13.3
Impact Factor
Theranostics 2019; 9(9):2439-2459. doi:10.7150/thno.31017 This issue Cite
Research Paper
Desktop-stereolithography 3D printing of a radially oriented extracellular matrix/mesenchymal stem cell exosome bioink for osteochondral defect regeneration
1. Department of Orthopaedic Surgery, Sir Run Run Shaw Hospital, Medical College of Zhejiang University, Hangzhou, China
2. Key Laboratory of Musculoskeletal System Degeneration and Regeneration Translational Research of Zhejiang Province
3. Department of Orthopedics, 5th Affiliated Hospital, Lishui Municipal Central Hospital, Wenzhou Medical University, Lishui, China
*These authors have contributed equally
Received 2018-10-28; Accepted 2019-3-3; Published 2019-4-13
Abstract
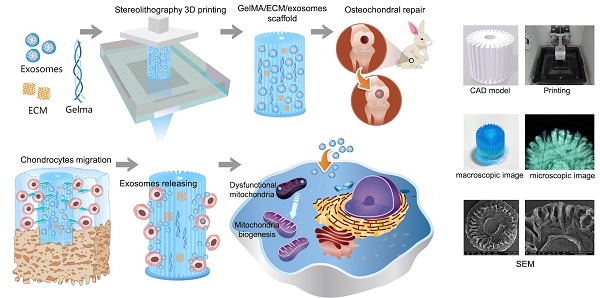
Mitochondrial dysfunction and oxidative stress damage are hallmarks of osteoarthritis (OA). Mesenchymal stem cell (MSC)-derived exosomes are important in intercellular mitochondria communication. However, the use of MSC exosomes for regulating mitochondrial function in OA has not been reported. This study aimed to explore the therapeutic effect of MSC exosomes in a three dimensional (3D) printed scaffold for early OA therapeutics.
Methods: We first examined the mitochondria-related proteins in normal and OA human cartilage samples and investigated whether MSC exosomes could enhance mitochondrial biogenesis in vitro. We subsequently designed a bio-scaffold for MSC exosomes delivery and fabricated a 3D printed cartilage extracellular matrix (ECM)/gelatin methacrylate (GelMA)/exosome scaffold with radially oriented channels using desktop-stereolithography technology. Finally, the osteochondral defect repair capacity of the 3D printed scaffold was assessed using a rabbit model.
Results: The ECM/GelMA/exosome scaffold effectively restored chondrocyte mitochondrial dysfunction, enhanced chondrocyte migration, and polarized the synovial macrophage response toward an M2 phenotype. The 3D printed scaffold significantly facilitated the cartilage regeneration in the animal model.
Conclusion: This study demonstrated that the 3D printed, radially oriented ECM/GelMA/exosome scaffold could be a promising strategy for early OA treatment.
Keywords: 3D printing, cartilage extracellular matrix, exosome, osteochondral defect
1. Introduction
Osteoarthritis (OA) is regarded as the most prevalent chronic joint disease [1]. Articular cartilage damage is regarded as a significant cause for early OA in adolescents and young adults [2]. The implantation of a scaffold or cartilage repair surgery represents a good choice after failure of conservative treatment for early OA patients [3, 4]. Under normal nutritional conditions, there is a dynamic balance of anabolic and catabolic activities of chondrocytes, which mainly depend on glycolysis to supply their basic energy requirements [5]. During periods of nutrient stress, chondrocytes exhibit flexible metabolism and can maintain cell viability and extracellular matrix (ECM) synthesis by enhancing mitochondrial respiration through the tricarboxylic acid cycle. In OA, chondrocytes lose their metabolic flexibility, decrease their cellular mitochondrial biogenesis, and increase their mitochondrial DNA (mtDNA) damage. This mitochondrial damage in OA chondrocytes further reduces ATP synthesis and impairs the anabolic metabolism of chondrocytes [5, 6].
Bone marrow-derived mesenchymal stem cells (BMSCs) are widely applied in cartilage regeneration studies. However, this type of therapy has limitations such as tumorigenicity, immune incompatibility, and chromosomal aberrations [7, 8]. Therefore, it is important to develop strategies that can take full advantage of stem cell properties and avoid these shortcomings. Exosomes (30-150 nm in diameter) are extracellular vesicles that possess analogous functions to the cells from which they are derived [9]. Studies have reported that MSCs have the potential for possible immunogenicity and tumorigenicity [10, 11]. However, MSC-derived exosomes do not exhibit such harmful potential [12, 13]. It has been suggested that the therapeutic effects of MSCs can be replicated by MSC-derived exosomes [14]. Recent studies have found that exosomes can maintain expression of the cartilage matrix protein COL2A1 in chondrocytes and decrease cartilage matrix degradation marker ADAMTS-5 expression under human recombinant interleukin-1β (IL-1β) treatment [9, 15]. The evidence is now building that exosomes play a significant role in the intercellular mitochondria communication. The information transportation between cells can be mediated as the mitochondrial genome and even whole mitochondria are mobile [16]. This rising new area will provide further insights into the significance of exosomes in mitochondrial homeostasis. However, the mechanism by which exosomes regulate chondrocyte mitochondria remains unknown. Thus, we attempted to explore the effect of MSC-derived exosomes on mitochondrial homeostasis.
Recent studies have indicated that chondrocyte migration is a major element in the healing process of osteochondral defect [17-19]. Therefore, cell recruitment ability must be considered in scaffolds for cartilage tissue engineering [20-22]. Previously, we fabricated a collagen scaffold with radially oriented channels by freeze-drying and thermal-crosslinking. Results indicated that the scaffold possessed superior cell recruitment ability compared with a random structure. However, the scaffold production process is based on physical freeze-drying, and an optimized fabrication protocol needs to be established [17]. The decellularized technology can preserve natural tissue ECM while removing specific tissue or organ cells to minimize in vivo immunologic rejections [23]. The decellularized ECM contains abundant peptides, such as collagen, fibrin, and proteoglycans, and the matricryptic site exposed by these peptides can promote cell adhesion and migration [24]. Moreover, studies have shown an increased number of macrophages in 3-4 days after decellularized ECM was implanted in the body and this immune response could facilitate tissue regeneration at a later stage [25]. It is difficult to pattern the traditional photo-crosslinked techniques with natural polymers such as fibrin, gelatin or hyaluronic acid, especially when difficult 3D models are employed [26]. In the present study, we applied desktop-stereolithography (SLA) technology for high resolution and photo-crosslinked biomaterial printing to achieve improved cartilage regeneration. The bioink was composed of MSC-derived exosomes, decellularized cartilage ECM, and gelatin methacrylate (GelMA) hydrogel.
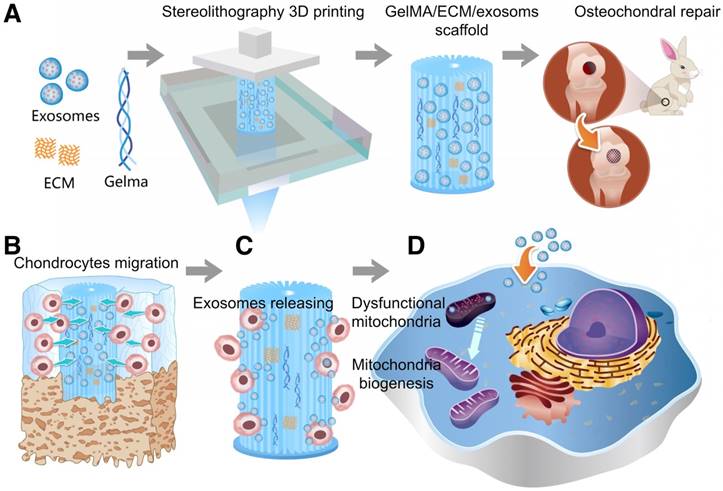
As shown in Scheme 1, we successfully fabricated a 3D printed ECM/GelMA/exosome scaffold with radially oriented channels by the SLA technology. The scaffold could promote the chondrocytes migration in the defect regions and sustainably release exosomes and the MSC-derived exosomes could be internalized by chondrocytes. The scaffolds could enhance the genesis of mitochondria and restore the dysfunctional mitochondria in chondrocytes by supplementing mitochondria-related proteins in exosomes. Finally, the promising osteochondral defects regeneration could be achieved through this one-step operation system.
2. Materials and Methods
2.1 Exosomes isolation and identification
2.1.1 Isolation of exosomes
When BMSCs reached 50-60% confluence they were washed and incubated for another 48 h in serum-free medium. The medium was collected and centrifuged at 300 × g for 15 min at 4 °C and then at 2,500 × g for 15 min. After centrifugation, the supernatant was filtered using a 0.22-μm filter (Merck Millipore, Burlington, MA) to remove remaining cells and debris. Next, a 15-mL Amicon Ultra-15 Centrifugal Filter Unit (Merck Millipore) was used for the filtered solution transfer. The solution was centrifuged at 4,000 × g until the volume in the upper compartment was concentrated to approximately 200 μL. The ultrafiltered solution was washed by phosphate-buffered saline (PBS) and centrifugation was repeated three times. The washed ultrafiltration liquid containing exosomes was laid on a 30% sucrose/D2O cushion in a Ultra-Clear™ tube (Beckman Coulter, Brea, CA) and ultra-centrifuged for 1 h at 100,000 × g. The pellets were resuspended in PBS and centrifuged at 4,000 × g to concentrate the volume to approximately 200 μL. Measurement of the exosome particle number was performed using a CD63 ExoELISA™ kit (System Biosciences, Palo Alto, CA).
Schematic illustration of the one-step operation system for facilitating osteochondral defect regeneration. (A) Stereolithography-based ECM/GelMA/exosome bioprinting and osteochondral defect implantation. (B) Migration of chondrocytes to the defect regions. (C) Controlled administration of exosomes by the 3D printed scaffolds. (D) Enhanced chondrocyte mitochondrial biogenesis by the scaffolds.
2.1.2 Characterization of exosomes
Dynamic light scattering (DLS) analysis using Nanosizer™ technology (Malvern Instruments, Malvern, UK) was applied for the exosome size distribution measurement. Then the exosome size was analyzed using Zetasizer software (Malvern). Transmission electron microscopy (TEM; H-9500; Hitachi, Tokyo, Japan) was used for exosome morphology observation. The Total Exosome RNA & Protein Isolation Kit (Invitrogen, Carlsbad, CA) was used for proteins extract. The proteins were subsequently used in Western blotting to detect exosome components.
