathway it works in.
Although the relationship between circRNAs and cancers has been widely studied, the underlying mechanisms remain largely obscure. The best-known function of circRNAs in cancers is to act as a miRNA sponge and compete for mRNA-binding sites on miRNAs [7, 9]. For example, circRNA hsa_circ_000984 can facilitate the growth and metastasis of colon cancer by sponging miR-106b [29]. However, the ability of circRNAs to modulate malignant behavior via RBPs has not previously been reported. RBPs play a major role in the post-transcriptional control of RNAs, such as in splicing, polyadenylation, and in mRNA stabilization, localization, and translation. RBPs are therefore likely to play a part in the regulation of circRNAs in cancers. The results of this study extend our understanding of their role by demonstrating that circZKSCAN1 inhibited stemness in HCC by acting as an RBP (FMRP) sponge, rather than a miRNA sponge.
FMRP mainly functions in the nervous system, where it acts as an RBP and controls the translation of its target mRNAs [30]. FMRP1 gene knockdown causes the relatively common genetic intellectual disability, fragile X syndrome [31]. Although FMRP has mostly been studied in the field of neuroscience, its expression is ubiquitous, suggesting that it may also play important roles in other organs and diseases, such as cancer [30]. Previous studies showed that FMRP expression was upregulated in HCC cells [32]. Furthermore, the risk of cancer was shown to be significantly lower in patients with fragile X syndrome [33]. However, the precise regulatory mechanism of FMRP in cancer biology remains unknown. The current results suggest that circZKSCAN1 and FMRP act to control HCC cell stemness in opposite directions, such that increased stemness induced by circZKSCAN1 knockout can be ameliorated by FMRP deficiency. We also demonstrated that FMRP enhanced stemness in HCC cells by regulating the expression of CCAR.
CCAR1 plays a crucial role in cell proliferation and apoptosis pathways and is originally identified as a perinuclear phosphoprotein triggering apoptosis signaling in breast cancer in a retinoid-dependent manner [34]. Other studies reported that CCAR1 participated in cell proliferation and apoptosis in various cell lines, including cancer cells [35, 36]. The Wnt/β-catenin signaling pathway is a key pathway involved in CSC regulation. Furthermore, CCAR1 has been confirmed as a functional β-catenin-binding protein, which binds directly with lymphoid enhancer binding factor 3/T cell factor family members and supports β-catenin in the transcriptional activation of Wnt target genes, thus further enhancing cell stemness in colon cancer [20]. Based on this information, we performed gain- and loss-of-function experiments in vitro and demonstrated that CCAR1 was positively correlated with cell stemness in HCC through upregulating levels of active β-catenin. Interestingly, CCAR1 was expressed almost exclusively in HCC compared with normal liver tissues, based on 20 sets of paired cancerous and para-cancerous tissues, suggesting that CCAR1 may play a more complex role in hepatocarcinogenesis, in addition to regulating cell stemness. Ha et al. also reported that overexpression of CCAR1 in HCC was associated with a poor prognosis [37].
Despite the fact that several circRNAs have been shown to be dysregulated in cancers, there is currently no evidence for the mechanisms underlying this phenomenon. Conn et al. discovered that the RBP and alternative splicing factor QKI5 regulated the formation of thousands of circRNAs, including circZKSCAN1, during epithelial-mesenchymal transition [21]. We further identified QKI5 as significantly downregulated in HCC tissues compared with paired paracancerous tissues, leading to the differential expression of circZKSCAN1, as confirmed in our further experiments. QKI5 may thus act as a tumor suppressor in HCC. However, previous studies demonstrated that QKI protein might play dual roles in cancers, by promoting proliferation and invasion in colon cancer but inhibiting tumor progression in prostate cancer [38, 39]. QKI itself might thus function as an oncogene in HCC, independent of the function of circZKSCAN1. Further experiments are needed to clarify the relationship between QKI and HCC.
Although most circRNAs are classified as non-coding RNAs, some possess coding potential [40]. However, the coding potential of circZKSCAN1 was not considered in the current study and should thus be addressed in future research.
Overall (Figure 6H), this study demonstrated that circZKSCAN1 is downregulated by QKI in HCC and inhibits multiple malignant behaviors by suppressing cell stemness. Mechanistically, circZKSCAN1 acts as an RBP sponge, competing against the FMRP target gene, CCAR1, and subsequently deactivating the Wnt/β-catenin signaling pathway. Furthermore, circZKSCAN1 induced tumor quiescence in vivo, thus indicating its potential for precision targeted treatment. These results thus provide novel insights into the potential role of the QKI5-circZKSCAN-FMRP-CCAR1 axis in the therapeutic management of HCC.
