13.3
Impact Factor
Theranostics 2019; 9(11):3262-3279. doi:10.7150/thno.31885 This issue Cite
Review
DNA-supramolecule conjugates in theranostics
1. Molecular Science and Biomedicine Laboratory (MBL), State Key Laboratory for Chemo/Bio-Sensing and Chemometrics, College of Chemistry and Chemical Engineering, College of Life Sciences, and Aptamer Engineering Center of Hunan Province, Hunan University, Changsha 410082, China;
2. Institute of Molecular Medicine (IMM), Renji Hospital Shanghai Jiao Tong University School of Medicine, and College of Chemistry and Chemical Engineering, Shanghai Jiao Tong University Shanghai (P. R. China).
3. Department of Chemistry, Department of Physiology and Functional Genomics, Center for Research at Bio/Nano Interface, UF Health Cancer Center, UF Genetics Institute and McKnight Brain Institute, University of Florida, Gainesville, FL 32611-7200, USA.
*Equal contribution
Received 2018-11-29; Accepted 2019-3-28; Published 2019-5-18
Abstract
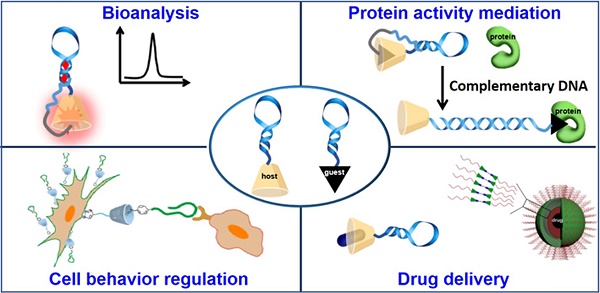
The elegant properties of deoxyribonucleic acid (DNA), such as accurate recognition, programmability and addressability, make it a well-defined and promising material to develop various molecular probes, drug delivery carriers and theranostic systems for cancer diagnosis and therapy. In addition, supramolecular chemistry, also termed "chemistry beyond the molecule", is a promising research field that aims to develop functional chemical systems by bringing discrete molecular components together in a manner that invokes noncovalent intermolecular forces, such as hydrophobic interaction, hydrogen bonding, metal coordination, and shape or size matching. Thus, DNA-supramolecule conjugates (DSCs) combine accurate recognition, programmability and addressability of DNA with the greater toolbox of supramolecular chemistry. This review discusses the applications of DSCs in sensing, protein activity regulation, cell behavior manipulation, and biomedicine.
Keywords: DNA-supramolecule conjugate, aptamer, supramolecular chemistry, DNA nanotechnology, host-guest interaction
1. Introduction
The American Association for Cancer Research states the following: "The mission of the Chemistry in Cancer Research Working Group (CICR) is to advocate the critical role of chemistry in the treatment of cancer through increasing chemistry awareness, knowledge, and capabilities of those invested in cancer research." Indeed, in accordance with this precept, chemists are collaborating with scientists representing a broad spectrum of disciplines, including, for example, biomedicine, materials engineering, biochemistry and cell biology, to investigate potential therapeutic targets and develop strategies for accurate disease diagnosis. Past decades are replete with instances of achievements made by chemists engaged in cancer research. For instance, on the basis of high-fidelity molecular recognition, easy synthesis, accurate chemical modification, programmability and addressability [1,2], deoxyribonucleic acid (DNA) has been used as a promising material with which to develop smart systems for cancer diagnosis and therapy. Furthermore, to endow DNA-based systems with diverse functions, various molecular elements, such as supramolecular host/guest molecules [3], lipid [4], organometallic complexes [5], peptides [6], proteins [7] and even nanoparticles [8], have been tethered to DNA. Herein, the applications of DNA-supramolecule conjugates (DSCs) in bioanalysis and biomedicine will be discussed based on combining the unique properties of DNA and supramolecular chemistry.
1.1 DNA supramolecular chemistry
DNA is an essential macromolecule of life, and it is composed of four different nucleotides (A, T, G, and C). All four nucleotides consist of a five-carbon sugar called deoxyribose, a phosphate group attached to the 5' carbon of deoxyribose, and a corresponding nitrogenous base (adenine, thymine, guanine or cytosine) attached to the 1' carbon of the deoxyribose. Nucleotides are joined together to form well-defined polynucleotide strands by phosphodiester bond. Founded on base pairing rules (A with T and C with G) between nitrogenous bases, two separate polynucleotide strands can form duplex helix responsible for carrying and retaining hereditary information. In 1953, James Watson and Francis Crick marked a milestone with their discovery of the double helix of DNA [9], followed by such groundbreaking discoveries as Central Dogma and polymerase chain reaction (PCR). Based on their precise recognition ability via Watson-Crick base pairing, easy synthesis and enzymatic manipulation, various DNA-based probes, such as the molecular beacon [10], TaqMan probe [11], and antisense oligonucleotide [12], have been developed for disease detection and therapy. DNA is the carrier of genetic information, but when genetic contexts are stripped, DNA remains a functional material with which to construct diverse molecular recognition probes and functional systems for disease diagnosis and therapy.
1.1.1 DNA aptamer - a promising chemical antibody
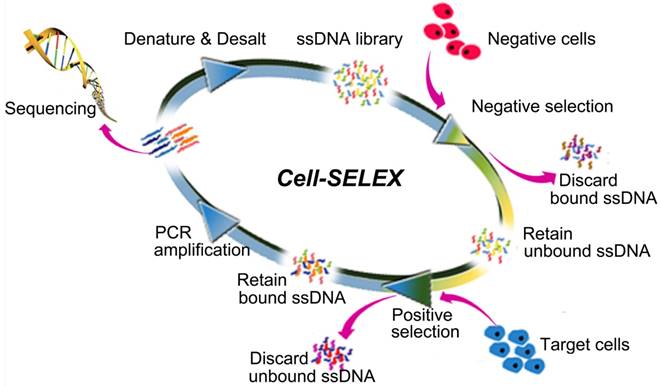
Systematic Evolution of Ligands by Exponential enrichment (SELEX) is now a common technique by which DNA aptamers, a kind of short single-stranded oligodeoxynucleotide molecule, can be selected from a DNA library [13,14]. The ability to fold into secondary and tertiary structures gives aptamers their ability to recognize such target molecules as metal ions [15,16], organic molecules [17-19], peptides [20] and protein biomacromolecules [21-24] with dissociation constants (Kds) down to picomolar values. In particular, aptamers can be selected to specifically recognize target molecules in their native conformations on the cell surface without prior knowledge of the molecular signatures of target cells when a SELEX procedure is directed against live cells (cell-SELEX) [25]. A typical cell-SELEX procedure is shown in Figure 1. Further specificity can be achieved by performing an additional subtractive selection to produce aptamers recognizing a given cell phenotype, e.g., differentiating tumor cells from normal cells. Up to now, many aptamers have been selected by cell-SELEX against various types of cancer cells, such as leukemia [26,27], pancreatic duct adenocarcinoma [28], glioblastoma [29], liver cancer [30], small cell lung cancer [31], non-small cell lung cancer [32,33], ovarian cancer [34], and breast cancer [35]. As a versatile chemical antibody, aptamers can be easily synthesized and precisely modified at specific sites for conjugation of fluorescent dyes, radionuclides, drugs and pharmacokinetic modifying agents [36-41]. Thus, aptamers can be expected to hold considerable promise for use in disease detection and targeted therapy, as well as imaging and biomarker discovery.
The scheme of Systematic Evolution of Ligands by EXponential enrichment of aptamers against live cells (Cell-SELEX). Adapted with permission from ref. [28]. Copyright (2015) Ivyspring.
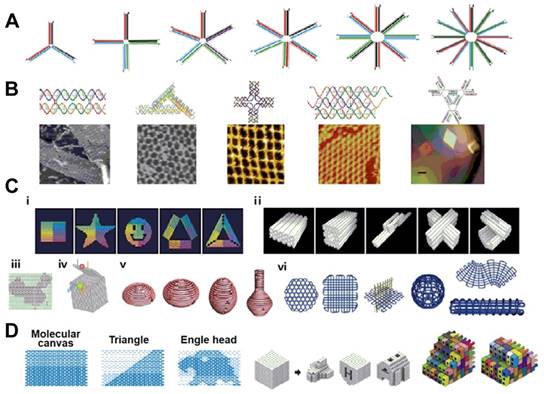
Examples of DNA nanostructures. (a) nucleic acid junctions; (b) DNA tiles with different shapes; (c) DNA origami; (d) DNA canvas. Adapted with permission from ref. [47]. Copyright (2015) Elsevier.
1.1.2 DNA - functional building blocks for smart systems
In the eukaryotic nucleus, the DNA duplex helix is tightly packaged with histone proteins into chromosomes, comprising the genetic architecture. However, by its programmability and addressability, DNA can also be used as functional building blocks for the construction of different smart nanostructures and nanosystems with nanoscale precision. Ever since the concept of DNA nanotechnology was postulated by Ned Seeman [42], various approaches, such as DNA tile assembly [43], DNA origami [44,45] and brick assembly [46], have been used to construct hierarchical and intelligent nanosystems [47] (Figure 2). Such DNA-based nanosystems have been extensively utilized in biophysics, medical diagnostics, nanoparticle and protein assembly, drug delivery and synthetic biology [48-51].