2.1.3 Internalization of exosomes by chondrocytes
BMSCs were labeled with the fluorescent lipophilic tracer Vybrant DiO (green) dye (Molecular Probes, Carlsbad, CA). Briefly, BMSCs were digested using trypsin-EDTA before being resuspended in 1 mL culture medium. After adding 5 μL DiO solution to the suspension, the mix was incubated in 5% humidified CO2 for 15 min. After the mix was centrifugated at 300 × g for 10 min and washed, cells were cultured in complete medium until 50-60% confluence. Exosomes were isolated using the protocol described in section 2.1.1 and incubated with chondrocytes for 6 h. After washing three times in PBS, the chondrocytes were fixed with formalin for 15 min. Then the chondrocytes were stained with DAPI for 5 min. Finally, the samples were imaged using confocal microscopy (FV300; Olympus, Tokyo, Japan). As for the colocalization of exosomes and mitochondria, labeled exosomes were incubated with chondrocytes for 48 h. Prior to DAPI staining, the mitochondria were stained with the MitoTracker Red CMXRos fluorescent probe (Life Technologies, Carlsbad, CA). The samples were again imaged using confocal microscopy.
2.2 Preparation of the 3D printed, radially oriented ECM/GelMA/exosome bioink
2.2.1 Synthesis of GelMA hydrogel
GelMA was synthesized as previously described [27]. Briefly, gelatin (Type A; 300 Bloom from porcine skin; Sigma-Aldrich) was mixed into PBS at a concentration of 10% (w/v). Methacrylic anhydride (MA; Sigma-Aldrich) was added to the solution and allowed to react with gelatin for 1 h. After the reaction was terminated by diluting with PBS, the mixture was dialyzed for 1 week, and then the solution was lyophilized for another week and stored at -80 °C.
2.2.2 Preparation of decellularized cartilage ECM hydrogel
Healthy and mature pigs of either sex were used for cartilage tissue samples harvest. Samples were extracted as solid cylinders using an 8-mm perforator and decellularized [28]. Briefly, cartilage tissue samples were treated with liquid nitrogen for three freeze/thaw cycles and 1% (v/v) Triton X-100 (Sigma-Aldrich) for 24 h. Then the samples were treated with 1% (w/v) sodium dodecyl sulfate (SDS; Sigma-Aldrich) for 24 h. Samples were digested by 200 U/mL DNase I (Sigma-Aldrich) at 37 °C for 12 h and washed in PBS. Next, hematoxylin and eosin (HE) and DAPI staining were applied for evaluation of cell obliteration. We then used scanning electron microscopy (SEM; SU-70; Hitachi, Tokyo, Japan) and TEM to visualize the 3D microstructures of the native and decellularized cartilage ECM.
2.2.3 Preparation and characterization of ECM/GelMA hydrogel
Cartilage ECM was lyophilized and then pulverized into a fine powder. Subsequently, 1-3% ECM solution was prepared by digesting approximately 1-3 g of pulverized cartilage ECM in 0.5 M acetic acid with 30 mg pepsin for 48 h. The solution was centrifuged at 500 × g for 10 min to remove residues after complete ECM solubilization. The ECM hydrogel was mixed with 10% (w/v) GelMA and 0.25% (w/v) lithium acylphosphinate photo-initiator (LAP) and then the ECM/GelMA pre-gel was crosslinked for 30 seconds at 405 nm of visible light exposure. Atomic force microscopy (AFM; Multimode 8; Bruker, Germany) was employed for high-resolution hydrogel imaging, and SEM was used to visualize the 3D microstructure of the lyophilized hydrogel. Porosity was analyzed using Image J software (NIH, Bethesda, MD). The live and dead cell assay was applied for cell status observation. Calcein-AM (2 μM; Dojindo, Kumamoto, Japan) and propidium iodide (10 μg/mL) were applied for live and dead cells staining, respectively.
2.2.4 Biofabrication of ECM/GelMA/exosome bioink using dynamic projection stereolithography
The fabrication system contained a digital light processing (DLP) chip (Discovery 4000; Texas Instruments, Dallas, TX), a visible light source with 405 nm wavelength (OmniCure S2000; EXFO, Quebec, Canada), and XYZ stages. The source output was 30 W/cm2. User-defined computer-aided-design (CAD) files were handled by the DLP chip. Virtual masks were then generated by the chip. The images modulated by DLP chip were projected onto the bioink through a visible light optical lens (Edmunds Optics, Barrington, NJ). The bioink illuminated by visible light was quickly crosslinked and this process was repeated according to the virtual masks. An intensity of 11 mW/cm2 for 30 s was used for irradiation. The 3D printed scaffold was then observed by microscopy (CX31; Olympus, Tokyo, Japan) and by SEM (after lyophilizing).
2.2.5 Exosomes retention ability of the 3D printed scaffold
The initial concentration of exosome in the hydrogel was 200 μg/mL. A particle analyzer (qNano platform; Izon Science, Christchurch, New Zealand) was used for exosome retention ability examination. The fabricated 3D printed ECM/GelMA/exosome scaffolds were placed in 12-well cell culture plate with 2 mL PBS. PBS was picked up daily and changed by 2 mL fresh PBS. The number of leaching exosomes was detected on the particle analyzer. The 3D printed ECM/GelMA/exosome scaffolds were implanted in subcutaneous mouse models and 200 μg exosomes dissolved in 200 μL PBS were injected into the mouse subcutis as a control. Both groups of exosomes were labeled with Vybrant DiO (green) dye before implantation. The IVIS Spectrum In Vivo Imaging System (bioluminescence imaging [BLI]: Xenogen, USA) was used to monitor exosome retention in vivo during day 0 and 7.
2.3 Osteochondral defect models
New Zealand white rabbits (6-month-old, female; Slaccas Company, Shanghai, China) were used for the animal studies. All animal experiments followed the ARRIVE guidelines and were conducted in accordance with the U.K. Animals (Scientific Procedures) Act, 1986 and related guidelines. Animals were anesthetized and a cylindrical defect 4 mm in diameter and 4 mm in height was created in the patellar groove of both limbs using a punch tool. The left defect was not treated (Group I) or received a 3D printed ECM/GelMA scaffold implant (Group III), whereas the right defect received a 3D printed GelMA (Group II) or 3D printed ECM/GelMA/exosome scaffold (Group IV) implant. The incisions were closed and penicillin (70 mg/kg) was administered to the rabbit periodically by intramuscular injection. At 6 or 12 weeks post-surgery, animals were sacrificed and their knee joints including synovial membranes and synovial fluid (n = 9 per group) were collected for further analysis. The samples were fixed overnight in 10% (v/v) formalin (Sigma-Aldrich). Each group of samples was taken and photographed according to the International Cartilage Repair Society (ICRS) macroscopic assessment scale [29]. Then, the samples were assessed using magnetic resonance imaging (MRI; Discovery MR 750W; General Electric Company, Boston, USA) and microcomputed tomography (μCT; SCANCO μCT 100; SCANCO Medical AG, Switzerland) scanning.
2.4 Determination of malondialdehyde
Malondialdehyde (MDA) was determined by reacting with 2-thiobarbituric acid (TBA) (Sigma-Aldrich, St. Louis, USA) at 95°C [30]. MDA reacted with TBA to produce a pink chromogen with absorbance at 532 nm. A standard curve was established by purified MDA (Sigma-Aldrich, St. Louis, USA).
2.5 Assessment of chondrocyte mitochondria
The relative mitochondrial copy number was represented by the ratio of mtDNA-encoded cytochrome c oxidase subunit I (COX1) or subunit II (COX2) DNA copies to nucleus-encoded 18S ribosomal DNA (18S rDNA; detailed in the Supplementary Materials). The MitoTracker Green FM and MitoSox Red fluorescent probes (Life Technologies) were used to detect the mitochondrial mass and reactive oxygen species (ROS) production, respectively. Mitochondria morphology was observed by TEM. Intracellular ATP was quantified by the ATP Colorimetric/Fluorometric Assay Kit (BioVision, Milpitas, CA).
2.6 Statistical analysis
Quantitative results are expressed as the means ± standard deviation (SD). Statistical data were analyzed by one-way analysis of variance (ANOVA) using SPSS 16.0 software (SPSS Inc, Chicago, IL). p < 0.05 was set to be statistically significant.
Other materials and methods are detailed in the Supplementary Materials.
3. Results
3.1 Energy metabolism imbalance in degenerated cartilage
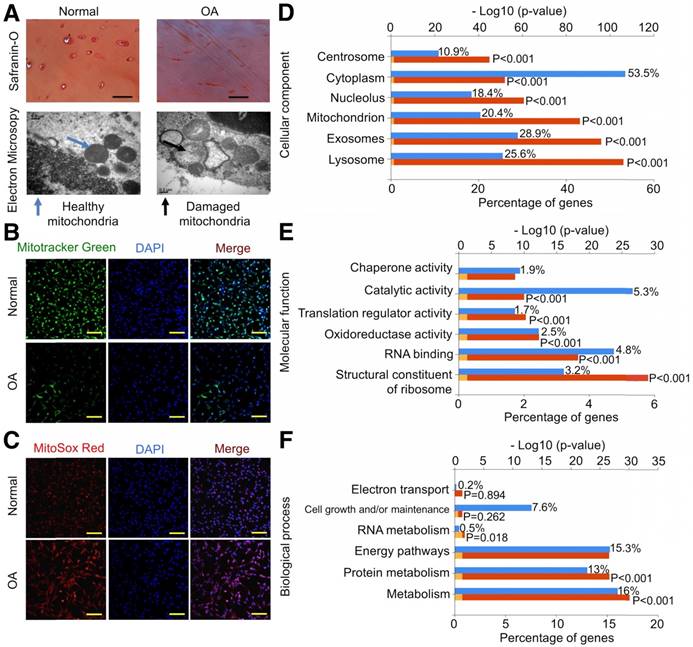
We first examined the mitochondria-related proteins in human cartilage samples. Safranin O staining confirmed the decrease in proteoglycan levels in OA samples (Fig. 1A). We analyzed mitochondria morphology using TEM and found that OA samples showed abnormal mitochondria with the appearance of rarefaction and vacuoles as well as breakage and disappearance of mitochondrial cristae (Fig. 1A). We then isolated chondrocytes for further evaluation and found that mitochondrial mass in normal chondrocytes was significantly higher than in OA chondrocytes (Fig. 1B). In parallel, ROS production indicated by MitoSOX Red staining in normal chondrocytes was significantly lower than in OA chondrocytes (Fig. 1C).
