13.3
Impact Factor
Theranostics 2018; 8(22):6149-6162. doi:10.7150/thno.29303 This issue Cite
Research Paper
Iron oxide nanozyme suppresses intracellular Salmonella Enteritidis growth and alleviates infection in vivo
1. Department of Feed and Nutrition, Poultry Institute, Chinese Academy of Agricultural Sciences, Yangzhou, Jiangsu 225125, China
2. College of Animal Science and Technology, Hunan Agricultural University, Changsha, Hunan 410128, China
3. Jiangsu Key Laboratory of Experimental & Translational Non-coding RNA Research, Institute of Translational Medicine , School of Medicine, Yangzhou University, Yangzhou, Jiangsu 225000, China
4. Department of Poultry Science, Texas A&M AgriLife Research, Texas A&M University, College Station, TX 77843, USA
5. Institute of Effective Evaluation of Feed and Feed Additive (Poultry institute), Ministry of Agriculture, Yangzhou, Jiangsu 225125, China
6. Jiangsu Co-Innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou University, Yangzhou, Jiangsu 225000, China
Received 2018-8-17; Accepted 2018-10-23; Published 2018-11-29
Abstract

Rational: Salmonella Enteritidis (S. Enteritidis) is a globally significant zoonotic foodborne pathogen which has led to large numbers of deaths in humans and caused economic losses in animal husbandry. S. Enteritidis invades host cells and survives within the cells, causing resistance to antibiotic treatment. Effective methods of elimination and eradication of intracellular S. Enteritidis are still very limited. Here we evaluated whether a new intracellular antibacterial strategy using iron oxide nanozymes (IONzymes) exerted highly antibacterial efficacy via its intrinsic peroxidase-like activity in vitro and in vivo.
Methods: The antibacterial activities of IONzymes against planktonic S. Enteritidis, intracellular S. Enteritidis in Leghorn Male Hepatoma-derived cells (LMH), and liver from specific pathogen free (SPF) chicks were investigated by spread-plate colony count method and cell viability assay. Changes in levels of microtubule-associated protein light chain 3 (LC3), a widely used marker for autophagosomes, were analyzed by immunoblotting, immunofluorescence, and electron microscopy. Reactive oxygen species (ROS) production was also assessed in vitro. High-throughput RNA sequencing was used to investigate the effects of IONzymes on liver transcriptome of S. Enteritidis-infected chicks.
Results: We demonstrated that IONzymes had high biocompatibility with cultured LMH cells and chickens, which significantly inhibited intracellular S. Enteritidis survival in vitro and in vivo. In addition, co-localization of IONzymes with S. Enteritidis were observed in autophagic vacuoles of LMH cells and liver of chickens infected by S. Enteritidis, indicating that IONzymes mediated antibacterial reaction of S. Enteritidis with autophagic pathway. We found ROS level was significantly increased in infected LMH cells treated with IONzymes, which might enhance the autophagic elimination of intracellular S. Enteritidis. Moreover, orally administered IONzymes decreased S. Enteritidis organ invasion of the liver and prevented pathological lesions in a chicken-infection model. Non-target transcriptomic profiling also discovered IONzymes could change hepatic oxidation-reduction and autophagy related gene expressions in the S. Enteritidis infected chickens.
Conclusion: These data suggest that IONzymes can increase ROS levels to promote the antibacterial effects of acid autophagic vacuoles, and thus suppress the establishment and survival of invading intracellular S. Enteritidis. As a result, IONzymes may be a novel alternative to current antibiotics for the control of intractable S. Enteritidis infections.
Keywords: Salmonella Enteritidis, iron oxide nanozyme, autophagy, reactive oxygen species, intracellular bacteria
Introduction
Salmonella is one of the major foodborne pathogens of worldwide importance. It is estimated that Salmonella causes over 1.3 billion illness and 200,000 deaths in humans annually worldwide [1]. Although more than 2500 different serotypes have been discovered, serotype Salmonella Enteritidis (S. Enteritidis) is one of the most important serotypes of Salmonella transmitted from animals to humans in most parts of the world [2]. More seriously, as a facultative intracellular pathogen, S. Enteritidis can invade and survive in both phagocytic and nonphagocytic cells [3-5]. This limits the treatment efficiency of traditional antibiotics that fail to penetrate into cells, such as chloramphenicol, ampicillin, and trimethoprim-sulfamethoxazole (co-trimoxazole). Additionally, S. Enteritidis tends to be highly resistant to multiple antibiotics due to the inadequate use of antibiotics. Therefore, it is critical to develop effective and environmentally-friendly antibiotic-alternatives that are capable of intracellular delivery to treat infectious diseases caused by intracellular S. Enteritidis so as to protect both animal and human health.