1.2 Host-guest supramolecular chemistry
Supramolecular chemistry, otherwise known as "chemistry beyond the molecule", is a promising research field and has yielded such discoveries as host-guest interaction [3,52,53] and the construction of molecular machines [54]. Supramolecular chemistry aims to develop functional chemical systems by bringing discrete molecular components together through invoking noncovalent intermolecular forces, such as hydrophobic interaction, hydrogen bonding, metal coordination, and shape or size matching [55,56]. In supramolecular chemistry, host-guest interaction based on macrocyclic molecules has been extensively investigated. Up to now, a series of biocompatible macrocyclic molecules, such as cyclodextrins (CDs), cucurbit[n]urils (CBs), calixarenes (CAs), and pillar[n]arenes, have been successfully developed as host molecules to specially encapsulate guest molecules via noncovalent forces. Such special host-guest inclusion can result in the formation of various smart supramolecular systems via the high Kds between hosts and guests. For example, the K values of CD-based host-guest complex and CB-based host-guest complex can reach 104 and 1015 M-1, respectively. Some fluorescent dyes can also be included in the cavity of various host macrocycles [57]. Once the fluorescent dyes are encapsulated, their photophysical properties will undergo significant changes owing to the change in environment, resulting in fluorescence increase or decrease. Host-guest inclusion is a facile and reversible process, enabling the development of stimuli-responsive supramolecular systems. In addition, macrocyclic molecules, such as CAs, CDs, and CBs, exhibit good biocompatibility and are friendly to biological environments. Therefore, the applications of host-guest systems are rapidly increasing and can be found in areas such as disease detection and therapy [57, 59-61].
1.3 The structure-activity of DSCs
The unique properties of DNA and host molecules, including self-assembly ability and target-recognition ability, have encouraged researchers to explore the properties and applications of DSCs. DSCs herein include DNA-host conjugates and DNA-guest conjugates. The preparation and key applications focusing on host-DNA conjugates was previously reviewed [62]. Here, we review DSCs where both host and/or guest molecules are conjugated to DNA. In addition, our focus is directed more towards biomedical and theranostic applications.
The study of DSCs can be traced back to Agrawal's 1995 work [63]. In this report, oligonucleotide-amino-β-cyclodextrin conjugates (DNA-β-CD) and oligonucleotide-adamantane conjugates (DNA-Ad) were synthesized by conjugating β-CD or Ad to the 3'-end of DNA and then examined for nuclease stability, hybridization properties, and cellular uptake. Agrawal et al. found that DNA-β-CD had increased nuclease resistance compared to its parent oligonucleotides. However, DNA-β-CD destabilized duplex formation with complementary RNA as a result of 1) steric hindrance and 2) the inclusion of nucleotides caused by β-cyclodextrin at the 3'-end of DNA. Agrawal et al. also found that the conjugated amino-β-CD did not facilitate the cellular uptake of DNA.
However, in 2009, the Komiyama group [64] revealed that the host-guest interaction between β-CD attached at the 5'-end of a DNA strand and Ad attached at the 3'-end of its complementary strand could stabilize the duplex. In particular, the melting temperature (Tm) values of 10-bp or 7-bp duplex containing β-CD/Ad were 22.1 °C and 34.0 °C higher than those of the corresponding natural duplexes. Furthermore, Inouye et al. constructed linear end-to-end assemblies of short DNA duplex based on β-CD-Ad complexation as a bio-orthogonal sticky end motif. The Tm value of the oligomer was 18 °C higher than that of the DNA duplex without β-CD/Ad modification, revealing that intermolecular host-guest interactions could drive the formation of the higher-order structure. Interestingly, free Ad or β-CD competitors could break the end-to-end assemblies and decrease the Tm, providing a strategy to manipulate the assembly of DSCs.
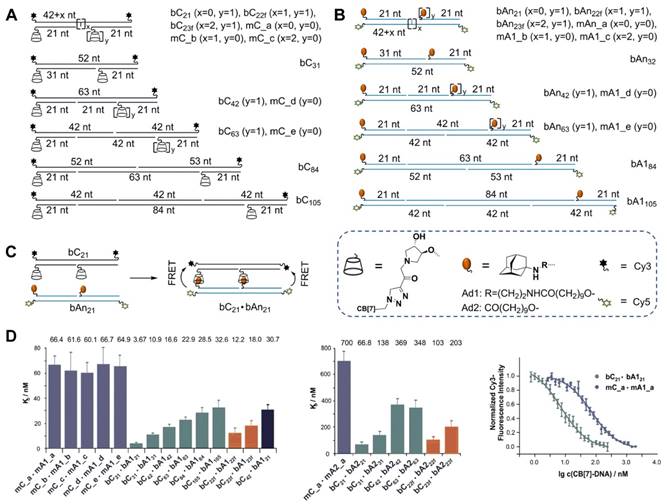
Very recently, Seitz et al. [65] developed a DSC-based approach to explore the distance limits of bivalent interactions. In their approach, monovalent (mCy) and bivalent (bCy) CB[7] receptors were prepared by assembling CB[7]-DNA conjugates on Cy5-labeled templates (Figure 3A), and monovalent (mAny) and bivalent (bAny) ligand displays were prepared by assembling Ad-DNA conjugates on Cy3-labeled templates (Figure 3B). The distances between bivalent receptors/ligands were accurately controlled within 70-360 Å based on the length of the helix in B-DNA, which is determined by 34 Å per 10 base-pairs. Two different Ad derivatives, Ad1 and Ad2, which differ in their affinity for the CB[7] host, were used in this study. Based on the Kds between each display of CB[7] and its distance-matched display of adamantane determined by measuring the fluorescence resonance energy transfer (FRET) between Cy3 and Cy5 (Figure 3C-D), Seitz et al. clearly revealed that the distance limits of bivalency were controlled by the distance between recognition modules, scaffold flexibility, and the strength of monovalent interaction. They also revealed that the lower the affinity of the receptor-ligand interaction, the lower the concentration threshold that needed to be surpassed to trigger crosslinking. These findings are expected to form a foundation for the design of potent bivalent probes and inhibitors. Taken together, these basic studies convincingly demonstrated that DSCs integrating the unique properties of DNA and supramolecular host-guest chemistry could be used in the design and preparation of nanostructures for versatile applications.
2. DSC-based sensors for bioanalysis
In DSCs, the DNA domain can be rationally designed to recognize various analytes [66,67], and the supramolecular domain can be rationally designed to recognize the desired analytes via complementarity in size, shape and charge between host and guest [68]. Therefore, a primary application of DSCs involves the sensing of various analytes. In this section, we will discuss the applications of DSCs in bioanalysis.
Identification and quantification of unsaturated fatty acid isomers in a biological system inform the study of lipid metabolism and catabolism, membrane biophysics, and pathogenesis of diseases [69], but their implementation is challenging. By using cyclodextrin as the sensing moiety, Inouye et al. reported a general sensor for fatty acid analysis [70]. Their sensor was composed of a pair of 7-nt complementary oligonucleotides (ODNs) with pyrene and α/β-CD at the opposite termini. To demonstrate the sensing mechanism and signal transduction of the as-prepared sensor, the sensor based on β-CD was titrated with a bis-adamantyl guest molecule, which could be strongly and specifically encapsulated by β-CD in a 1:2 binding model. Upon addition of the bis-adamantyl guest molecule, the two 7-nt complementary sequences were brought together to form a stable duplex structure with Tm value increasing from 33 to 50 °C. The stable duplex, in turn, resulted in gradual decrease of the fluorescence signal of pyrene monomer, while that of pyrene excimer gradually increased, revealing that pyrene-emission switching had been induced by the cooperation of DNA hybridization and host-analyte interaction. The β-CD-based sensor could discriminate three unsaturated fatty acids (arachidonic, oleic, and elaidic) from saturated fatty acids (stearic acid). To improve the discrimination ability of the sensor, Inouye et al. further developed the sensor based on α-CD, which has a smaller cavity than β-CD, for analysis of the three unsaturated fatty acids under the same experimental conditions. In these analyses, arachidonic acid could not be detected since the smaller cavity of α-CD could not incorporate it. However, oleic acid was detected more sensitively than elaidic acid. These results illustrated that the sensor based on α-CD had better selectivity and could differentiate cis/trans species. The strategy was highly modular and could be adapted to other analytes by modulating the nature of the host, e.g., porphyrin derivatives [71].