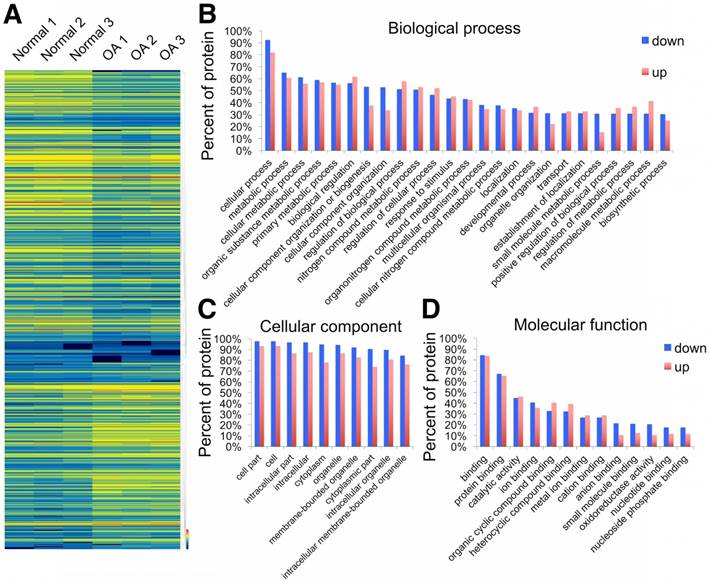
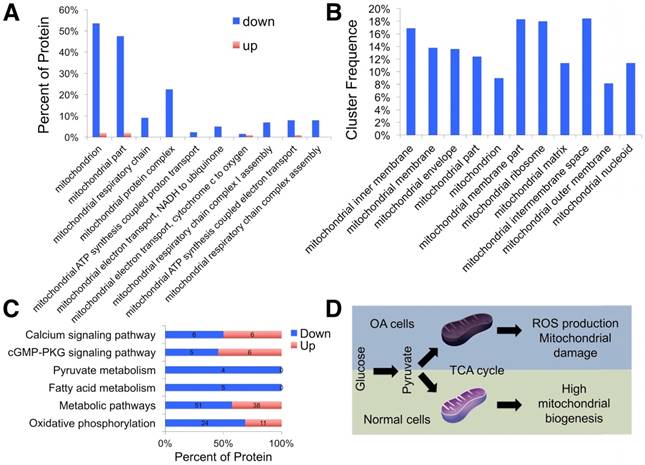
Using the label-free proteomics approach, we further explored protein changes in normal and OA chondrocytes and identified 4856 proteins (Fig. S1). The identified proteins were classified into three categories (biological process, cellular component, and molecular function) by gene ontology (GO) classification and annotated with the Kyoto Encyclopedia of Genes and Genomes (KEGG; Fig. 1D-F and Fig. S2A-C). For the expressed variation, the cutoff value was 1.5-fold and we found 480 differentially expressed proteins (p < 0.05), including 289 downregulated and 191 upregulated proteins in OA samples compared with normal samples (Fig. 2A and Table S1). The differentially expressed proteins are shown in Figure 2B-D through GO classification. The data indicated that there were more aberrations in the cellular component organization, organelle organization, and small molecular metabolic process, which were related to mitochondria. Thus, we focused on organelle organization-related differentially expressed proteins. We found that all mitochondria-related proteins, including the mitochondrion (141 proteins), mitochondrial part (125 proteins), and mitochondrial respiratory chain (25 proteins), were enriched in normal samples (Fig. 3A). As shown by GO pathway enrichment analysis, various components of the mitochondria, including the mitochondrial inner membrane, membrane, envelope, ribosome, matrix, intermembrane space, outer membrane, and nucleoid were also enriched in normal samples (Fig. 3B).
Furthermore, protein-protein interaction networks for differentially expressed proteins were analyzed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; Fig. S2D, E). In normal samples, STRING revealed that a lot of binary interactions could be found in the single-organism metabolic process, oxidoreductase activity acting on NAD(P)H, and mitochondrial part protein-protein interaction network. KEGG annotation analysis was also used to evaluate the potential processes (Fig. 3C). In normal samples, pyruvate metabolism, fatty acid metabolism, metabolic pathways, and oxidative phosphorylation were significantly higher than in OA samples.
Based on these findings, we demonstrated that mitochondrial metabolism was impaired and ROS accumulated in human OA chondrocytes, damaging mitochondria (Fig. 3D).
Energy metabolism imbalance in degenerated cartilage. (A) Safranin O staining and mitochondria morphology in human cartilage samples. Scale bar = 100 μm. (B, C) MitoTracker Green FM staining and MitoSox Red staining in chondrocytes. Scale bar = 200 μm. (D-F) GO classification of all quantified proteins in the cellular component, molecular function, and biological process.
3.2 Attenuation of chondrocyte degeneration by MSC exosomes
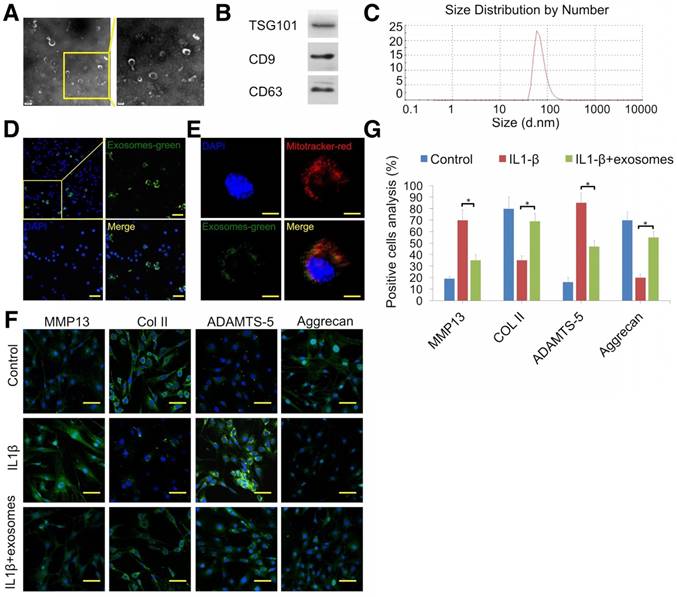
TEM, Western blotting, and DLS were employed to identify the vesicles secreted by MSCs. Hollow spherical microvesicles were observed (Fig. 4A) and expression of exosome markers (TSG101, CD9, and CD63; Fig. 4B) was significantly enriched in exosomes. Moreover, the widely used exosome markers CD9 and CD63 were also found by exosome proteomics (Table S2). DLS analysis indicated that the diameters of these vesicles ranged from 40 to 110 nm (Fig. 4C). Thus, the data confirmed that we successfully isolated exosomes. Next, we examined whether chondrocytes could take up MSC exosomes. Prior to exosome isolation, MSCs were first marked with a green fluorescent dye and then chondrocytes were treated for 6 h with DiO-labeled exosomes. Using confocal microscopy, we found that DiO-labeled exosomes were localized in the perinuclear region of chondrocytes (Fig. 4D), confirming exosome internalization by chondrocytes. Quantitative analysis showed 34.9% of chondrocytes internalized exosomes. We also observed colocalization of exosomes with mitochondria 48 h after administration of chondrocytes, suggesting that exosomes could fuse with the mitochondria of recipient cells (Fig. 4E).
To explore the therapeutic effect of MSC-derived exosomes on degenerated cartilage, an IL-1β-stimulated chondrocyte model was employed. Immunofluorescence results showed that exosomes decreased MMP13 (cartilage degradation marker) and ADAMTS-5 protein levels and increased COL2A1 and aggrecan expression (Fig. 4F, G). These results indicated that MSC exosomes could attenuate chondrocyte degeneration.
Differentially expressed proteins in normal and OA chondrocytes. (A) Heat map of 289 downregulated and 191 upregulated proteins in OA samples compared with normal samples. (B-D) GO classification of differentially expressed proteins.
Proteomic analysis of differentially expressed proteins. (A) GO classification of differentially expressed proteins related to organelle organization. (B) Significantly enriched GO terms in mitochondrial functions. (C) KEGG annotation analysis of differentially expressed proteins. (D) Schematic illustration of the energy metabolism imbalance in degenerated cartilage.
Attenuation of chondrocyte degeneration by MSC exosomes. (A) Morphology of exosomes imaged by TEM. (B) Western blotting of the exosome markers TSG101, CD9, and CD63. (C) Particle size distribution of exosomes. (D) Representative immunofluorescence micrograph of DiO (green)-labeled exosomes taken up by chondrocytes. Scale bar = 100 μm. (E) Confocal microscopy of chondrocytes incubated with DiO-labeled exosomes (green) for 48 h, MitoTracker-labeled mitochondria (red), and exosomes colocalized with mitochondria (yellow). Scale bar = 20 μm. (F, G) Immunofluorescence staining and quantification of MMP13, COL2A1, ADAMTS-5, and aggrecan in chondrocytes treated with IL-1β or IL-1β + exosomes. n = 5. *p < 0.05; **p < 0.01.
3.3 Mitochondrial dysfunction in degenerative chondrocytes rescued by exosomes
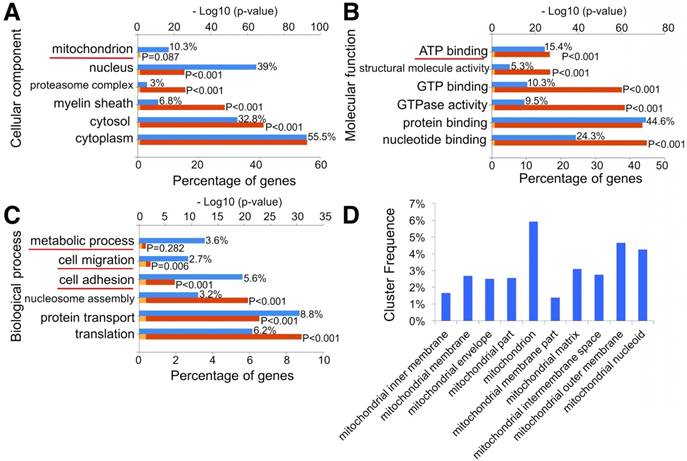
To investigate whether MSC exosomes could enhance mitochondrial biogenesis, we analyzed the exosomes through proteomics (Fig. S3 and Table S2). The data indicated that exosome proteins mainly originated from the cytoplasm (55.5%), nucleus (39%), and cytosol (32.8%). Notably, 10.3% of exosome proteins originated from mitochondria (Fig. 5A). Molecular function analysis showed that 15.4% of exosome proteins were involved in ATP binding (Fig. 5B). Moreover, 3.6% of exosome proteins were found to participate in metabolic processes through biological process analysis (Fig. 5C). GO pathway enrichment analysis also indicated that various segments of the mitochondria, including mitochondrial inner membrane, membrane, envelope, matrix, intermembrane space, outer membrane, and nucleoid, were enriched in MSC exosomes (Fig. 5D). STRING was then used to understand the interaction between the potential responses and the protein-protein interaction network of exosome mitochondrial proteins (Fig. S4). Results showed that lots of binary interactions were involved in these mitochondria-associated proteins.