One of promising treatments of S. Enteritidis involves the use of nanoparticles, that have been shown to induce autophagic pathways in mammalian cells [6]. Autophagy is a highly conserved cellular process in eukaryotes, which degrades unnecessary cellular components or invading pathogens by fusion of autophagosome with lysosome [7]. Many pathogens have been proven to be targeted by autophagic responses after entering host cells [8]. Among them, some bacteria can be killed by autophagy directly, such as group A Streptococcus [9]. Other bacteria are able to antagonize the autophagy pathway or exploit autophagosomes for their benefit, such as Listeria monocytogenes [10]. It has been reported that autophagy could protect several cell culture models from Salmonella infection, such as HeLa cells, mouse embryonic fibroblasts and macrophages [11-14].
Nanozymes are nanomaterials that possess instrinsic enzyme-like properties. They can catalyze biological reactions under physiological conditions and perform the biofunctions which are often complemented by natural enzymes, thus representing a new generation of enzyme mimics/artificial enzymes [15-19]. The enzyme-like catalytic activities come from their nanoscale effects of nanozymes and no extra natural enzymes or catalysts are required to modify or functionalize them. Compared to natural enzymes or other traditional artificial enzymes, nanozymes exert high catalytic efficiency, versatile tunability, multiple functionality, high stability and low cost for scale up production, showing great promise in biomedical applications [20,21]. Currently, there are three main classes of nanozymes according to the materials, including metal oxide nanozymes, metal nanozymes and carbon nanozymes. Iron oxide (Fe3O4 or Fe2O3) nanoparticles are one of typical metal oxide nanozymes which possess peroxidase-like activity under acidic pH and catalase-like activity under neutral pH [22-24], showing the ability to regulate ROS level. In addition, Fe3O4 nanoparticles often perform higher enzyme-like activities than those composed of Fe2O3, as ferrous iron provides more contribution in oxidation-reduction catalysis [22]. Therefore, most researches focus on Fe3O4 to develop iron oxide nanozymes (IONzymes). These properties allow IONzymes (Fe3O4 if no specific declaration) to regulate cell proliferation and differentiation, such as promoting the proliferation of human mesenchymal stem cell [25] and cancer stem cells from U251 glioblastoma multiform [26]. In addition, IONzymes have been reported to induce autophagy responses in several types of cells, such as human umbilical vein endothelial cells, leukemia cell lines and multiple myeloma cell lines [27,28]. Importantly, IONzymes can inhibit the growth of various bacteria, including Escherichia coli [29], methicillin-resistant Staphylococcus aureus [30] and Streptococcus mutans [31]. Our previous work also found that IONzymes possessed an intrinsic peroxidase-like activity that decomposed hydrogen peroxide to generate free radicals, which could be used to kill bacteria and disrupt biofilm formation by inducing high levels of ROS under acidic conditions [32-34]. Considering the acidic condition of autophagolysosomes, therefore we proposed that IONzymes could promote autophagy-mediated elimination of S. Enteritidis through its peroxidase-like activity. The purpose of our studies were to evaluate the effects of IONzymes on growth of planktonic cultures of S. Enteritidis, intracellular survivability of S. Enteritidis in vitro and in vivo and to investigate the mechanism of action of IONzymes against intracellular growth of S. Enteritidis.
Epidemiological studies show that S. Enteritidis infections are usually caused by ingestion of contaminated food or water, especially contaminated poultry products [35-38]. Chickens are the single largest reservoir host for S. Enteritidis. Source attribution studies have determined that contaminated poultry and poultry products are the major sources of human infection with S. Enteritidis, which colonize internal organs including the intestines, liver, and spleen [39,40]. In this study, we used cultured cells (Leghorn Male Hepatoma, LMH) and specific pathogen free (SPF) chicks to construct S. Enteritidis infection models, and experimentally verified that biocompatible IONzymes alleviated S. Enteritidis infection in vitro and in vivo through an autophagic pathway. We demonstrated that IONzymes could significantly inhibit the growth of extracellular and intracellular S. Enteritidis by generating reactive oxygen species (ROS) to intensify autophagic elimination of intracellular S. Enteritidis. The detail bacteriostatic principle based on IONzymes was presented in Scheme 1. More importantly, the S. Enteritidis loads of infected chickens were decreased by orally administrating IONzymes significantly.