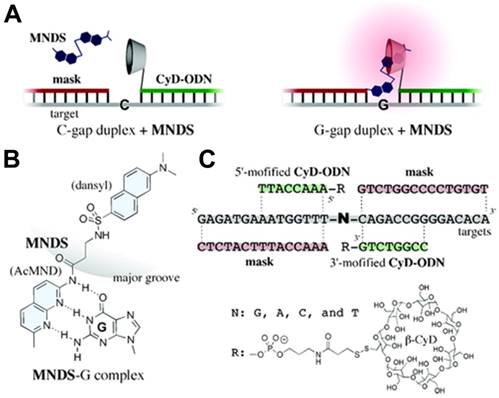
Moreover, based on the precise base-pairing rule of the DNA domain, DSCs could undoubtedly be used for gene analysis. For example, Ihara and co-workers developed a smart method for genotyping single nucleotide polymorphisms (SNP) in a homogeneous solution based on the cooperation of multiple weak interactions [72]. As shown in Figure 4, the strategy involved three components: an 8-nt DNA conjugated with β-CD (CyD-ODN), a 15-nt ''mask'' DNA strand, which simultaneously hybridized with both adjacent sequences to the SNP base of the target DNA strand, and a nucleobase-specific fluorescent DNA ligand (MNDS), which consisted of 2-acetoamide-7-methyl-1,8-naphthyridine (AcMND) and 2,6-dansyl fluorophore. In MNDS, the AcMND moiety could bind with guanine (G) by complementary hydrogen bonding, and dansyl moiety could be included with nearby cyclodextrin to form a luminous inclusion complex. Therefore, fluorescent SNP analyses could be performed by adding MNDS after the CyD-ODN strand, while the “mask” DNA strand hybridized with the target DNA strand to form N-gap duplex. Thus, when G was displayed in the gap of the ternary duplex, the specific interaction between G and AcMND in MNDS would bring dansyl moiety close to β-CD moiety in CyD-ODN, favoring the formation of the luminous inclusion complex and thus producing a strong fluorescence signal. The fluorescence signal was 13.3-fold, 25.6-fold and 23.8-fold compared to A, C and T in the gap, respectively. Since small ligands that recognize any specific nucleobases could be designed in consideration of their complementarity for hydrogen bonding, the method could be expanded for fluorescent SNP analysis in a homogeneous solution.
(A) Schematic preparation of monovalent and bivalent cucurbituril (CB[7]) receptor displays bCy. (B) Schematic preparation of monovalent and bivalent adamantane (An) ligand display b Any. (C) Schematic illustration of the bimolecular interaction between a bivalent CB[7] receptor display bC21 and a distance-matched Adn ligand display bAn21, which resulted in fluorescence resonance energy transfer (FRET). In bCn·bAn, “b” and “C” denominated bivalency and CB[7], respectively. “An” indicated the adamantane derivative where n=1 (Ad1) or n=2 (Ad2). (D) Kds for bCy receptor displays and A1y- or A2y-based ligand displays. Subscript numbers represented the number of nucleotides between hosts or guests. Cited from ref. [65].
DSC-based fluorescent sensor for detecting single nucleotide polymorphisms (SNP). (A) Sensor in the OFF state (left) where the gap nucleobase was cytosine (C) and in the ON state (right) where the gap nucleobase was guanine (G). (B) Schematic illustration of complementary hydrogen bonding between MNDS and G. (C) The structure and sequences of the components in the sensor. Adapted with permission from ref. [72]. Copyright (2009) American Chemical Society.
By integrating the recognition ability of aptamer, DSCs could be used to construct sensors with high sensitivity and selectivity to sense any target, especially low molecular weight target, for which an aptamer could be selected. It is well known that low molecular weight targets cannot typically be measured by sandwich assays because of their small size. Usually, low molecular weight targets are detected with less sensitive and less specific competitive methods [73]. On the other hand, a split aptamer-based sandwich assay, in which the split fragments of aptamer can specifically form a ternary assembly with low molecular weight targets, has now been developed for sensing low molecular weight targets [74]. However, such splitting can still compromise the aptamer's target affinity, resulting in low sensitivity of these split aptamer-based sensors.
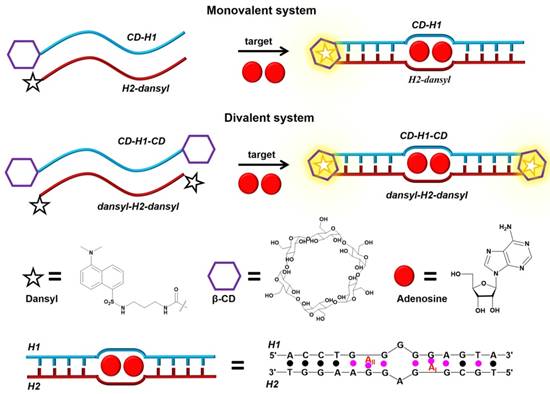
Nevertheless, based on strong host-guest interaction, DSCs can be developed to circumvent this drawback. For example, Peyrin's group [75] developed an exquisite and sensitive split aptamer-based sandwich assay for sensing adenosine triphosphate (ATP) by using host-guest interaction to stabilize aptamer-analyte ternary complex (Figure 5). In their strategy, two DSCs, CD-H1-CD and dansyl-H2-dansyl, were designed. The former was one fragment of ATP aptamer with modified β-CD at both ends, and the latter was another fragment of ATP aptamer functionalized with dansyl dye at both ends. The presence of ATP would bring CD-H1-CD and dansyl-H2-dansyl together to form a sandwich-like duplex-type structure, which would be further stabilized by host-guest interactions between β-CD and dansyl dye. Concomitantly, the formation of β-CD-dansyl dye luminous inclusion complex would, in turn, produce a high signal-to-background fluorescence signal for quantitative analysis of ATP. The strategy efficiently enhanced the sensitivity of the split aptamer-based sensor. Moreover, the strategy could be extended to multiplexed analysis based on the diversity of available responsive dye-macrocycle systems [76,77].
3. DSCs for protein activity mediation
Proteins are the executors of life activities and involved in various cellular processes [78], such as communication, differentiation, proliferation, migration and apoptosis [79]. Therefore, accurately mediating protein activities will facilitate elucidating pathogenic mechanisms and developing theranostic technologies. Mediating protein activities in a spatiotemporal manner has been a consistent goal in many scientific fields, especially biomedicine. Based on the input oligonucleotide or target-selective structural transformation, DNA serves as an efficient and functional material to construct molecular tools for regulating protein activities. In addition, intramolecular host-guest interactions arising from a macrocycle host accommodating some protein-binding inhibitors within its cavity can provide a steric barrier against inhibitors accessing their targeted proteins. By integrating the features of DNA and host-guest supramolecular chemistry, DSCs can control the encapsulation of inhibitors via input stimuli-selective structural transformation of DNA, providing a kind of efficient molecular switch to mediate protein activities.
Schematic illustration of the host-dye reporter sandwich-type aptamer sensing platform for adenosine detection. Adapted with permission from ref. [75]. Copyright (2015) American Chemical Society.
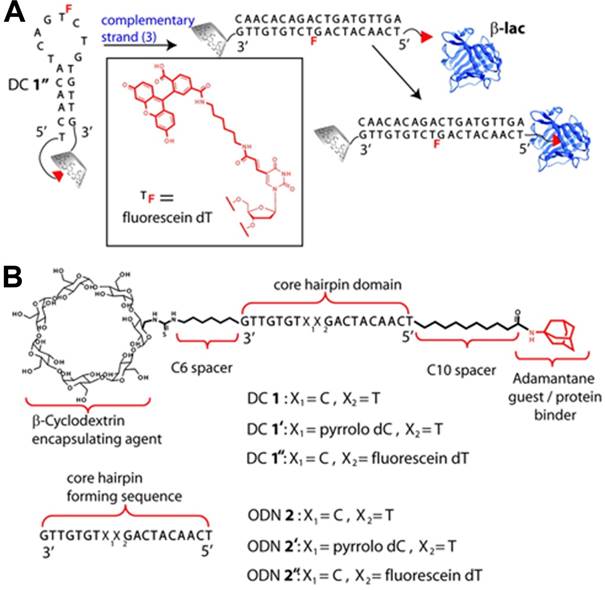
DNA molecular beacons can undergo input oligonucleotide-selective structural transformation from a stem-loop conformation to a duplex. Based on their smart molecular design, Jayawickramarajah et al. constructed a DSC-based molecular switch by tethering Ad and β-cyclodextrin to the 5' and 3' termini of 19-base hairpin-forming DNA [80], respectively, to modulate protein activity. As shown in Figure 6, the 19-base DNA was composed of a five base pair stem, an unhybridized eight nucleotide loop region, and a non-base-paired thymidine residue at the 5'-end serving as an external toehold. The Ad in the molecular switch served as a prototype hydrophobic protein-binder and a well-established guest for β-CD. Within the hairpin-forming molecular switch, a robust Ad/β-CD inclusion complex formed from the synergism between the two binding partners of DNA hybridization and supramolecular host-guest interaction. Thus, the protein-binder Ad in the hairpin-forming molecular switch could not bind to β-Lactoglobulin (β-lac) and was inactive. However, in the presence of a cognate input ODN, the hairpin undergoes a conformational change to a duplex, forcing Ad/β-CD inclusion complex apart and resulting in a freely accessible Ad. Thus, the protein-binder Ad in the duplex molecular switch became active and can strongly bind to β-lac. Moreover, the input ODN containing one or two base pair mismatches was unable to effectively switch the hairpin form to the duplex state, indicating that the system could perfectly control protein activity through accurate DNA recognition.