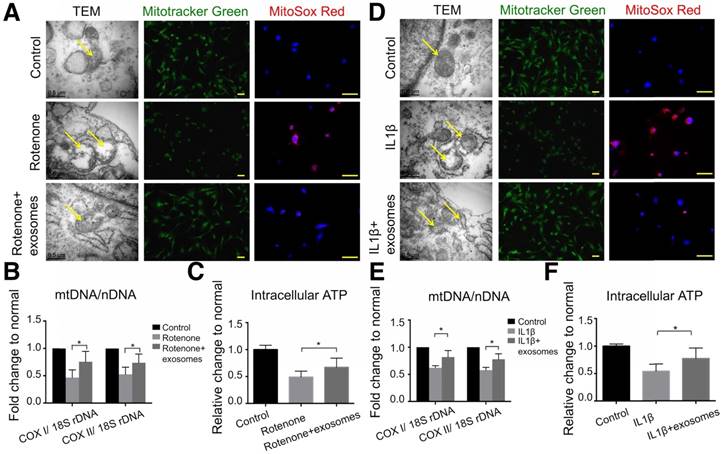
To reveal the mechanism of the chondroprotective effect of exosomes and whether it is via mitochondrial dysfunction inhibition, we used rotenone, the mitochondrial electron transport chain complex I inhibitor, to induce mitochondrial damage. In 2.5 μM rotenone-stimulated chondrocytes, exosomes restored mitochondria exhibiting a normal appearance (Fig. 6A). Exosome-treated chondrocytes also demonstrated increased mitochondrial mass and mtDNA content as evidenced by increased MitoTracker Green FM staining and a significant increase in the mtDNA-encoded COX1 or COX2 to nucleus-encoded 18S rDNA ratio (p < 0.05), respectively (Fig. 6A, B). Furthermore, rotenone treatment increased mitochondrial ROS production in chondrocytes which was significantly suppressed by exosome treatment (Fig. 6A). In functional analyses, intracellular ATP levels in chondrocytes treated with exosomes were increased by 21% compared with that of rotenone-stimulated chondrocytes (Fig. 6C). Similar changes in appearance, DNA content, mass, ROS production, and intracellular ATP levels of mitochondria were detected in the IL-1β-stimulated chondrocyte-degeneration model (Fig. 6D-F). Altogether, these results suggest that MSC exosomes provided mitochondrial proteins to chondrocytes thus restoring the damaged mitochondria.
Proteomic analysis of exosomes. (A-C) GO classification of exosome proteins. (D) Significantly enriched GO terms in mitochondria functions.
Mitochondrial dysfunction in degenerative chondrocytes rescued by exosomes. (A) TEM images of mitochondria with MitoTracker Green FM and MitoSox Red staining in chondrocytes treated with 2.5 μM rotenone or 2.5 μM rotenone + exosomes. Scale bar = 0.5 or 100 μm. (B, C) mtDNA content and intracellular ATP levels in chondrocytes treated with rotenone or rotenone + exosomes. n = 5. *p < 0.05; **p < 0.01. (D) TEM images of mitochondria with MitoTracker Green FM and MitoSox Red staining in chondrocytes treated with 10 ng/mL IL-1β or 10 ng/mL IL-1β + exosomes. Scale bar = 0.5 or 100 μm. (E, F) mtDNA content and intracellular ATP levels in chondrocytes treated with IL-1β or IL-1β + exosomes. n = 5. *p < 0.05; **p < 0.01.
3.4 Cartilage ECM/GelMA bioink preparation
The cartilage ECM scaffold was first prepared and comprehensively assessed (see Supplementary Results for more details; Fig. S5). For applying cartilage ECM to the 3D printing system, the ECM was solubilized at a final concentration of 1, 2, and 3 wt%. Subsequently, the ECM hydrogel was mixed with the unpolymerized GelMA solution followed by visible light-induced crosslinking in the presence of LAP (Fig. S6). There was no apparent change in the gelation time of 1-3% ECM incorporated into 10 wt% GelMA hydrogel relative to GelMA alone and all mixtures were successfully gelatinized (Fig. 7A). SEM showed a reticular structure of GelMA with a pore size of 91.6 ± 7.0 μm (Fig. 7B and Fig. S7) while analogous reticular structures with pore sizes of 116.9 ± 9.4, 124.6 ± 8.8, and 106.8 ± 6.9 μm were found for each ECM/GelMA hydrogel at 1, 2, or 3% ECM, respectively (Fig. 7B and Fig. S7). It is widely acknowledged that a suitable pore size (100-500 μm) is crucial for cell proliferation in biomaterial design [31-33]. At the pore size range examined, the larger porosity scaffold had the superior cytocompatibility. Thus, our results demonstrated that the porosity of 2% ECM was better than 1% or 3%.
Cartilage ECM/GelMA bioink preparation. (A) Images of the hydrogel before and after crosslinking. (B) SEM images of the hydrogel with 1-3 wt % ECM in different magnifications (scale bar = 100 μm or 50 μm). (C) 2D and 3D AFM images of the hydrogel with different ECM concentrations. (D) Live/dead cell staining. Scale bars = 100 μm.
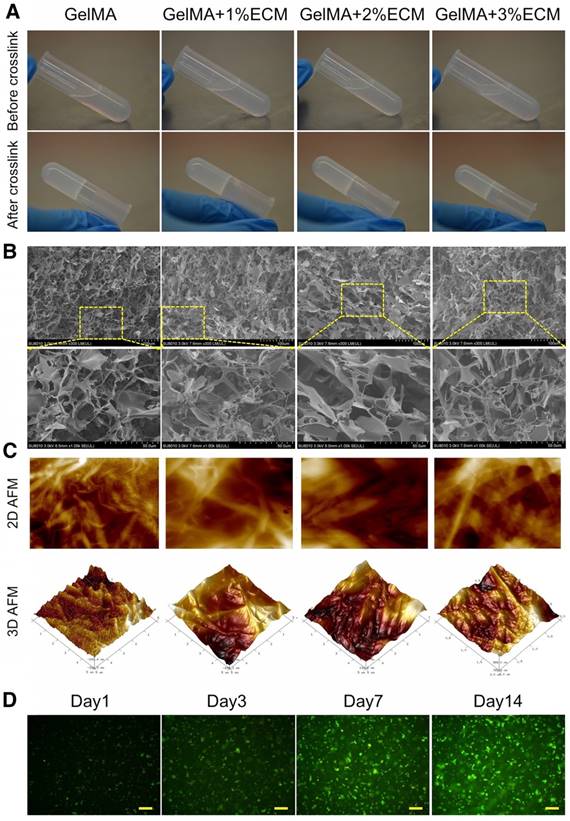
Next, we employed the AFM technique for high-resolution soft tissue imaging. With increasing ECM concentrations, Young's modulus and surface roughness of the hydrogel gradually increased, whereas the adhesion ability of the hydrogel gradually decreased. We also observed decreasing fibril length indicated by D-spacing values and constricted gaps between each fibril with increasing ECM concentrations (Fig. 7C and Table 1). Considering the scaffold biomechanical characteristics (the ideal bioink for SLA 3D printing should have satisfactory biomechanical properties), it was more efficient to use 2% ECM in 10% GelMA for SLA printing. We, therefore, chose 2% ECM in 10% GelMA for further study as it showed the most appropriate characteristics; specifically, we used GelMA + 2% ECM to test cell viability of the scaffold. The live and dead cell assay showed normal viability of BMSCs during 14 days of culture with the hydrogel (Fig. 7D). Also, more cells were observed at Day 14 compared to Day 1 or Day 3 due to cell proliferation.
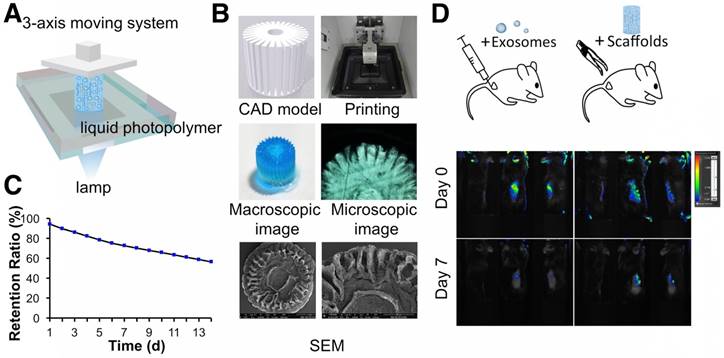
3D printing of radially oriented ECM/GelMA/exosome bioink. (A) Illustration of the SLA printing process. The printing resolution was set to 0.05 mm. (B) CAD file, printing process, and scaffold (stained with methylene blue dye) printed by the 3D printer. The scaffold was observed under microscopy and SEM. (C) The exosome retention ratio of the scaffold for 14 days. (D) In vivo fluorescence imaging of exosomes in PBS and in the 3D printed scaffold. Both groups of exosomes were labeled with Vybrant DiO (green) dye before implantation.
Physical properties of hydrogel with different ECM ratios as measured by AFM techniques, n = 5 in each group.
| Physical Properties | GelMA | GelMA+1%ECM | GelMA+2%ECM | GelMA+3%ECM |
|---|---|---|---|---|
| Collagen fibril D-Spacing (nm) | 64.03±2.23 | 55.83±1.78 | 46.1±2.01 | 42.7±3.27 |
| Average Roughness (nm) | 5.41±0.9 | 8.95±0.92 | 15.53±4.09 | 17.62±1.77 |
| Young's Modulus (Kpa) | 16.09±0.56 | 18.38±1.63 | 33.24±8.8 | 37.72±2.82 |
| Adhesion (nN) | 17.39±0.36 | 11.86±0.97 | 8.98±1.38 | 7.04±0.17 |
3.5 3D printing of radially oriented ECM/GelMA/exosome bioink
Following the successful preparation of the ECM/GelMA hydrogel and exosomes, we employed a stereolithography-based 3D printer for ECM/GelMA/exosome bioprinting (Fig. 8A). CAD files, as shown in Figure 8B, were used to produce the virtual mask and transmit visible light to the ECM/GelMA/exosome bioink to print scaffolds with radially oriented 3D architectures. The stereolithography-based technique successfully yielded a 4 × 4 mm radially oriented scaffold with radially oriented channels that could be observed under a microscope (Fig. 8B). These channels were also visible by SEM after lyophilizing the scaffold for 12 h (Fig. 8B). We next investigated whether the scaffold could sustainably release exosomes and found that it could effectively retain exosomes for 14 days in vitro (> 56%; Fig. 8C). Furthermore, in vivo fluorescence imaging indicated that, compared with the control (exosomes in PBS), the 3D printed scaffold could better retain exosomes for at least 7 days (Fig. 8D). These results indicated that the 3D printed scaffold could effectively control the administration of exosomes both in vitro and in vivo.