Schematic illustration of the bacteriostatic effects of IONzymes and mechanism responsible for the antibacterial activity. The autophagy pathway starts with the formation of phagophores, which engulf S. Enteritidis. IONzymes were internalized to cells by endocytosis. The phagophore expanded and fused with the IONzymes containing endosomes into amphisome. It then fused with lysosomes to form acid autolysosomes for subsequently digestion of bacteria. IONzymes enhance the autophagic responses on the elimination of intracellular S. Enteritidis by producing intracellular reactive oxygen species (ROS).

Results and discussion
Preparation and characterization of IONzymes
We prepared IONzymes using a typical solvothermal method for the formation of iron oxide nanoparticles described previously [32,41-43]. The prepared iron oxide nanoparticles showed a fairly uniform size with an average diameter of 200 nm (Figure S1A,B). X-ray diffraction (XRD) showed that the products were composed of Fe3O4 (Figure S2) and energy dispersive spectrometry (EDS) analysis showed that the atomic ratio of Fe/O was 48.42/51.58 (Figure S3). These iron oxide nanoparticles had good dispersibility in the water and the characterization by dynamic light scattering (DLS) showed their hydrodynamic diameter was around 350nm (Figure S4). Importantly, these nanoparticles showed very high peroxidase-like activity as demonstrated by the colorimetric reaction of 3,3',5,5'-tetramethylbenzidine (TMB) and hydrogen peroxide (H2O2) under acidic condition. The enzyme-catalysed reaction followed the Michaelis-Menten kinetics either H2O2 amount or TMB was variable (Figure S5A,B), which was consistent with our previous study [32]. The catalytic kinetics followed Michaelis-Menten model (Figure S5). The Vmax values to H2O2 and TMB were 96.4 nM/s and 128.5 nM/s, respectively (Table S1 and S2). These kinetics and parameters demonstrated the prepared iron oxide nanoparticles performed peroxidase-like activity by themselves and thus were a peroxidase-mimicking nanozymes. To verify their ability to generate ROS under acidic condition, electron spin-resonance spectroscopy (ESR) was used to monitor hydroxyl radicals and the results showed that our IONzymes successfully induced hydroxyl radicals generation from H2O2 in sodium acetate (NaAc) buffer (pH 4.5) (Figure S6). In addition, we found that our IONzymes after sterilization in an autoclave (121 oC, 30 min) still showed comparable catalytic activity compared to IONzymes (without sterllization treatment). The Kcat values for sterilized IONzymes to H2O2 and TMB were 1.5 × 105/s and 2.1 × 105/s, respectively (Table S1 and S2), suggesting that IONzymes were quite stable and could be readily used for the following antibacterial experiments after autoclave disinfection.
IONzymes inhibit planktonic and intracellular S. Enteritidis growth in vitro
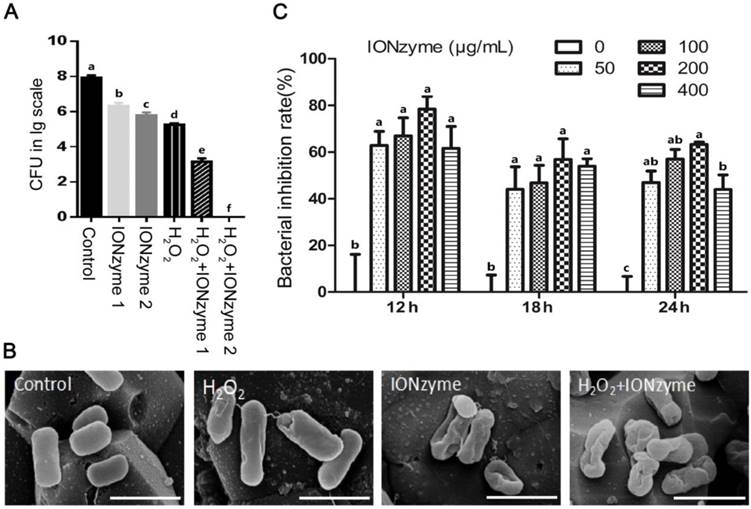
Then, we examined the growth suppressive effect of IONzymes on S. Enteritidis in liquid culture and Leghorn Male Hepatoma (LMH) cells in vitro, especially the intracellular S. Enteritidis. First, we used sodium acetate (NaAc) buffer (pH 4.5) to mimic the acidic condition in autophagic vacuoles to test the effects of IONzymes on planktonic cultures of S. Enteritidis. The results showed that, compared with the control group without IONzyme, either IONzymes (0.25 mg/mL) or H2O2 alone caused a significant reduction in S. Enteritidis colony forming units (CFU) numbers (Figure 1A). Moreover, synergistic inhibitory effects were shown in the combination of hydrogen peroxide (0.05%) and IONzymes treatment (0.25 mg/mL) resulting a 4-log reduction (from 7.2 to 3.1) of viable bacteria within 30 min (Figure 