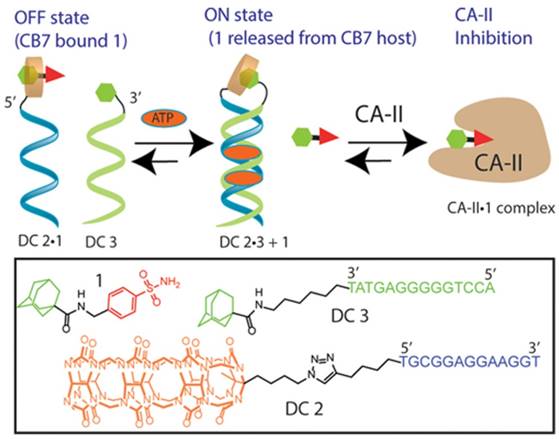
With the development of functional DNA [81,82], e.g., aptamer, such DSC-based molecular switches could also be tuned to other input stimuli, such as proteins, small molecules or metal ions, to regulate protein activity. For example, Jayawickramarajah et al. developed a smart adenosine triphosphate (ATP)-selective molecular switch via an ATP-binding aptamer (ABA) to regulate the activity of carbonic anhydrase-II (CA-II) [83]. In brief, the DNA switch had three main components: a CB7-binding Ad-benzenesulfonamide (a prototype CA-II inhibitor) Janus molecule 1, DNA (residues 1-13 of ABA)-CB7 chimera (DC2), and DNA (residues 14-27 of ABA)-Ad chimera (DC3). Owing to the strong binding between Ad in Janus molecule 1 and CB7 in DC2, the host-guest complex DC2•1 would be formed, preventing the benzenesulfonamide moiety of the Janus molecule 1 from penetrating to the deep (15 Å) and conical active site of CA-II. The DNAs of DC2•1 and DC3 were not complementary to each other. In addition, the electrostatic repulsion between the negatively charged DNA backbones would significantly impede the intermolecular CB7/Ad host-guest interaction between DC2•1 and DC3. Therefore, in the absence of ATP, the CA-II inhibitor in Janus molecule 1 was expected to remain inactive as a result of the formation of DC2•1. However, in the presence of ATP, DC2•1 and DC3 would form an ATP-templated noncanonical duplex with the 5′ terminus of DC2•1 positioned in proximity to the 3′ terminus of DC3. This duplex architecture would, in turn, drive stronger host-guest interaction between the CB7 moiety of DC2•1 and the Ad moiety of DC3, evicting the Janus molecule 1 for optimal inhibition of CA-II (Figure 7). The molecular switch was highly modular and could be easily adapted to regulate various protein activities through biologically/clinically relevant stimuli.
A protein regulator (DC 1) based on the DNA hairpin integrated with host-guest system responsive to an oligonucleotide trigger. (A) Schematic illustration of response mechanism of DC 1 in the presence of a cognate complementary oligonucleotide (ODN). (B) The structures and sequences of DC 1, ODN 2, and their respective derivatives. Adapted with permission from ref. [80]. Copyright (2011) American Chemical Society.
A DSC-based transducer for input-responsive protein inhibition. Adapted with permission from ref. [83]. Copyright (2017) American Chemical Society.
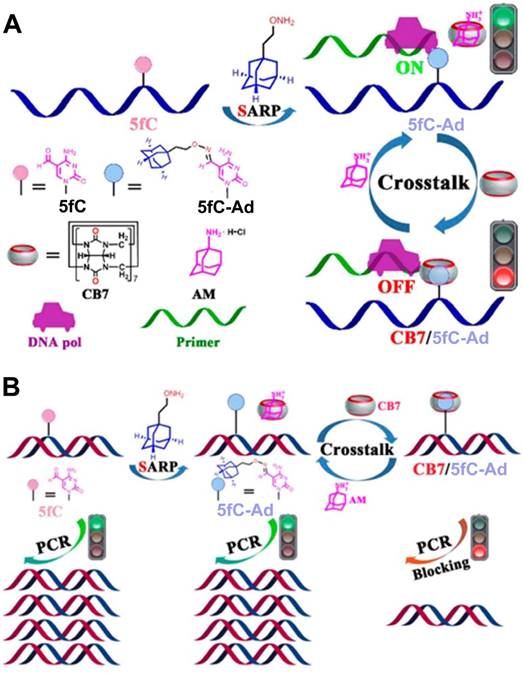
5-Formylcytosine (5fC), one of the key intermediate DNA methylation and demethylation processes, is an epigenetic mark in mammals [84]. Thus, 5fC can serve as an attractive target for chemical intervention to precisely manipulate 5fC-associated biochemical reactions and help gain insight into its important role in biological processes. Photochemical activation and deactivation on the basis of installing a photolabile protecting group on DNA substrate have provided a temporospatial intervention [85]. However, such intervention was achieved in an irreversible manner. Therefore, Zhou et al. developed a reversible intervention strategy to control the related bioactivity occurring at 5fC sites by manipulating the host-guest interactions between CB7 and 5fC-AD [86], as described in Figure 8. In the reversible strategy, a supramolecular aldehyde reactive probe (SARP), synthesized by functionalizing Ad moiety with the hydroxylamine group, served as a site selective tag for converting the 5fC to 5fC-Ad. Based on host-guest interaction, CB7 could selectively target the 5fC-AD nucleotide in DNA. The interaction between CB7 and 5fC-AD had no effect on the hydrogen bonding properties of natural nucleobases in duplex DNA, but through steric hindrance, it did create obstructions that prevented the enzyme from binding to the substrate and caused a variety of 5fC-targeted biochemical reactions, such as restriction endonuclease digestion, DNA polymerase elongation, and polymerase chain reaction. Importantly, the intervention caused by CB7/5fC-AD host-guest interactions could be reversed via treatment with adamantanamine. Based on the unique features of reversibility, the strategy showed promising potential for host-guest chemistry for future DNA/RNA epigenetics.
(A) Schematic illustration of the design and working principle of host-guest interaction-based chemical intervention. (B) Schematic illustration of the reversible regulation of DNA pol elongation and PCR reaction of 5fC-targeted DNA by CB7-adamantanamine. Adapted with permission from ref. [86]. Copyright (2017) American Chemical Society.
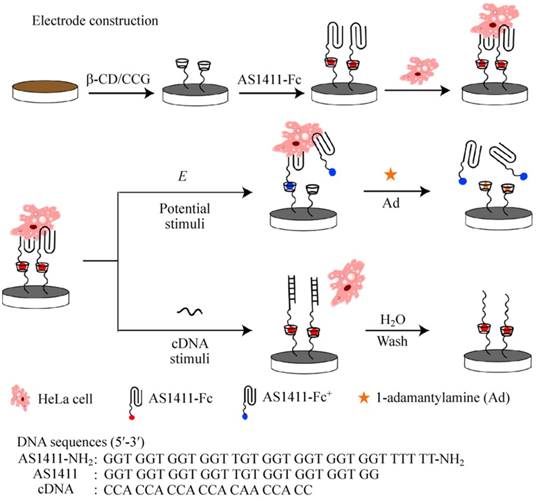
Schematic illustration of electrode construction and cell catch/release on AS1411-Fc/β-CD/CCG-Nafion-modified electrode mediated by different stimuli. Adapted with permission from ref. [94]. Copyright (2015) Springer.
4. DSCs for cell behavior regulation
Dynamic regulation of cell behavior is important to biomedical research because it facilitates understanding of the fundamental principles of life science [87], as well as developing the next generation of theranostic technology. Aptamers, also called chemical antibodies, as noted above, can recognize various cell lines with high affinity and selectivity, providing an effective kind of tool to capture and manipulate cells. Supramolecular host-guest interaction is usually based on various noncovalent bonding modes, such as hydrogen bonding, cationic-anionic electrostatic interactions, aromatic interactions, metal-ligand bonding, hydrophobic interaction, and charge-transfer interaction. Owing to the dynamic and reversible nature of these noncovalent bonds, the interaction between host-guest is dynamic and reversible, and it can be affected by external stimuli, such as temperature [88], voltage [89], enzymatic activity [90], light [91] and pH [92,93]. Again, therefore, aptamer-based DSCs that integrate the target specificity of aptamers and supramolecular chemistry are expected to act as effective molecular tools to regulate cell behavior.
For example, on the basis of supramolecular ferrocene/β-cyclodextrin (Fc/β-CD) interaction and reversible AS1411 aptamer structural transition, Qu's group constructed a novel reusable dual-functionalized graphene substrate to realize electrochemical and DNA-triggered cell capture/release [94]. As shown in Figure 9, graphene modified with β-CD was immobilized on glassy carbon electrode or ITO substrate with nafion. Then Fc-labeled AS1411 (AS1411-Fc) was assembled on the substrate via Fc/β-CD host-guest interaction. Since AS1411 has high binding affinity to nucleolin protein, nucleolin-expressing HeLa cells as models could be efficiently captured on the substrate. On one hand, applying an oxidizing potential onto the modified surface would result in the electrochemical oxidation of ferrocenyl moieties, decrease the binding affinity of Fc to β-CD, and trigger the release of HeLa cells from the substrate. The addition of competitor 1-adamantylamine could further enhance the release of HeLa cells because of its higher binding affinity to β-CD cavity than that of Fc. On the other hand, applying a complementary DNA sequence of AS1411 could also trigger the release of HeLa cells from the substrate because the formation of duplex between complementary DNA and AS1411 would eliminate the binding ability of AS1411.