3.6 Increased chondrocyte migration by the 3D printed scaffold
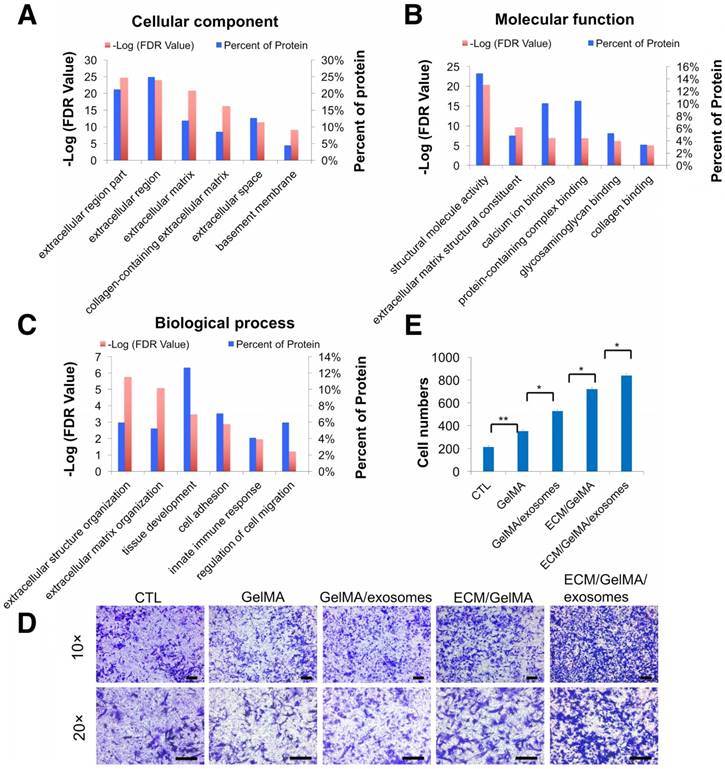
To investigate whether the 3D printed ECM/GelMA/exosome scaffold can recruit cells, we first performed proteomic analysis of ECM and divided the identified proteins into three classes (Fig. 9A-C, Fig. S3, and Table S3). GO cellular component analysis confirmed the successful retention of ECM region proteins (24.9%) after decellularization (Fig. 9A). GO biological process analysis found that ECM proteins were participated in cell adhesion and regulation of cell migration (Fig. 9C) and that MSC exosome proteins were participated in cell adhesion and cell migration (Fig. 5C). Thus, the results indicated that the 3D printed scaffold probably had a positive effect on chondrocyte migration. Transwell migration assays showed that chondrocyte migration was significantly increased in GelMA, GelMA/exosome, ECM/GelMA, and ECM/GelMA/exosome groups compared to the control group without scaffolds (p < 0.01). Cell numbers were significantly increased in the order GelMA < GelMA/exosomes (p < 0.05), GelMA/exosomes < ECM/GelMA (p < 0.05), and ECM/GelMA < ECM/GelMA/exosomes (p < 0.05; Fig. 9D, E and Fig. S9). These findings indicated that ECM had the largest effect on cell migration and MSC exosomes could further enhance this effect.
Increased chondrocyte migration by the 3D printed scaffold. (A-C) GO classification of cartilage ECM proteins. (D) Transwell migration assay of different treatments. Scale bars = 100 µm. (E) Quantitative analysis of migrating cells. n = 5. *p < 0.05; **p < 0.01.
3.7 In vivo biosecurity of the 3D printed ECM/GelMA/exosome scaffold
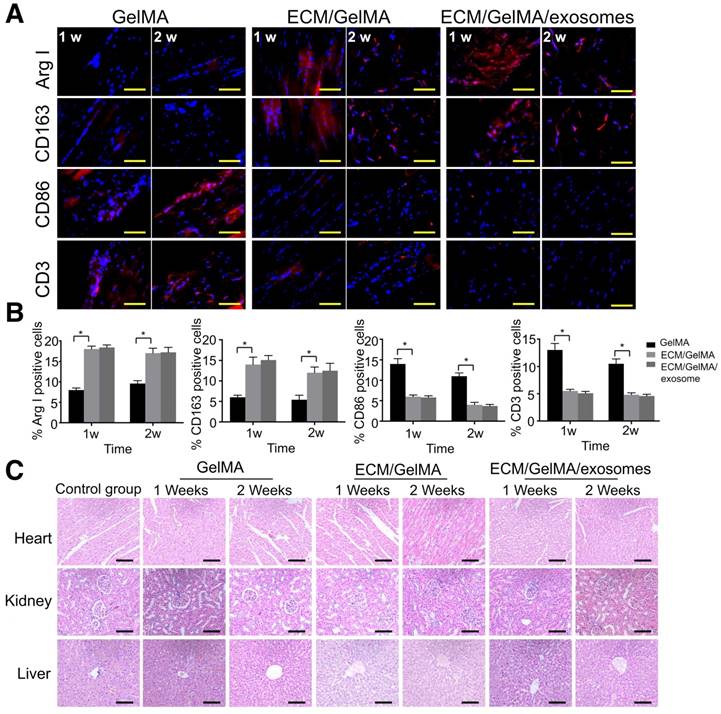
Immune response and scaffold toxicity were investigated in a rat subcutaneous model. All rats survived the 2 weeks of implantation and the scaffold could not be found at 3 weeks, indicating that the scaffold degradation time was less than 3 weeks. Macrophage markers were examined by immunofluorescence and results showed that, compared with the GelMA scaffold, the ECM/GelMA and ECM/GelMA/exosome scaffolds showed significantly increased ARG-I+ and CD163+ (M2 macrophage markers) cells and decreased CD86+ (M1 macrophage marker) cells in the surrounding tissues after 1 and 2 weeks (p < 0.05; Fig. 10A, B). Studies revealed that a classical Th2 response (associated with transplant acceptance) could be induced by M2 macrophages, which was beneficial for tissue repair [34]. We further evaluated the T cell-mediated immune response by CD3+ cell staining and found decreased CD3+ cells in the ECM/GelMA scaffold with or without exosomes during the implantation period compared to the GelMA group (p < 0.05; Fig. 10A, B). Also, HE staining of the myocardium, liver, and kidney exhibited no apparent pathological changes among all groups (Fig. 10C).
In vivo biosecurity of the 3D printed ECM/GelMA/exosome scaffold. (A) Immunofluorescence staining of ARG-I, CD163, CD86, and CD3 of scaffolds after 1 and 2 weeks. Scale bars = 200 μm. (B) Quantification of ARG-I, CD163, CD86, and CD3 protein levels. n = 5. *p < 0.05. (C) Host systemic toxicity of the 3D printed scaffold. HE staining of heart, kidney, and liver at 1 and 2 weeks after implanting the 3D printed scaffold. Scale bar = 200 μm.
3.8 Simulation of M2 macrophage polarization in vivo by the 3D printed, radially oriented ECM/GelMA scaffold
The osteochondral defect repair capacity of the 3D printed scaffold was assessed in a rabbit osteochondral defect model. The scaffold was implanted into the defect immediately after surgery. We then investigated the immune response induced by the 3D printed ECM/GelMA/exosome scaffold. Specifically, we evaluated the differences in M1 and M2 type macrophage infiltration in various groups.
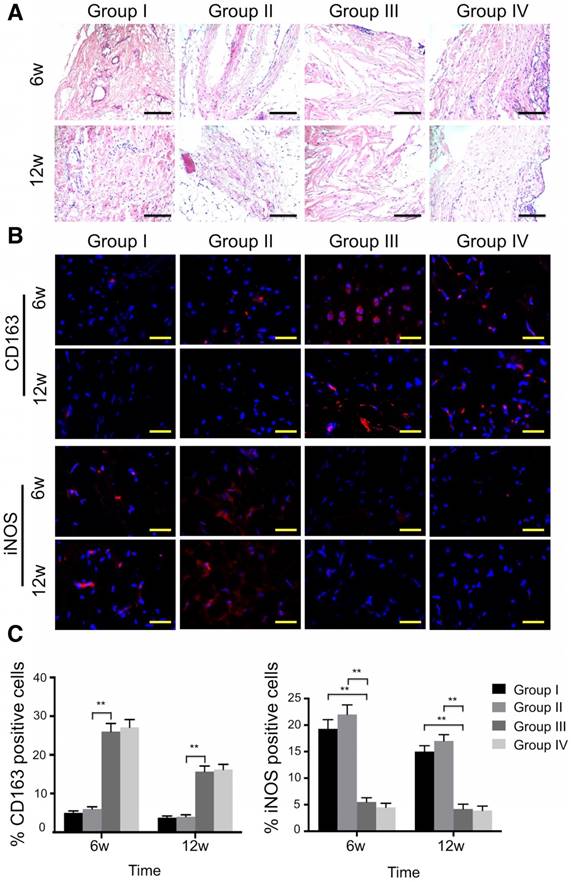
Histological analysis of the synovial membrane showed that synovial vascular hyperplasia could be found in the control group at 6 weeks, while there was no apparent synovitis in all other groups at both time points (Fig. 11A). In the ECM/GelMA group with or without exosomes, an abundance of M2 macrophages was observed as indicated by the presence of CD163+ cells in the synovium (Fig. 11B) at 6 and 12 weeks. Compared to the control and 3D printed GelMA groups, the number of CD163+ cells in the other two groups increased markedly (p < 0.01; Fig. 11C); however, no significant difference was found between the ECM/GelMA scaffold with or without exosomes. Furthermore, iNOS+ (M1 macrophage marker) cells were significantly reduced in the ECM/GelMA group with or without exosomes compared with control or 3D printed GelMA groups at both time points (p < 0.01; Fig. 11B, C). Thus, our results indicated that the addition of cartilage ECM to the scaffold may have a positive effect in M2 macrophage polarization.
Effects of 3D printed scaffold on macrophage infiltration during cartilage repair. (A) Histological appearance of the synovium at both time points. Scale bars = 100 μm. (B, C) Immunofluorescence staining of CD163+ and iNOS+ cells and quantitative analyses of CD163+ and iNOS+ cells in the reparative synovium at 6 and 12 weeks. Scale bars = 100 μm. n = 6. *p < 0.05; **p < 0.01. Group I, control group without treatment; Group II, defect with 3D printed GelMA implant; Group III, defect with 3D printed ECM/GelMA implant; Group IV, defect with 3D printed ECM/GelMA/exosome implant.