Abbreviations
HCC, hepatocellular carcinoma; circRNA, Circular RNA; CCAR1, cell cycle and apoptosis regulator 1; FMRP, fragile X mental retardation protein; CSC, cancer stem cells; EpCAM, epithelial cell adhesion molecule; RBPs, RNA-binding proteins; ZKSCAN1, Zinc-finger protein with KRAB and SCAN domains 1.
Supplementary Material
Supplementary methods, figures and tables 4-6.
Supplementary table 1.
Supplementary table 2.
Supplementary table 3.
Supplementary table 7.
Supplementary table 8.
Supplementary table 9.
Acknowledgements
This work was supported by the state Key project for liver cancer (2018ZX10732202-001-001/013), the National Research Program of China (2017YFA0505803, 2017YFC0908100), National Natural Science Foundation of China (81790633, 91729303, 81672860, 81702298 and 81422032), National Natural Science Foundation of Shanghai (17ZR143800).
Author Contributions
Yan-Jing Zhu, Bo Zheng, Xu-Kai Ma and Xin-Yuan Lu performed all the experiments. Xi-Meng Lin, Gui-Juan Luo and Shuai Yang provided human specimens, clinical information and data analysis. Qing Zhao, Xin Chen, Ying-Cheng Yang, Xiao-Long Liu, Rui Wu, Jing-Feng Liu, Yang Ge and Li Yang provided support with experimental materials and techniques. Hong-Yang Wang and Lei Chen designed research and wrote the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Marquardt JU, Andersen JB, Thorgeirsson SS. Functional and genetic deconstruction of the cellular origin in liver cancer. Nat Rev Cancer. 2015;15:653-67
2. Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY. et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012-24
3. Li Q, Li S, Wu Y, Gao F. miRNA-708 functions as a tumour suppressor in hepatocellular carcinoma by targeting SMAD3. Oncol Lett. 2017;14:2552-8
4. Zhang J, Li Z, Liu L, Wang Q, Li S, Chen D. et al. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology. 2018;67:171-87
5. Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B. et al. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nat Commun. 2016;7:13608
6. Li L, Tang J, Zhang B, Yang W, LiuGao M, Wang R. et al. Epigenetic modification of MiR-429 promotes liver tumour-initiating cell properties by targeting Rb binding protein 4. Gut. 2015;64:156-67
7. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-8
8. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-57
9. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-8
10. Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919-31
11. Han D, Li J, Wang H, Su X, Hou J, Gu Y. et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151-64
12. van Dekken H, Tilanus HW, Hop WC, Dinjens WN, Wink JC, Vissers KJ. et al. Array comparative genomic hybridization, expression array, and protein analysis of critical regions on chromosome arms 1q, 7q, and 8p in adenocarcinomas of the gastroesophageal junction. Cancer Genet Cytogenet. 2009;189:37-42
13. Yao Z, Luo J, Hu K, Lin J, Huang H, Wang Q. et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11:422-37
14. Panda AC, Dudekula DB, Abdelmohsen K, Gorospe M. Analysis of Circular RNAs Using the Web Tool CircInteractome. Methods Mol Biol. 2018;1724:43-56
15. Muthu M, Cheriyan VT, Rishi AK. CARP-1/CCAR1: a biphasic regulator of cancer cell growth and apoptosis. Oncotarget. 2015;6:6499-510
16. Lazarini M, Traina F, Machado-Neto JA, Barcellos KS, Moreira YB, Brandao MM. et al. ARHGAP21 is a RhoGAP for RhoA and RhoC with a role in proliferation and migration of prostate adenocarcinoma cells. Biochim Biophys Acta. 2013;1832:365-74
17. Addou-Klouche L, Adelaide J, Finetti P, Cervera N, Ferrari A, Bekhouche I. et al. Loss, mutation and deregulation of L3MBTL4 in breast cancers. Mol Cancer. 2010;9:213
18. Yamada K, Ogata-Kawata H, Matsuura K, Kagawa N, Takagi K, Asano K. et al. ZHX2 and ZHX3 repress cancer markers in normal hepatocytes. Front Biosci (Landmark Ed). 2009;14:3724-32