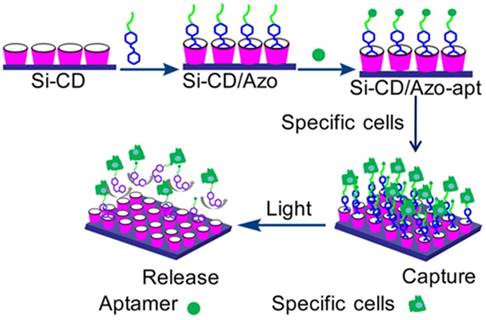
Light was also chosen as an external stimulus to regulate cell capture and release based on its outstanding temporospatial property. Trans-azobenzene (trans-Azo) [95] is an excellent guest molecule and can spontaneously form a host-guest complex with β-CD via van der Waals force and hydrophobic interactions, whereas cis-Azo cannot form an inclusion complex with β-CD because of the size mismatch between them. Based on the conformation change of Azo induced by light, Wang et al. developed a novel photoresponsive surface with specific aptamer S2.2 to trigger MCF-7 cell capture/release via host-guest interactions between Azo and β-CD [96], which was described in detail in Figure 10. In this study, aptamer S2.2 was first covalently linked with guest molecule Azo, and then immobilized on silicon (Si) substrate modified with host molecule β-CD which via host-guest interaction. On the basis of accurate recognition of aptamer, the Si-CD/Azo surface (Si-CD/Azo-apt) efficiently captured the specific cells (MCF-7). Once irradiated by UV light, Azo switched from trans- to cis-isomers, which could not be recognized by β-CD because of the unmatched host-guest size, thus releasing the captured MCF-7 cells. Moreover, the surface could selectively capture MCF-7 cells from a binary cell mixture.
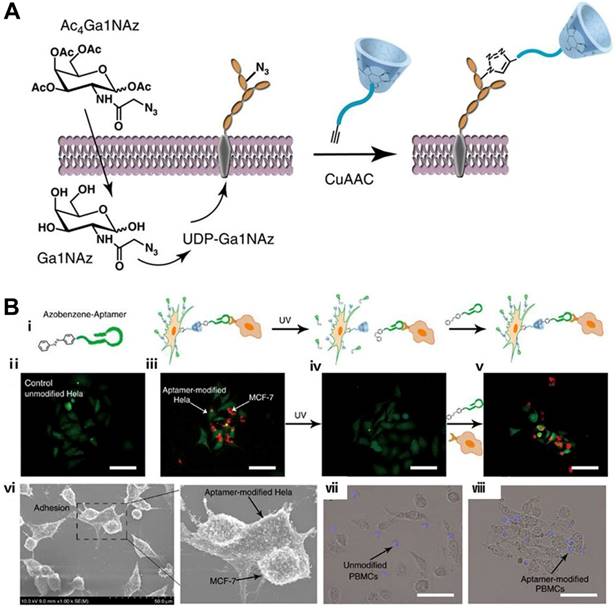
Cell-cell interaction is a basic cellular behavior in the living organism. Undoubtedly, cell biology would be informed by our ability to observe the flow of communication or material exchange between cells. Therefore, controlling intercellular interaction via nongenetic engineering techniques would be desirable. Based on a metabolic labeling approach, bioorthogonal click reaction and aptamer-based DSC, Qu's group developed an efficient strategy to realize spatiotemporal control of cell-cell reversible interactions [97]. In this strategy, β-CD was engineered on cell surface by feeding cells with peracetylated N-azidoacetylgalactosamine (Ac4GalNAz), followed by conjugating with alkynyl and poly(ethylene glycol)-modified β-CD (alkynyl-PEG-β-CD) via a bioorthogonal copper(I)-catalyzed click reaction (Figure 11A). By using an Azo-incorporated homobifunctional crosslinking agent as a switchable recognition component, the group first demonstrated the reversible control of cell assembly and disassembly through light manipulation of conformation changes from trans- to cis-isomer of Azo. Furthermore, based on the aptamer's high binding ability to target cells, the authors developed Azo-labeled mucin 1 aptamer, which could recognize mucin 1 protein expressed on epithelial cancer cells, e.g., MCF-7 cells. After Azo-labeled-aptamers (Azo-aptamer) were anchored on the surface of β-CD-labeled HeLa cells through host-guest interaction, aptamer-modified HeLa cells could specifically recognize target MCF-7 cells and form cell-cell heterotypic adhesion. When the system was exposed to UV radiation, Azo changed its conformation from trans- to cis-isomer and detached from the cavity of cyclodextrin, resulting in cell separation. After cell separation, β-CD-labeled HeLa cells could once again be modified with Azo-aptamer for cell-cell adhesion. Therefore, the strategy could enable light-controllable reversible assembly of cell-cell adhesion, exhibiting obvious advantages over the previously reported irreversible effects. The strategy was further successfully applied in cell-based therapy. Peripheral blood mononuclear cells (PBMCs) modified with aptamer could be effectively redirected towards target cells (Figure 11B), resulting in enhanced cell apoptosis. The DSC-based strategy allowed control of cell behavior and thus benefited studies of contact-dependent cell-cell communication.
Schematic illustration of cell capture/release based on the UV light-mediated interaction between azobenzene and β-CD. Adapted with permission from ref. [96]. Copyright (2016) American Chemical Society.
Schematic illustration of β-CD modification (a) and photocontrolled host-guest recognition (B, i) on cell surfaces. (ii) Unmodified HeLa cells. (iii) Mucin 1 aptamer-modified HeLa cells (green) could specifically recognize MCF-7 cells (red) and form heterotypic cell adhesion. (iv) Ultraviolet irradiation followed by washing with PBS induced the release of MCF-7 cells from the surface of HeLa cells. (v) Aptamer-modified HeLa cells could be formed again for cell-aptamer-cell assembly. (vi) Scanning electron microscope images of cell interactions. Microscope images of heterotypic cell adhesion between unmodified PBMCs (vii), modified PBMCs (viii) and MCF-7 cells. Adapted with permission from ref. [97]. Copyright (2016) Springer Nature.
5. DSCs-based nanostructure for drug delivery
DSCs, combining the unique properties of DNA and host-guest supramolecular chemistry, provide a kind of functional building block to construct various nanosystems for disease detection and therapy. For instance, the Varghese group [98] assembled a DSC-based nanogel as an efficient drug carrier for tumor therapy by host-guest interaction. As shown in Figure 12, DSCs with an appending β-CD at the 5' ends were first assembled to form branched X-shaped DNA (β-CD-X-DNA) or Y-shaped DNA (β-CD-Y-DNA), which served as multivalent host DNA in nanogel formation. The host DNA was assembled with adamantyl terminated 8-arm PEG polymer to form nanogel by multivalent host-guest interaction between β-CD and Ad. DLS and AFM revealed that DNA nanogel with size ranging from 100 nm to 300 nm could be formed at very low concentrations of the host and guest. Furthermore, the nanogels would be hierarchically self-assembled to form a hydrogel at high concentration of the host and guest. Interestingly, the as-prepared DNA gel presented excellent cell permeability and doxorubicin-loading ability, providing a promising candidate for drug delivery applications. The interaction between host and guest could be easily manipulated by external stimuli; therefore, stimuli-responsive DNA nanogel could be expected for cancer therapy.
In addition, Varghese et al. reported a self-assembled DNA vesicle via host-guest recognition between β-CD-functionalized DNA and Ad-modified hydrophobic guests [99]. As shown in Figure 13, DSCs were prepared by conjugation of a 5'-alkyne-modified DNA with azide-functionalized β-CD via a copper-assisted click chemistry reaction. Ad-modified alkyl chains tethered phenylene ethynylene and porphyrin that served as the guests. Supramolecular DNA amphiphiles were prepared by assembling DNA-β-CD with Ad-modified hydrophobic guests by host-guest interaction. The resultant DNA amphiphiles could spontaneously undergo amphiphile-driven self-assembly to form thermally stable nano- to microsized DNA vesicles, which were like the phospholipid bilayer of a cell. Varghese et al. further demonstrated the DNA-directed surface addressability of the vesicles by assembling these vesicles on Au-NPs modified with DNA via hybridization. In contrast to covalent DNA amphiphiles, the noncovalent approach provided high coupling efficiency (~77%) to prepare DNA amphiphiles. The reversibility of host-guest interactions and the modular assembly made it a promising method of fabricating not only smart nanocarriers, but also responsive vesicles.
Schematic illustration of the self-assembly of β-CD-tethered branched X- or Y-DNA and the formation of doxorubicin (DOX)-loaded DNA nanogels via multivalent host-guest interactions between β-CD with 8-arm star PEG polymer carrying adamantane (G8). Chemical structures of β-CD, DOX and G8 are shown in the box. Adapted with permission from ref. [98]. Copyright (2017) the Royal Society of Chemistry.