3.9 Enhancement of cartilage regeneration by the 3D printed, radially oriented ECM/GelMA/exosome scaffold
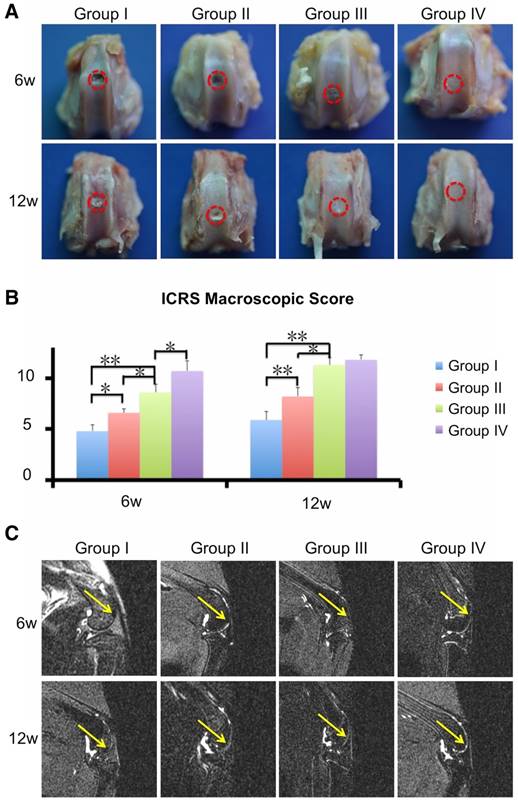
Next, we evaluated the cartilage regenerative capacity of the 3D printed scaffolds. Macroscopic observation at 6 and 12 weeks showed intact and smooth tissues in the ECM/GelMA/exosome group, whereas the GelMA and control groups exhibited rough and friable tissues (Fig. 12A). Furthermore, the ECM/GelMA groups had higher scores based on the ICRS macroscopic grading system compared with the GelMA or control groups at both time points (p < 0.05; Fig. 12B). ICRS scoring also indicated that ECM/GelMA/exosome scaffolds had the highest scores among all groups at both time points (Fig. 12B). Through MRI scanning, the ECM/GelMA scaffold groups with or without exosome exhibited smoother neo-cartilage and enhanced defect filling compared with those of the other two groups (Fig. 12C).
Efficacy of the 3D printed, radially oriented ECM/GelMA/exosome scaffolds for osteochondral defect repair. (A) Macroscopic evaluation of the osteochondral defect regions at 6 and 12 weeks. (B) ICRS macroscopic scores of different groups. n = 6. *p < 0.05; **p < 0.01. (C) MRI imaging of repaired knees at 6 and 12 weeks after surgery in various groups (yellow arrow indicates the repaired sites).
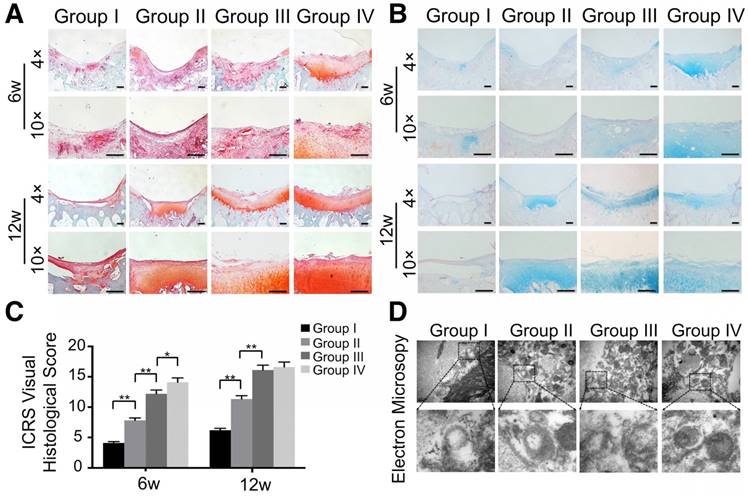
Besides macroscopic and imaging evaluations, we employed safranin O, alcian blue, and HE staining for proteoglycan and morphology evaluation. Six weeks post-surgery, the defect was covered with fibrous tissue instead of hyaline cartilage in the control group (Fig. 13A, B and Fig. S10). In contrast, the GelMA and ECM/GelMA groups showed a mixture of fibrous tissue and fibrocartilage in the defect regions. In the ECM/GelMA/exosome group, however, the defect area was regenerated with a mixture of fibrocartilage and hyaline-like cartilage (Fig. 13A, B and Fig. S10). We then evaluated the regenerated cartilage by the ICRS visual histological score system. The score in the ECM/GelMA group (12.2 ± 0.61) was significantly higher than the GelMA group (7.8 ± 0.39; p < 0.05, Fig. 13C). Meanwhile, the score of the GelMA group was markedly higher than the control group (4.1 ± 0.21; p < 0.05, Fig. 13C). The score was 14.1 ± 0.71 in the ECM/GelMA/exosome group, markedly higher than the ECM/GelMA group (p < 0.05, Fig. 13C). After 12 weeks, the defect area in the control group was still covered with fibrous tissue (Fig. 13A, B and Fig. S10), whereas hyaline cartilage-like tissue in the defect area was observed for the ECM/GelMA and ECM/GelMA/exosome groups (Fig. 13A, B and Fig. S10). The ICRS visual histological score was 6.2 ± 0.31, 11.3 ± 0.57, 16.1 ± 0.81, and 16.1 ± 0.83 in the control, GelMA, ECM/GelMA, and ECM/GelMA/exosome groups, respectively (Fig. 13C). No significant difference was found in the ICRS score between the ECM/GelMA scaffold groups (with or without exosomes). Compared to the GelMA group, the ICRS score was significantly higher in the ECM/GelMA group (p < 0.05, Fig. 13C) and the score of the GelMA group was markedly higher than the control group (p < 0.01, Fig. 13C).
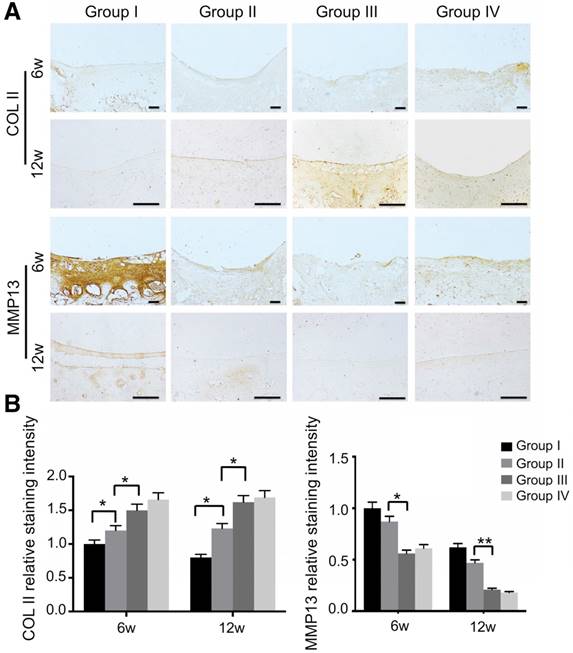
We also evaluated mitochondria morphology on cartilage adjacent to the defect after 12 weeks by TEM and quantified the percentage of mitochondrial vacuoles. The results showed that the percentage of mitochondrial vacuoles/mitochondria in ECM/GelMA/exosome group was markedly lower than in other groups (p<0.05, Fig. 13D and Fig. S12). We also measured the MDA levels in the synovial fluid at 12 weeks post-surgery. The results showed that the MDA level in the ECM/GelMA/exosome group was lower than in other groups (p<0.05; Fig. S13), indicating the anti-oxidative stress ability of exosomes. We then evaluated the protein expression of MMP13 and COL2A1 by immunohistochemistry. At both time points, expression levels of COL2A1 were up-regulated while MMP13 levels were down-regulated in the ECM/GelMA and ECM/GelMA/exosome groups than in the other two groups (p < 0.01; Fig. 14A, B). These findings demonstrated that the 3D printed, radially oriented ECM/GelMA/exosome group has the best therapeutic effect in cartilage regeneration in the rabbit model.
3.10 Enhancement of subchondral bone regeneration by the 3D printed, radially oriented ECM/GelMA/exosome scaffold
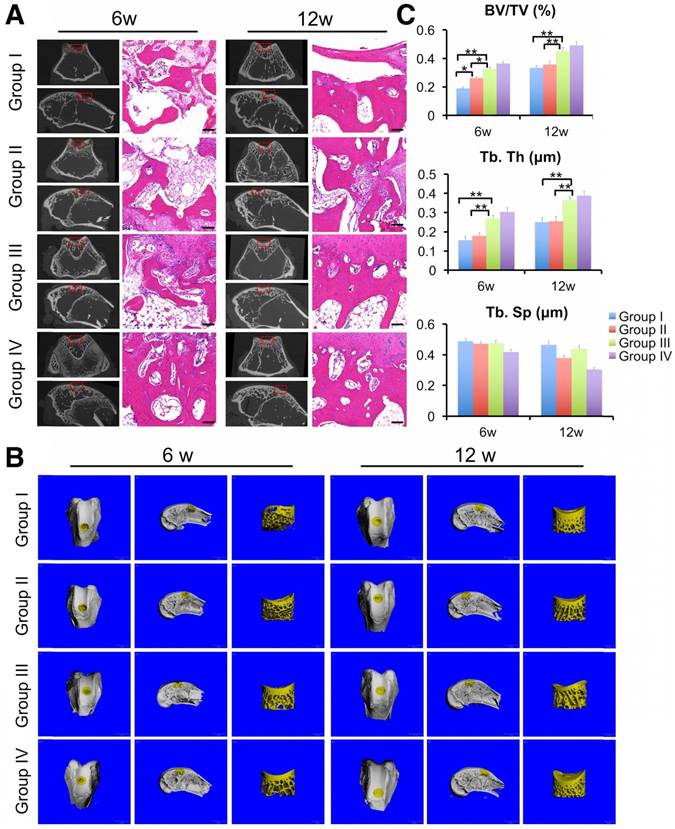
Finally, we evaluated the regeneration of subchondral bone by μCT scanning and HE staining. μCT results showed the generation of more ossified tissues at the defect regions implanted with the ECM/GelMA scaffold with or without exosomes compared with those implanted with the GelMA or control groups (Fig. 15A, B). Compared with the control or GelMA groups, the increase in ossified tissue was also confirmed by the significantly increased ratio of bone volume to tissue volume (BV/TV) and trabecular thickness (Tb.Th) in the ECM/GelMA scaffolds with or without exosomes (p < 0.05; Fig. 15C,). Additionally, the BV/TV ratio of the GelMA group was higher than the control group (p < 0.05; Fig. 15C). However, no significant difference was found between the ECM/GelMA scaffold groups with or without exosomes, and trabecular spacing (Tb.Sp) did not exhibit any significant differences among all groups (Fig. 15C). Consistent with this data, HE staining showed more subchondral bone trabecular integrity in the ECM/GelMA scaffold groups with or without exosomes (Fig. 15A).