1A). When 0.5 mg/mL IONzymes and H2O2 (0.05%) were combined, the total viable S. Enteritidis numbers were significantly decreased to the undetectable level (Figure 1A). Meanwhile, we investigated the effect of IONzymes on morphology changes of S. Enteritidis. As shown in Figure 1B, untreated S. Enteritidis were typically rod-shaped with smooth surface. After exposure of S. Enteritidis to IONzymes for 30 min, the cell walls of bacteria became wrinkled indicating that markable damage on bacterial viability was induced by IONzymes (Figure 1B). We next investigated the effects of IONzymes on intracellular growth of S. Enteritidis on S. Enteritidis infected LMH cells. At 12 to 24 h post infection, compared with the control group, all concentrations of IONzymes have reduced intracellular S. Enteritidis loads significantly ranging from 43.95% to 78.42% inhibition (Figure 1C). In addition, a time course and concentration-response study showed that 100 μg/mL IONzymes had minimal effect on infected-LMH cell viability at 12 h (Figure S7). We also constructed green fluorescent protein (GFP)-expressing S. Enteritidis and investigated the antibacterial activities of IONzymes on infected LMH cells (Figure S8). IONzymes (100 μg/mL) at 12 h significantly decreased counts of GFP-S. Enteritidis as shown in Figure S9. These results suggest that IONzymes not only inhibited the survival of planktonic S. Enteritidis, but also effectively suppressed intracellular growth of S. Enteritidis.
The inhibition effects of IONzymes on S. Enteritidis. (A) The inhibition effects of IONzymes to planktonic S. Enteritidis in the absent or presence of H2O2. IONzyme 1: 0.25 mg/ml IONzyme; IONzyme 2: 0.5 mg/ml IONzyme; H2O2: 0.05% H2O2; H2O2+IONzyme 1: 0.05% H2O2+0.25 mg/ml IONzyme; H2O2+IONzyme 2: 0.05% H2O2+0.5 mg/ml IONzyme. (B) SEM micrographs of S. Enteritidis. S. Enteritidis were treated with vehicle control, H2O2 alone (0.05%), IONzymes alone (0.5 mg/mL), and the combination treatment of IONzymes (0.5 mg/mL) and H2O2 (0.05%). Scale bar: 10 µm. (C) The effect of dose- and time-response of IONzymes on S. Enteritidis infected-LMH cells. LMH cells were infected with 5×107 CFU/mL S. Enteritidis and treated with indicated concentrations of IONzymes (0, 50, 100, 200, and 400 μg/mL) to 12, 18 and 24 h. Bacterial inhibition rates were expressed as a percentage of the control group without IONzymes treatment. Data represented as Mean ± SEM (n=3). Different letters indicate statistically significant difference (P < 0.05).
IONzymes inhibit the growth of S. Enteritidis by increasing ROS to promote the antibacterial effects of acid autophagic vacuoles
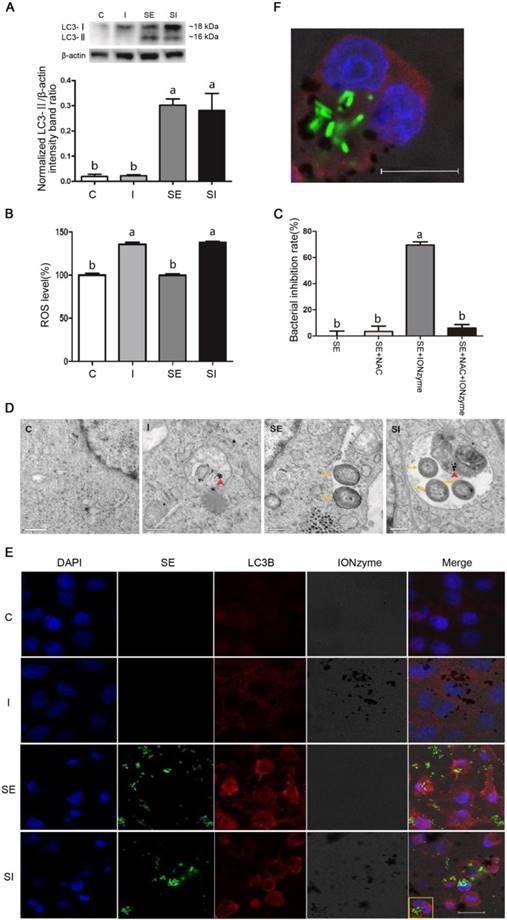
We subsequently sought to determine the mechanism by which the IONzymes were able to inhibit the growth of S. Enteritidis in vitro. We hypothesized that IONzymes would inhibit intracellular growth of S. Enteritidis in infected LMH cells by promoting autophagic activities and inducing ROS levels. To test the hypothesis, we firstly used rapamycin (RAPA, a common agonist of autophagy) as a positive control [44], to study whether increased autophagy could help eradicate intracellular S. Enteritidis in infected LMH cells. The results showed that compared with the S. Enteritidis-infected group, the intracellular S. Enteritidis cell counts were significantly decreased in LMH cells treated with RAPA (Figure S10), indicating that induced autophagy can help eliminate intracellular S. Enteritidis in LMH cells. This was consistent with several previous studies which demonstrated that Salmonella infection induced autophagy to help eliminating intracellular pathogens [11-14]. Next, we investigated that whether the autophagy level further