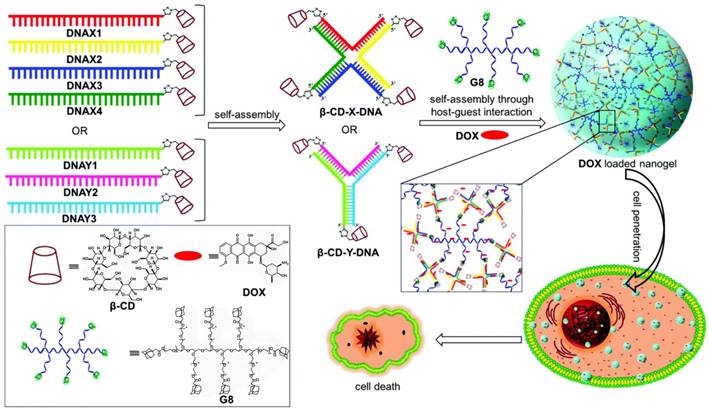
(A) Schematic illustration of structures of β-CD-functionalized DNA hosts (DNA1-β-CD and DNA2-β-CD) and hydrophobic guests (1, adamantane-modified phenylene ethynylene; 2, adamantane-modified porphyrin). (B) Schematic illustration of the preparation of β-CD- functionalized DNA hosts via click chemistry and the amphiphile-driven self-assembly of β-CD-functionalized DNA hosts/hydrophobic guests into DNA-decorated vesicles. Adapted with permission from ref. [99]. Copyright (2017) The Royal Society of Chemistry.
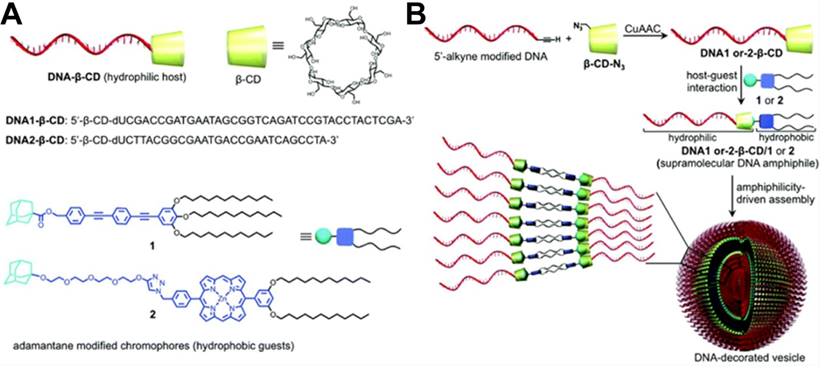
(A) Schematic illustration of controlled-release azobenzene-modified antisense oligonucleotide (ASO) from β-CD-modified nanocarriers in living cells based on cooperative interaction of UV light and competitor Fc. (B) Silencing efficiency (%) of β-CD-AuNP/antisense oligonucleotide under UV-light irradiation for different times with (a) and without competitor Fc (b) addition. Adapted with permission from ref. [100]. Copyright (2014) American Chemical Society.
Apart from facilitating the formation of DNA nanostructures for drug delivery, host/guest molecules also provide a good platform for delivery of biomolecules, including antisense oligonucleotides and toxic proteins. For instance, Yang et al. [100] successfully delivered azobenzene-modified antisense oligonucleotides to green fluorescence protein-expressing lung cancer A549 (A549-GFP) cells with gold nanoparticles (AuNPs) modified with mercapto-β-CD as a carrier (Figure 14). Based on the host-guest interaction, azobenzene-conjugated antisense oligonucleotide could be efficiently loaded on the carrier. By simultaneously applying UV light and competitor Fc to shift the equilibrium of the inclusion-exclusion process between trans-Azo and β-CD, Yang et al. realized the remote-controlled release of antisense oligonucleotides and achieved about 62.4% gene silence of GFP in A549-GFP cells. The strategy might show potential for orthogonal DNA or siRNA delivery and therapeutic activation.
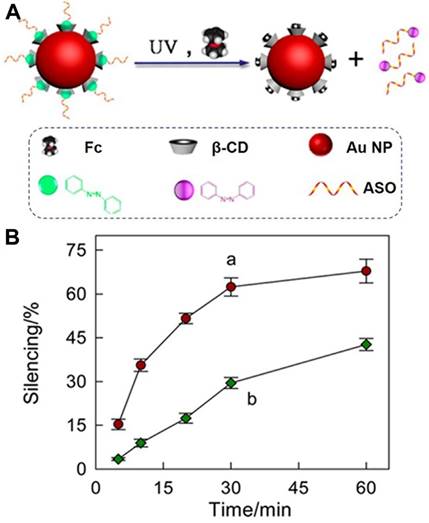
Based on high affinity, specific recognition ability, easy synthesis, precise chemical modification and designability of aptamer, our group reported a β-CD-modified circular bivalent aptamer (cb-apt-β-CD) as a delivery platform for intracellular delivery of toxic proteins and small molecules via host-guest interaction [101], as described in Figure 15. In contrast to monoaptamer, the circular bivalent aptamer (cb-aptamer or cb-apt) presented improved thermal stability, nuclease resistance and binding affinity [102]. Therefore, cb-aptamer could efficiently recognize and accumulate in tumor. In this study, we first attached a drug container, β-CD, to monoaptamer sgc8 that targets protein tyrosine kinase-7 on the cell surface. After cyclizing the as-prepared mono-apt-β-CD with mono-aptc, a complementary sequence, cb-apt-β-CD, was formed. Incorporating β-CD with cb-apt not only retained the high serum stability of the circular bivalent aptamer, but also enabled encapsulation of a hydrophobic small-molecule, or Ad-modified protein, into cb-apt, forming a supramolecular ensemble for enhanced intracellular delivery. We demonstrated that cb-apt-β-CD could efficiently deliver N-heterocyclic carbene-gold(I) and cytotoxic macromolecule saporin to inhibit tumor cell growth, highlighting the potency of using the supramolecularly engineered cb-apt as a general platform for the delivery of various therapeutics and for new biomedical applications.
(A) Schematic illustration of the synthesis of cb-apt-β-CD and the assembly of cb-apt-protein based on host-guest interaction. (B) Schematic illustration of cb-apt-β-CD for enhanced protein delivery. Adapted with permission from ref. [101]. Copyright (2018) American Chemical Society.
6. Perspectives of DSCs in disease theranostics
By integrating the unique properties of DNA and host/guest supramolecules, e.g., programmed self-assembly, precise recognition, and input-responsive features, DSCs have provided well-defined and promising materials to construct smart systems, such as a) sensors capable of detecting small biomolecules and even target DNA, b) regulators that accurately control protein activity, c) operators capable of regulating cell behavior via relevant inputs, and d) nanostructures that specially deliver drugs and biomacromolecules in cancer cells.
Along with rapid advancement of nucleic acid chemistry, supramolecular chemistry, nanotechnology, and conjugation techniques, DSCs have shown exceptional potential for biomedical applications. However, it is worth noting that the use of DSC-based smart systems still faces many practical challenges. In particular, since the preparation and the application of major DSCs are based on the use of β-CD as the host molecule, applications of DSCs in biomedicine depend on developing diverse guest-host systems, which is challenging. For instance, developing diverse dye-macrocycle systems will be expected to facilitate achieving multiplexed analysis of proteins or SNPs. To ensure biological safety, along with biomedical applications of DSC-based systems, exploring the interaction between the existing supramolecular systems and living organisms and developing biocompatible and biodegradable supramolecular systems are also important tasks. The long-term toxicity, immunological reaction, biodegradation, excretion of host-guest system and the introduced external functional moieties all need to be systematically investigated in living organisms. To improve the synthetic efficiency of DSCs and develop intelligent DSC-based systems, developing diverse artificial bases, especially host- and guest-based artificial bases, is also an important research field. The introduction of artificial bases not only increases the efficiency of synthesis and the diversity of DSCs, but also endows DSCs with new properties with which to develop DSC-based intelligent systems responsive to external or intracellular stimulus. In summary, the current applications of DSCs remain in the preliminary stage. However, DSCs can be expected to impact future biomedical applications, including those involving theranostics, as researchers from different disciplines, including chemists, join hands, as suggested by the CICR mission statement quoted above.
Abbreviations
A: Adenine; ABA: ATP-binding aptamer; AcMND: 2-acetoamide-7-methyl-1,8-naphthyridine; Ac4GalNAz: azidoacetylgalactosamine; Ad: adamantane; ASO: antisense oligonucleotide; ATP: adenosine triphosphate; AuNP: gold nanoparticle; Azo: azobenzene; C: cytosine; CA-II: carbonic anhydrase-II; CA: calixarene; CB: cucurbit[n]uril; cb-apt: circular bivalent aptamer; CD: cyclodextrin; α-CD: α-cyclodextrin; β-CD: β-cyclodextrin; CICR: cancer research working group; DC2: DNA-CB7 chimera; DC3: DNA-Ad chimera; DNA: deoxyribonucleic acid; DSC: DNA-supramolecule conjugate; Fc: ferrocene; 5fC: 5-formylcytosine; FRET: fluorescence resonance energy transfer; G: guanine; GFP: green fluorescence protein; Kd: dissociation constant; β-lac: β-Lactoglobulin; MNDS: nucleobase-specific fluorescent DNA ligand; ODN: oligonucleotide; PBMC: peripheral blood mononuclear cell; PEG: poly(ethylene glycol); SELEX: systematic evolution of ligands by exponential enrichment; Si: silicon; SNP: single nucleotide polymorphisms; T: thymine; Tm: melting temperature.