Histological assessment of repaired cartilage in vivo. (A, B) Safranin O and alcian blue staining of repaired cartilage at both time points. Scale bars = 100 μm. (C) Macroscopic evaluation based on ICRS visual histological scores in cartilage samples at 6 and 12 weeks. n = 6. *p < 0.05; **p < 0.01. (D) TEM of mitochondria morphology in various groups at 12 weeks.
Immunohistochemical assessment of repaired cartilage in vivo. (A) Immunohistochemical staining for COL2A1 and MMP13. Scale bars = 100 μm. (B) Quantification of COL2A1 and MMP13 in cartilage samples at 6 and 12 weeks. n = 6. *p < 0.05; **p < 0.01.
The enhanced therapeutic outcomes of 3D printed ECM/GelMA scaffolds with or without exosomes compared with the other two groups is consistent with our finding that ECM/GelMA scaffolds increased M2 macrophage polarization. Taken together, our findings demonstrated that the 3D printed, radially oriented ECM/GelMA/exosome scaffold was effective in osteochondral regeneration in the rabbit osteochondral defect model.
4. Discussion
Mitochondria are regarded as the powerhouse of the cell, providing energy in the form of ATP and participating in many important biological processes, such as cell differentiation, cell death, and the cell cycle [35]. Mitochondrial dysfunction and oxidative stress damage are hallmarks of OA [36]. Alterations in mitochondrial structure, dynamics, and genome stability are found in degenerative OA cartilage leading to reduced mitochondrial respiration and immoderate ROS production. Compared with chondrocytes from healthy individuals, chondrocytes from OA patients exhibit increased mtDNA damage [37]. In our study, we found that chondrocytes from OA patients showed abnormal mitochondria with structure rarefaction and cristae disappearance. Furthermore, we found that chondrocyte treatment with MSC exosomes could recover mitochondrial dysfunction. Considering the results of the proteomic analysis, this effect may be caused by the 10.3% mitochondria-related proteins in exosomes. GO pathway enrichment analysis also indicated that various regions of the mitochondria were enriched in MSCs exosomes; therefore, further investigation of the most relevant proteins involved during the mitochondrial dysfunction recovery process is warranted.
Efficacy of the 3D printed, radially oriented ECM/GelMA/exosome scaffolds for subchondral bone repair in the rabbit model. (A) μCT reconstruction and histomorphological analysis of the osteochondral defect regions at both time points. Scale bars = 100 μm. (B) µCT reconstruction imaging of repaired knees at 6 and 12 weeks after surgery in various groups. (C) BV/TV, Tb.Th and TB.Sp values in various groups. n = 6. *p < 0.05; **p < 0.01.
ECM-based biomaterials consisting of native tissues have been widely researched in tissue engineering. One study showed that ECM might promote cell recruitment, cell infiltration, and cell differentiation without additional growth factors [38]. Recently, the hydrogel-based system of cartilage ECM has shown chondro-inductive potential; however, natural material-derived hydrogels possess disadvantages such as being mechanically inferior to synthetic materials, limiting the application of this type of hydrogel in load-bearing tissue [39]. In this study, we printed bioink based on photo-crosslinked cartilage ECM and GelMA hydrogels, which exhibited proper mechanical properties (33.24 ± 8.8 kPa) compared with that of un-crosslinked hydrogels. We also showed that cartilage ECM hydrogels could enhance cell migration which was consistent with a previous report [40]. In the ECM proteomics analysis, the cell migration-related proteins such as prelamin-A/C, chemerin, vinculin, apolipoprotein E, and decorin were detected. These proteins may be the reason why the addition of ECM could enhance cell migration. Except for chondro-inductive capacity, cartilage ECM has been reported to support endochondral ossification [41]. Visser et al. fabricated a template composite with GelMA hydrogel, MSCs, and cartilage-derived ECM particles and found that the scaffold was remodeled into mineralized bone tissue in a subcutaneous rat model [41]. We did not observe this phenomenon in our rat subcutaneous experiment; nevertheless, we found that more calcified tissue was produced at the defect regions of a rabbit osteochondral defect model implanted with the 3D printed ECM/GelMA scaffold compared with the 3D printed GelMA or control groups, implicating the possible osteogenesis effect of cartilage ECM.
Compared with traditional 3D printing techniques, SLA has many advantages for bioprinting [42]. Rather than printing in straps or droplets, SLA can print photo-crosslinked hydrogels in a layer-by-layer assembly. Moreover, the time needed for printing each layer is the same independent of how small or large the layer is, as the total printing time depends only on the number of layers [43]. Thus, compared with traditional 3D printing techniques such as fused deposition modeling, SLA can markedly reduce printing time. In addition, SLA is a nozzle-free printing technique that offers higher resolution than other techniques [44]. In this study, we applied SLA technology to print cartilage ECM, GelMA, and exosome bioink. Our results indicated that this low-cost technique could support bioink printing with subtle resolution. Moreover, our findings showed that this 3D-printed scaffold can effectively control exosome administration in vitro and in vivo, providing a one-step operation system that is more convenient than repetitive intra-articular injections of exosomes.
We have previously reported that the radially oriented scaffold can enhance osteochondral repair [17]. Numerous studies on osteochondral repair have also adopted this kind of architecture [45-48]. Compared to a random structure, oriented channels are more efficient for cells and tissues ingrowth, nutrients and waste exchange, ECM deposition, and cell interaction [48-50]. However, the in-depth molecular mechanism of how the structure of the scaffold affects cell behavior remains to be further explored. In this study, we showed that the scaffold could effectively retain exosomes for 2 weeks and used the AFM technique to explain this phenomenon. We found the collagen fibril D-Spacing to be 46.1±2.01 nm in the GelMA+2%ECM hydrogel which was smaller than the size of exosomes retaining most of the exosomes inside ECM/GelMA hydrogel. With increasing ECM concentrations, we also observed reduced gaps between fibrils indicated by decreasing collagen fibril D-spacing values. Thus, we hypothesized that the higher ECM concentration helps to improve the sustained releasing ability of the scaffold. However, the link between the collagen fibril D-Spacing and scaffold's controlled releasing ability remains to be investigated and might be related to the classical rubber theory [51].
The mechanisms of the immune response towards allografts have not been entirely determined [52]. Previously, Vas et al. evaluated the immunocompatibility of decellularized cartilage ECM scaffolds in mice and found a predominantly M2 macrophage-mediated immune response towards the decellularized ECM, which is indicative of a favorable long-term outcome and tissue-specific remodeling [53]. This is consistent with our results that the 3D printed ECM/GelMA scaffold can effectively induce M2 macrophages. Given our proteomics analysis results, which showed 4.1% of ECM proteins were involved in innate immune response, this effect may be caused by immune-regulated proteins such as CD59 glycoprotein, complement C3, protein S100-A12, and serum amyloid P-component in the ECM. Further work is needed to investigate the most abundant proteins involved in the immune-regulation process.
5. Conclusion
In the present study, we found that mitochondrial dysfunction and oxidative stress damage in degenerative cartilage could be restored by treatment with MSC exosomes by supplementing mitochondrial-related proteins. By applying MSC-derived exosomes to degenerative cartilage, the imbalance in energy metabolism was rescued, promoting cartilage regeneration. Using SLA, our 3D printed, radially oriented ECM/GelMA/exosome scaffolds were shown to effectively restore cartilage mitochondrial dysfunction, enhance chondrocyte migration, and polarize the synovial macrophage response towards an M2 phenotype. Finally, promising efficacy of the 3D printed, radially oriented scaffold on osteochondral defect repair was confirmed in an osteochondral defect model.
Supplementary Material
Supplementary materials and methods, figures and tables.