19. Liu WB, Han F, Jiang X, Yin L, Chen HQ, Li YH. et al. Epigenetic regulation of ANKRD18B in lung cancer. Mol Carcinog. 2015;54:312-21
20. Ou CY, Kim JH, Yang CK, Stallcup MR. Requirement of cell cycle and apoptosis regulator 1 for target gene activation by Wnt and beta-catenin and for anchorage-independent growth of human colon carcinoma cells. J Biol Chem. 2009;284:20629-37
21. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125-34
22. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852-6
23. Luo YH, Zhu XZ, Huang KW, Zhang Q, Fan YX, Yan PW. et al. Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed Pharmacother. 2017;96:892-8
24. Yan N, Xu H, Zhang J, Xu L, Zhang Y, Zhang L. et al. Circular RNA profile indicates circular RNA VRK1 is negatively related with breast cancer stem cells. Oncotarget. 2017;8:95704-18
25. Yang X, Yuan W, Tao J, Li P, Yang C, Deng X. et al. Identification of circular RNA signature in bladder cancer. J Cancer. 2017;8:3456-63
26. Sun H, Tang W, Rong D, Jin H, Fu K, Zhang W. et al. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. 2018;21:299-306
27. Gao D, Zhang X, Liu B, Meng D, Fang K, Guo Z. et al. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics. 2017;9:1175-88
28. Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275-91
29. Xu XW, Zheng BA, Hu ZM, Qian ZY, Huang CJ, Liu XQ. et al. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget. 2017;8:91674-83
30. Alpatov R, Lesch BJ, Nakamoto-Kinoshita M, Blanco A, Chen S, Stutzer A. et al. A chromatin-dependent role of the fragile X mental retardation protein FMRP in the DNA damage response. Cell. 2014;157:869-81
31. O'Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annu Rev Neurosci. 2002;25:315-38
32. Li Y, Tang Y, Ye L, Liu B, Liu K, Chen J. et al. Establishment of a hepatocellular carcinoma cell line with unique metastatic characteristics through in vivo selection and screening for metastasis-related genes through cDNA microarray. J Cancer Res Clin Oncol. 2003;129:43-51
33. Schultz-Pedersen S, Hasle H, Olsen JH, Friedrich U. Evidence of decreased risk of cancer in individuals with fragile X. Am J Med Genet. 2001;103:226-30
34. Rishi AK, Zhang L, Boyanapalli M, Wali A, Mohammad RM, Yu Y. et al. Identification and characterization of a cell cycle and apoptosis regulatory protein-1 as a novel mediator of apoptosis signaling by retinoid CD437. J Biol Chem. 2003;278:33422-35
35. Rishi AK, Zhang L, Yu Y, Jiang Y, Nautiyal J, Wali A. et al. Cell cycle- and apoptosis-regulatory protein-1 is involved in apoptosis signaling by epidermal growth factor receptor. J Biol Chem. 2006;281:13188-98
36. Jiang Y, Puliyappadamba VT, Zhang L, Wu W, Wali A, Yaffe MB. et al. A novel mechanism of cell growth regulation by Cell Cycle and Apoptosis Regulatory Protein (CARP)-1. J Mol Signal. 2010;5:7
37. Ha SY, Kim JH, Yang JW, Kim J, Kim B, Park CK. The Overexpression of CCAR1 in Hepatocellular Carcinoma Associates with Poor Prognosis. Cancer Res Treat. 2016;48:1065-73
38. He B, Gao SQ, Huang LD, Huang YH, Zhang QY, Zhou MT. et al. MicroRNA-155 promotes the proliferation and invasion abilities of colon cancer cells by targeting quaking. Mol Med Rep. 2015;11:2355-9
39. Zhao Y, Zhang G, Wei M, Lu X, Fu H, Feng F. et al. The tumor suppressing effects of QKI-5 in prostate cancer: a novel diagnostic and prognostic protein. Cancer Biol Ther. 2014;15:108-18
40. Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558-60
Author contact
![]() Corresponding author: Lei Chen, E-mail: chenleiedu.cn. Hong-Yang Wang, E-mail: hywangksina.com.
Corresponding author: Lei Chen, E-mail: chenleiedu.cn. Hong-Yang Wang, E-mail: hywangksina.com.
 Global reach, higher impact
Global reach, higher impact