Acknowledgements
This work is supported by NIH GM R35 127130 and NSF 1645215 and by NSFC grants (NSFC 21827811) and the Science and Technology Project of Hunan Province (2017XK2103). It is also supported by NSFC grants (21327009, 21804037 and 61527806), by the National Natural Science Foundation of China (31701249), and by Fundamental Research Funds for the Central Universities.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Beal PA, Dervan PB. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991;251:1360-3
2. Caruthers MH, Matteucci MD. Synthesis of deoxyoligonucleotides on a polymer support. J Am Chem Soc. 1981;103:3185-91
3. Yu G, Jie K, Huang F. Supramolecular amphiphiles based on host-guest molecular recognition motifs. Chem Rev. 2015;115:7240-303
4. Langecker M, Arnaut V, Martin TG, List J, Renner S, Mayer M. et al. Synthetic lipid membrane channels formed by designed DNA nanostructures. Biophys J. 2013;104:545a
5. Stewart KM, Rojo J, McLaughlin LW. Ru(II) tris(bipyridyl) complexes with six oligonucleotide arms as precursors for the generation of supramolecular assemblies. Angew Chem Int Ed. 2004;43:5808-11
6. Jiang T, Meyer TA, Modlin C, Zuo X, Conticello VP, Ke Y. Structurally ordered nanowire formation from co-assembly of DNA origami and collagen-mimetic peptides. J Am Chem Soc. 2017;139:14025-8
7. Brodin JD, Sprangers AJ, McMillan JR, Mirkin CA. DNA-mediated cellular delivery of functional enzymes. J Am Chem Soc. 2015;137:14838-41
8. Degliangeli F, Kshirsagar P, Brunetti V, Pompa PP, Fiammengo R. Absolute and direct microRNA quantification using DNA-gold nanoparticle probes. J Am Chem Soc. 2014;136:2264-7
9. Watson JD, Crick FH. Molecular structure of nucleic acids. Nature. 1953;171:737-8
10. Tyagi S, Kramer FR. Molecular beacons: probes that fluorescence upon hybridization. Nat Biotechnol. 1996;14:303-8
11. Boyle DG, Boyle DB, Olsen V, Morgan JA, Hyatt AD. Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis Aquat Organ. 2004;60:141-8
12. Crooke ST, Bennett CF. Progress in antisense oligonucleotide therapeutics. Annu Rev Pharmacol Toxicol. 1996;36:107-29
13. Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818-22
14. Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505-10
15. Ciesiolka J, Gorski J, Yarus M. Selection of an RNA domain that binds to Zn2+. RNA. 1995;1:538-50
16. Hofmann HP, Limmer S, Hornung V, Sprinzl M. Ni2+-binding RNA motifs with an asymmetric purine-rich internal loop and a G-A base pair. RNA. 1997;3:1289-300
17. Ellington AD, Szostak JW. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992;355:850-2
18. Mannironi C, Di Nardo A, Fruscoloni P, Tocchini-Valentini GP. In vitro selection of dopamine RNA ligands. Biochemistry. 1997;36:9726-34
19. Stojanovic MN, Landry DW. Aptamer-based colorimetric probe for cocaine. J Am Chem Soc. 2002;124:9678-9
20. Xu W, Ellington AD. Anti-peptide aptamers recognize amino acid sequence and bind a protein epitope. Proc Natl Acad Sci USA. 1996;93:7475-80
21. Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc Natl Acad Sci USA. 1993;90:3745-9
22. Green LS, Jellinek D, Jenison R, Ostman A, Heldin CH, Janjic N. Inhibitory DNA ligands to platelet-derived growth factor B-chain. Biochemistry. 1996;35:14413-24
23. Saito T, Tomida M. Generation of inhibitory DNA aptamers against human hepatocyte growth factor. DNA Cell Biol. 2005;24:624-33
24. Ng EW, Shima DT, Calias P, Cunningham ET Jr, Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov. 2006;5:123-32
25. Fang X, Tan W. Aptamers generated from cell-SELEX for molecular medicine: a chemical biology approach. Accounts Chem Res. 2009;43:48-57
26. Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P. et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci USA. 2006;103:11838-43
27. Mallikaratchy P, Tang Z, Kwame S, Meng L, Shangguan D, Tan W. Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt's lymphoma cells. Mol Cell Proteomics. 2007;6:2230-8
28. Wu X, Zhao Z, Bai H, Fu T, Yang C, Hu X. et al. DNA Aptamer Selected against Pancreatic Ductal Adenocarcinoma for in vivo Imaging and Clinical Tissue Recognition. Theranostics. 2015;5:985-94
29. Wu X, Liang H, Tan Y, Yuan C, Li S, Li X. et al. Cell-SELEX aptamer for highly specific radionuclide molecular imaging of glioblastoma in vivo. PLoS One. 2014;9:e90752
30. Shangguan D, Meng L, Cao ZC, Xiao Z, Fang X, Li Y. et al. Identification of liver cancer-specific aptamers using whole live cells. Anal Chem. 2008;80:721-8
31. Chen HW, Medley CD, Sefah K, Shangguan D, Tang Z, Meng L. et al. Molecular recognition of small-cell lung cancer cells using aptamers. ChemMedChem. 2008;3:991-1001
32. Zhao Z, Xu L, Shi X, Tan W, Fang X, Shangguan D. Recognition of subtype non-small cell lung cancer by DNA aptamers selected from living cells. Analyst. 2009;134:1808-14
33. Jiménez E, Sefah K, López-Colón D, Van Simaeys D, Chen HW, Tockman MS. et al. Generation of lung adenocarcinoma DNA aptamers for cancer studies. PLoS One. 2012;7:e46222
34. Van Simaeys D, López-Colón D, Sefah K, Sutphen R, Jimenez E, Tan W. Study of the molecular recognition of aptamers selected through ovarian cancer cell-SELEX. PLoS One. 2010;5:e13770
35. Zhang N, Bing T, Shen L, Song R, Wang L, Liu X. et al. Intercellular connections related to cell-cell crosstalk specifically recognized by an aptamer. Angew Chem Int Ed. 2016;55:3914-8
36. Shi H, He X, Wang K, Wu X, Ye X, Guo Q. et al. Activatable aptamer probe for contrast-enhanced in vivo cancer imaging based on cell membrane protein-triggered conformation alteration. Proc Natl Acad Sci USA. 2011;108:3900-5
37. Wu YR, Sefah K, Liu Haipeng, Wang RW, Tan WH. DNA aptamer-micelle as an efficient detection/delivery vehicle toward cancer cells. Proc Natl Acad Sci USA. 2010;107:5-10
38. Niu W, Chen X, Tan W, Veige AS. N-Heterocyclic Carbene-Gold(I) Complexes Conjugated to a Leukemia-Specific DNA Aptamer for Targeted Drug Delivery. Angew Chem Int Ed. 2016;55:8889-93
39. Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Discov. 2010;9:537-50
40. Zhou J, Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discovery. 2017;16:181-202
41. Tan WH, Donovan MJ, Jiang JH. Aptamers from cell-based selection for bioanalytical applications. Chem Rev. 2013;113:2842-62
42. Seeman NC. DNA in a material world. Nature. 2003;421:427-31
43. Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science. 2003;301:1882-4
44. Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H. DNA origami with complex curvatures in three-dimensional space. Science. 2011;332:342-6
45. Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297-302
46. Ke Y, Ong LL, Shih WM, Yin P. Three-dimensional structures self-assembled from DNA bricks. Science. 2012;338:1177-83
47. Chao J, Lin Y, Liu H, Wang L, Fan C. DNA-based plasmonic nanostructures. Mater Today. 2015;18:326-35
48. Aldaye FA, Palmer AL, Sleiman HF. Assembling materials with DNA as the guide. Science. 2008;321:1795-9
49. Bandy TJ, Brewer A, Burns JR, Marth G, Nguyen T, Stulz E. DNA as supramolecular scaffold for functional molecules: progress in DNA nanotechnology. Chem Soc Rev. 2011;40:138-48
50. Seeman NC, Sleiman HF. DNA nanotechnology. Nat Rev Mater. 2017;3:17068
51. Pinheiro AV, Han D, Shih WM, Yan H. Challenges and opportunities for structural DNA nanotechnology. Nat Nanotechnol. 2011;6:763-72
52. Stoddart JF. Mechanically Interlocked Molecules (MIMs)-Molecular Shuttles, Switches, and Machines (Nobel Lecture). Angew Chem Int Ed. 2017;56:11094-125
53. Yu Z, Zhang J, Coulston RJ, Parker RM, Biedermann F, Liu X. et al. Supramolecular hydrogel microcapsules via cucurbit[8]uril host-guest interactions with triggered and UV-controlled molecular permeability. Chem Sci. 2015;6:4929-33
54. Harada A. Cyclodextrin-based molecular machines. Accounts chem Res. 2001;34:456-64