Acknowledgements
The study was supported by the National Key R&D Program of China [grant numbers 2018YFC1105202]; the National Nature Science Fund of China [grant numbers 81802147, 81702143, 81772387, 81871797]; the Natural Science Fund of Zhejiang Province [grant numbers Y18H060001, LZ15H060002, LY16H060004]; and the Key research and development plan in Zhejiang Province [grant numbers 2018C03060]. The authors thank Mr. Nianhang Rong for SEM imaging, the Imaging Center of Zhejiang University School of Medicine for technical assistance.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Glyn-Jones S, Palmer AJ, Agricola R, Price AJ, Vincent TL, Weinans H. et al. Osteoarthritis. Lancet. 2015;386:376-87
2. Albright JC, Daoud AK. Microfracture and Microfracture Plus. Clin Sports Med. 2017;36:501-7
3. Di Martino A, Kon E, Perdisa F, Sessa A, Filardo G, Neri MP. et al. Surgical treatment of early knee osteoarthritis with a cell-free osteochondral scaffold: results at 24 months of follow-up. Injury. 2015;46(Suppl 8):S33-8
4. Jungmann PM, Gersing AS, Baumann F, Holwein C, Braun S, Neumann J. et al. Cartilage repair surgery prevents progression of knee degeneration. Knee Surg Sports Traumatol Arthrosc. 2018 [Epub ahead of print]
5. Mobasheri A, Rayman MP, Gualillo O, Sellam J, van der Kraan P, Fearon U. The role of metabolism in the pathogenesis of osteoarthritis. Nat Rev Rheumatol. 2017;13:302-11
6. Luo Z, Jiang L, Xu Y, Li H, Xu W, Wu S. et al. Mechano growth factor (MGF) and transforming growth factor (TGF)-beta3 functionalized silk scaffolds enhance articular hyaline cartilage regeneration in rabbit model. Biomaterials. 2015;52:463-75
7. Burand AJ, Gramlich OW, Brown AJ, Ankrum JA. Function of Cryopreserved Mesenchymal Stromal Cells With and Without Interferon- Prelicensing is Context Dependent. Stem Cells. 2017;35:1437-9
8. Castellone MD, Laatikainen LE, Laurila JP, Langella A, Hematti P, Soricelli A. et al. Brief Report: Mesenchymal Stromal Cell Atrophy in Coculture Increases Aggressiveness of Transformed Cells. Stem Cells. 2013;31:1218-23
9. Wang YF, Yu DS, Liu ZM, Zhou F, Dai J, Wu BB. et al. Exosomes from embryonic mesenchymal stem cells alleviate osteoarthritis through balancing synthesis and degradation of cartilage extracellular matrix. Stem Cell Res Ther. 2017;8:189
10. Veceric-Haler Z, Cerar A, Perse M. (Mesenchymal) Stem Cell-Based Therapy in Cisplatin-Induced Acute Kidney Injury Animal Model: Risk of Immunogenicity and Tumorigenicity. Stem Cells Int. 2017;2017:7304643
11. Barkholt L, Flory E, Jekerle V, Lucas-Samuel S, Ahnert P, Bisset L. et al. Risk of tumorigenicity in mesenchymal stromal cell-based therapies-bridging scientific observations and regulatory viewpoints. Cytotherapy. 2013;15:753-9
12. Burger D, Vinas JL, Akbari S, Dehak H, Knoll W, Gutsol A. et al. Human endothelial colony-forming cells protect against acute kidney injury: role of exosomes. Am J Pathol. 2015;185:2309-23
13. Xin HQ, Li Y, Chopp M. Exosomes/miRNAs as mediating cell-based therapy of stroke. Front Cell Neurosci. 2014;8:377
14. Zhang S, Chuah SJ, Lai RC, Hui JHP, Lim SK, Toh WS. MSC exosomes mediate cartilage repair by enhancing proliferation, attenuating apoptosis and modulating immune reactivity. Biomaterials. 2018;156:16-27
15. Zhang S, Chu WC, Lai RC, Lim SK, Hui JH, Toh WS. Exosomes derived from human embryonic mesenchymal stem cells promote osteochondral regeneration. Osteoarthritis Cartilage. 2016;24:2135-40
16. Singh B, Modica-Napolitano JS, Singh KK. Defining the momiome: Promiscuous information transfer by mobile mitochondria and the mitochondrial genome. Semin Cancer Biol. 2017;47:1-17
17. Chen P, Tao J, Zhu S, Cai Y, Mao Q, Yu D. et al. Radially oriented collagen scaffold with SDF-1 promotes osteochondral repair by facilitating cell homing. Biomaterials. 2015;39:114-23
18. Qi YY, Chen X, Jiang YZ, Cai HX, Wang LL, Song XH. et al. Local delivery of autologous platelet in collagen matrix simulated in situ articular cartilage repair. Cell Transplant. 2009;18:1161-9
19. Zhang W, Chen J, Tao J, Jiang Y, Hu C, Huang L. et al. The use of type 1 collagen scaffold containing stromal cell-derived factor-1 to create a matrix environment conducive to partial-thickness cartilage defects repair. Biomaterials. 2013;34:713-23
20. Hu X, Wang Y, Tan Y, Wang J, Liu H, Wang Y. et al. A Difunctional Regeneration Scaffold for Knee Repair based on Aptamer-Directed Cell Recruitment. Adv Mater. 2017;29:e1605235
21. Shi W, Sun M, Hu X, Ren B, Cheng J, Li C. et al. Structurally and Functionally Optimized Silk-Fibroin-Gelatin Scaffold Using 3D Printing to Repair Cartilage Injury In Vitro and In Vivo. Adv Mater. 2017;29:e1701089
22. Wang Y, Hu X, Dai J, Wang J, Tan Y, Yang X. et al. A 3D graphene coated bioglass scaffold for bone defect therapy based on the molecular targeting approach. J Mater Chem B. 2017;5:6794-800
23. Crapo PM, Gilbert TW, Badylak SF. An overview of tissue and whole organ decellularization processes. Biomaterials. 2011;32:3233-43
24. Brown BN, Badylak SF. Extracellular matrix as an inductive scaffold for functional tissue reconstruction. Transl Res. 2014;163:268-85
25. Valentin JE, Stewart-Akers AM, Gilbert TW, Badylak SF. Macrophage participation in the degradation and remodeling of extracellular matrix scaffolds. Tissue Eng Part A. 2009;15:1687-94
26. Knowlton S, Yu CH, Ersoy F, Emadi S, Khademhosseini A, Tasoglu S. 3D-printed microfluidic chips with patterned, cell-laden hydrogel constructs. Biofabrication. 2016;8:025019
27. Chen P, Xia C, Mei S, Wang J, Shan Z, Lin X. et al. Intra-articular delivery of sinomenium encapsulated by chitosan microspheres and photo-crosslinked GelMA hydrogel ameliorates osteoarthritis by effectively regulating autophagy. Biomaterials. 2016;81:1-13
28. Lin X, Chen J, Qiu P, Zhang Q, Wang S, Su M. et al. Biphasic hierarchical extracellular matrix scaffold for osteochondral defect regeneration. Osteoarthritis Cartilage. 2018;26:433-44
29. van den Borne MP, Raijmakers NJ, Vanlauwe J, Victor J, de Jong SN, Bellemans J. et al. International Cartilage Repair Society (ICRS) and Oswestry macroscopic cartilage evaluation scores validated for use in Autologous Chondrocyte Implantation (ACI) and microfracture. Osteoarthritis Cartilage. 2007;15:1397-402
30. Suantawee T, Tantavisut S, Adisakwattana S, Tanavalee A, Yuktanandana P, Anomasiri W. et al. Oxidative stress, vitamin e, and antioxidant capacity in knee osteoarthritis. J Clin Diagn Res. 2013;7:1855-9
31. Perez RA, Mestres G. Role of pore size and morphology in musculo-skeletal tissue regeneration. Mater Sci Eng C Mater Biol Appl. 2016;61:922-39
32. Murphy CM, Haugh MG, O'Brien FJ. The effect of mean pore size on cell attachment, proliferation and migration in collagen-glycosaminoglycan scaffolds for bone tissue engineering. Biomaterials. 2010;31:461-6
33. Hollister SJ. Porous scaffold design for tissue engineering. Nat Mater. 2005;4:518-24
34. Fishman JM, Lowdell MW, Urbani L, Ansari T, Burns AJ, Turmaine M. et al. Immunomodulatory effect of a decellularized skeletal muscle scaffold in a discordant xenotransplantation model. Proc Natl Acad Sci U S A. 2013;110:14360-5
35. Shutt TE, Shadel GS. A compendium of human mitochondrial gene expression machinery with links to disease. Environ Mol Mutagen. 2010;51:360-79
36. Loeser RF, Collins JA, Diekman BO. Ageing and the pathogenesis of osteoarthritis. Nat Rev Rheumatol. 2016;12:412-20
37. Grishko VI, Ho R, Wilson GL, Pearsall AWt. Diminished mitochondrial DNA integrity and repair capacity in OA chondrocytes. Osteoarthritis Cartilage. 2009;17:107-13
38. Rowland CR, Glass KA, Ettyreddy AR, Gloss CC, Matthews JRL, Huynh NPT. et al. Regulation of decellularized tissue remodeling via scaffold-mediated lentiviral delivery in anatomically-shaped osteochondral constructs. Biomaterials. 2018;177:161-75
39. Beck EC, Barragan M, Tadros MH, Gehrke SH, Detamore MS. Approaching the compressive modulus of articular cartilage with a decellularized cartilage-based hydrogel. Acta Biomater. 2016;38:94-105
40. Ghosh P, Gruber SMS, Lin CY, Whitlock PW. Microspheres containing decellularized cartilage induce chondrogenesis in vitro and remain functional after incorporation within a poly(caprolactone) filament useful for fabricating a 3D scaffold. Biofabrication. 2018;10:025007
41. Visser J, Gawlitta D, Benders KEM, Toma SMH, Pouran B, van Weeren PR. et al. Endochondral bone formation in gelatin methacrylamide hydrogel with embedded cartilage-derived matrix particles. Biomaterials. 2015;37:174-82
42. Wang ZJ, Abdulla R, Parker B, Samanipour R, Ghosh S, Kim K. A simple and high-resolution stereolithography-based 3D bioprinting system using visible light crosslinkable bioinks. Biofabrication. 2015;7:045009
43. Bhattacharjee N, Parra-Cabrera C, Kim YT, Kuo AP, Folch A. Desktop-Stereolithography 3D-Printing of a Poly(dimethylsiloxane)-Based Material with Sylgard-184 Properties. Adv Mater. 2018;30:e1800001
44. Wang Z, Kumar H, Tian Z, Jin X, Holzman JF, Menard F. et al. Visible Light Photoinitiation of Cell Adhesive Gelatin Methacryloyl Hydrogels for Stereolithography 3D Bioprinting. ACS Appl Mater Interfaces. 2018;10:26859-69
45. Zhao P, Liu S, Bai Y, Lu S, Peng J, Zhang L. et al. hWJECM-Derived Oriented Scaffolds with Autologous Chondrocytes for Rabbit Cartilage Defect Repairing. Tissue Eng Part A. 2018;24:905-14
46. Dai Y, Shen T, Ma L, Wang D, Gao C. Regeneration of osteochondral defects in vivo by a cell-free cylindrical poly(lactide-co-glycolide) scaffold with a radially oriented microstructure. J Tissue Eng Regen Med. 2018;12:e1647-e61
47. Dai Y, Gao Z, Ma L, Wang D, Gao C. Cell-Free HA-MA/PLGA Scaffolds with Radially Oriented Pores for In Situ Inductive Regeneration of Full Thickness Cartilage Defects. Macromol Biosci. 2016;16:1632-42
48. Jia S, Zhang T, Xiong Z, Pan W, Liu J, Sun W. In Vivo Evaluation of a Novel Oriented Scaffold-BMSC Construct for Enhancing Full-Thickness Articular Cartilage Repair in a Rabbit Model. PLoS One. 2015;10:e0145667
49. Zhang Y, Yang F, Liu K, Shen H, Zhu Y, Zhang W. et al. The impact of PLGA scaffold orientation on in vitro cartilage regeneration. Biomaterials. 2012;33:2926-35
50. Arora A, Kothari A, Katti DS. Pore orientation mediated control of mechanical behavior of scaffolds and its application in cartilage-mimetic scaffold design. J Mech Behav Biomed Mater. 2015;51:169-83
51. Liu X, Yang Y, Li Y, Niu X, Zhao B, Wang Y. et al. Integration of stem cell-derived exosomes with in situ hydrogel glue as a promising tissue patch for articular cartilage regeneration. Nanoscale. 2017;9:4430-8
52. Ma R, Li M, Luo J, Yu H, Sun Y, Cheng S. et al. Structural integrity, ECM components and immunogenicity of decellularized laryngeal scaffold with preserved cartilage. Biomaterials. 2013;34:1790-8
53. Vas WJ, Shah M, Blacker TS, Duchen MR, Sibbons P, Roberts SJ. Decellularized Cartilage Directs Chondrogenic Differentiation: Creation of a Fracture Callus Mimetic. Tissue Eng Part A. 2018;24:1364-76
Author contact
![]() Corresponding authors: Xianfeng Lin, email: xianfeng_linedu.cn; Pengfei Chen, email: pengfei_chenedu.cn; Shunwu Fan, email: shunwu_fanedu.cn.
Corresponding authors: Xianfeng Lin, email: xianfeng_linedu.cn; Pengfei Chen, email: pengfei_chenedu.cn; Shunwu Fan, email: shunwu_fanedu.cn.
 Global reach, higher impact
Global reach, higher impact