55. Zhu H, Shangguan L, Shi B, Yu G, Huang F. Recent progress in macrocyclic amphiphiles and macrocyclic host-based supra-amphiphiles. Mater Chem Front. 2018;2:2152-74
56. Zhao Y, Truhlar DG. Size-selective supramolecular chemistry in a hydrocarbon nanoring. J Am Chem Soc. 2007;129:8440-2
57. Loh XJ. Supramolecular host-guest polymeric materials for biomedical applications. Mater Horiz. 2014;1:185-95
58. Dsouza RN, Pischel U, Nau WM. Fluorescent dyes and their supramolecular host/guest complexes with macrocycles in aqueous solution. Chem Rev. 2011;111:7941-80
59. Ma X, Zhao Y. Biomedical applications of supramolecular systems based on host-guest interactions. Chem Rev. 2015;115:7794-839
60. Lim YB, Moon KS, Lee M. Recent advances in functional supramolecular nanostructures assembled from bioactive building blocks. Chem Soc Rev. 2009;38:925-34
61. Qu DH, Wang QC, Zhang QW, Ma X, Tian H. Photoresponsive host-guest functional systems. Chem Rev. 2015;115:7543-88
62. Zhou X, Pathak P, Jayawickramarajah J. Design, synthesis, and applications of DNA-macrocyclic host conjugates. Chem Commun. 2018;54:11668-80
63. Zhao Q, Temsamani J, Agrawal S. Use of cyclodextrin and its derivatives as carriers for oligonucleotide delivery. Antisense Res Dev. 1995;5:185-92
64. Kuzuya A, Ohnishi T, Wasano T, Nagaoka S, Sumaoka J, Ihara T. et al. Efficient guest inclusion by β-cyclodextrin attached to the ends of DNA oligomers upon hybridization to various DNA conjugates. Bioconjugate Chem. 2009;20:1643-9
65. Dubel N, Liese S, Scherz F, Seitz O. Exploring the limits of bivalency by DNA-based spatial screening. Angew Chem Int Ed. 2018;58:907-11
66. Dirks RM, Pierce NA. Triggered amplification by hybridization chain reaction. Proc Natl Acad Sci USA. 2004;101:15275-8
67. Husale S, Persson HH, Sahin O. DNA nanomechanics allows direct digital detection of complementary DNA and microRNA targets. Nature. 2009;462:1075-8
68. Hu WB, Yang HM, Hu WJ, Ma ML, Zhao XL, Mi XQ. et al. A pillar[5]arene and crown ether fused bicyclic host: synthesis, guest discrimination and simultaneous binding of two guests with different shapes, sizes and electronic constitutions. Chem Commun. 2014;50:10460-3
69. Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol. 2010;11:593-8
70. Fujimoto K, Yamada S, Inouye M. Synthesis of versatile fluorescent sensors based on click chemistry: detection of unsaturated fatty acids by their pyrene-emission switching. Chem Commun. 2009:7164-6
71. Fujimoto K, Muto Y, Inouye M. A DNA Duplex-Based, Tailor-made fluorescent sensor for porphyrin derivatives. Bioconjugate Chem. 2008;19:1132-34
72. Ihara T, Uemura A, Futamura A, Shimizu M, Baba N, Nishizawa S. et al. Cooperative DNA probing using a β-cyclodextrin-DNA conjugate and a nucleobase-specific fluorescent ligand. J Am Chem Soc. 2009;131:1386-7
73. Gosling JP. A decade of development in immunoassay methodology. Clin Chem. 1990;36:1408-27
74. Sharma AK, Kent AD, Heemstra JM. Enzyme-linked small-molecule detection using split aptamer ligation. Anal Chem. 2012;84:6104-9
75. Yang C, Spinelli N, Perrier S, Defrancq E, Peyrin E. Macrocyclic host-dye reporter for sensitive sandwich-type fluorescent aptamer sensor. Anal Chem. 2015;87:3139-43
76. Zhou Y, Gao L, Tong X, Li Q, Fei Y, Yu Y. et al. Supramolecularly multicolor DNA decoding using an indicator competition assay. Anal Chem. 2018;90:13183-7
77. Zhang S, Assaf KI, Huang C, Hennig A, Nau WM. Ratiometric DNA sensing with a host-guest FRET pair. Chem Commun. 2019;55:671-4
78. Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC. et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834-40
79. Yamada KM, Araki M. Tumor suppressor PTEN: modulator of cell signaling, growth, migration and apoptosis. J Cell Sci. 2001;114:2375-82
80. Harris DC, Saks BR, Jayawickramarajah J. Protein-binding molecular switches via host-guest stabilized DNA hairpins. J Am Chem Soc. 2011;133:7676-9
81. Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372-81
82. Lu Y, Liu J. Functional DNA nanotechnology: emerging applications of DNAzymes and aptamers. Curr Opin Biotechnol. 2006;17:580-8
83. Zhou X, Su X, Pathak P, Vik R, Vinciguerra B, Isaacs L. et al. Host-guest tethered DNA transducer: ATP fueled release of a protein inhibitor from cucurbit[7]uril. J Am Chem Soc. 2017;139:13916-21
84. Razin A, Riggs AD. DNA methylation and gene function. Science. 1980;210:604-10
85. Wang X, Feng M, Xiao L, Tong A, Xiang Y. Postsynthetic modification of DNA phosphodiester backbone for photocaged DNAzyme. ACS Chem Biol. 2016;11:444-51
86. Wang SR, Song YY, Wei L, Liu CX, Fu BS, Wang JQ. et al. Cucurbit[7]uril-driven host-guest chemistry for reversible intervention of 5-formylcytosine-targeted biochemical reactions. J Am Chem Soc. 2017;139:16903-12
87. Discher D, Dong C, Fredberg JJ, Guilak F, Ingber D, Janmey P. et al. Biomechanics: cell research and applications for the next decade. Ann Biomed Eng. 2009;37:847-59
88. Tayi AS, Shveyd AK, Sue AC, Szarko JM, Rolczynski BS, Cao D. et al. Room-temperature ferroelectricity in supramolecular networks of charge-transfer complexes. Nature. 2012;488:485-9
89. Yan Q, Yuan J, Cai Z, Xin Y, Kang Y, Yin Y. Voltage-responsive vesicles based on orthogonal assembly of two homopolymers. J Am Chem Soc. 2010;132:9268-70
90. Wiester MJ, Ulmann PA, Mirkin CA. Enzyme mimics based upon supramolecular coordination chemistry. Angew Chem Int Ed. 2011;50:114-37
91. Chen S, Chen LJ, Yang HB, Tian H, Zhu W. Light-triggered reversible supramolecular transformations of multi-bisthienylethene hexagons. J Am Chem Soc. 2012;134:13596-9
92. Zhang X, Rehm S, Safont-Sempere MM, Wurthner F. Vesicular perylene dye nanocapsules as supramolecular fluorescent pH sensor systems. Nat Chem. 2009;1:623-9
93. Yan X, Wang F, Zheng B, Huang F. Stimuli-responsive supramolecular polymeric materials. Chem Soc Rev. 2012;41:6042-65
94. Feng L, Li W, Ren J, Qu X. Electrochemically and DNA-triggered cell release from ferrocene/β-cyclodextrin and aptamer modified dualfunctionalized graphene substrate. Nano Res. 2014;8:887-99
95. Auernheimer J, Dahmen C, Hersel U, Bausch A, Kessler H. Photoswitched cell adhesion on surfaces with RGD peptides. J Am Chem Soc. 2005;127:16107-10
96. Bian Q, Wang W, Wang S, Wang G. Light-triggered specific cancer cell release from cyclodextrin/azobenzene and aptamer-modified substrate. ACS Appl Mater Inter. 2016;8:27360-7
97. Shi P, Ju E, Yan Z, Gao N, Wang J, Hou J. et al. Spatiotemporal control of cell-cell reversible interactions using molecular engineering. Nat Commun. 2016;7:13088
98. Thelu HVP, Albert SK, Golla M, Krishnan N, Ram D, Srinivasula SM. et al. Size controllable DNA nanogels from the self-assembly of DNA nanostructures through multivalent host-guest interactions. Nanoscale. 2017;10:222-30
99. Albert SK, Thelu HVP, Golla M, Krishnan N, Varghese R. Modular synthesis of supramolecular DNA amphiphiles through host-guest interactions and their self-assembly into DNA-decorated nanovesicles. Nanoscale. 2017;9:5425-32
100. Zheng J, Nie Y, Yang S, Xiao Y, Li J, Li Y. et al. Remote-controlled release of DNA in living cells via simultaneous light and host-guest mediations. Anal Chem. 2014;86:10208-14
101. Jiang Y, Pan X, Chang J, Niu W, Hou W, Kuai H. et al. Supramolecularly engineered circular bivalent aptamer for enhanced functional protein delivery. J Am Chem Soc. 2018;140:6780-4
102. Kuai H, Zhao Z, Mo L, Liu H, Hu X, Fu T. et al. Circular bivalent aptamers enable in vivo stability and recognition. J Am Chem Soc. 2017;139:9128-31
Author contact
![]() Corresponding author: tanufl.edu, xbzhangedu.cn, zlzhaoedu.cn.
Corresponding author: tanufl.edu, xbzhangedu.cn, zlzhaoedu.cn.
 Global reach, higher impact
Global reach, higher impact