increased by supplemental IONzymes through detecting LC3-II protein expression by western blotting. The level of LC3-II correlates with autophagosome formation, and is therefore traditionally used as a marker for the autophagosomes [45]. As shown in Figure 2A, compared to the control group (group C), the protein expression of LC3-II was not affected by IONzymes treatment alone (group I). The protein levels of LC3-II in the S. Enteritidis infection group in the absence (group SE) or presence of IONzymes (group SI) were significantly increased. These results indicated that IONzymes didn't increase the autophagy level of LMH cells regardless of S. Enteritidis infection. This was different with a common notion that nanoparticles are capable to induce autophagy in cells [6], it may be associated with the different cell types and nanoparticles. Briefly, our results suggested that, S. Enteritidis infection induced an antibacterial autophagic response which helped eliminate the invading bacteria by LMH cells, but IONzymes didn't promote the intracellular S. Enteritidis inhibition rate by further increasing the autophagy level of LMH cells infected by S. Enteritidis.
Recent studies reported that the generation of ROS plays a central role in antibacterial autophagy [46]. In addition, IONzymes have been reported to degrade H2O2 by generating the ROS, thus eradicating bacteria and eliminating formation of biofilms in acidic conditions [32]. Giving that autophagosomes are acid vacuoles, we examined that whether IONzymes induced intracellular ROS production. We found that IONzymes significantly increased intracellular ROS level (Figure 2B). To further confirm the role of ROS generated by IONzymes in suppressing intracellular S. Enteritidis proliferation, NAC, an ROS scavenger, was used to inhibit the generation of ROS. It was shown that the intracellular S. Enteritidis inhibition rate of IONzymes was significantly attenuated in the presence of NAC (10 nmol/L) (P < 0.05) (Figure 2C). These results suggest that IONzymes can increase ROS generation, and promote the antibacterial effects of acidic autophagic vacuoles, consequently inhibit the rate of survival of S. Enteritidis.
IONzymes co-localize with S. Enteritidis in autophagosomes
Next, we tried to verify the IONzymes would be exactly co-localized with S. Enteritidis in autophagosomes of S. Enteritidis-infected LMH cells by transmission electron microscopy (TEM) and immunofluorescence (IF). The TEM results (Figure 2D) revealed that a significant fraction of intracellular S. Enteritidis in infected LMH cells were restrained inside of the double membrane of autophagosomes, while such structures were not observed in uninfected cells. Furthermore, in the cytoplasm of infected LMH cells of group SI, some intracellular S. Enteritidis and IONzymes were co-located in the same autophagosomes. The IF results (Figure 2E) also showed that, compared with group C, punctuate LC3 proteins (red) significantly accumulated in group SE and group SI, while group I showed no significant difference, which was consistent with the protein expression results of LC3-II. In group SE, the co-localization of autophagosomes (LC3-postitive punctuate structures; red) and GFP-S. Enteritidis were observed. We showed that autophagosome structures (LC3-postitive punctuate structures; red) were associated with GFP- S. Enteritidis (green) and IONzymes (black dots) in group SI (Figure 2F). Therefore, we speculate that the accumulation of IONzymes inside of autophagosomes may mediate the interaction of S. Enteritidis with autophagic pathways. It should be noted that on entry into the cell, IONzymes may have different pathway compared with S. Enteritidis via autophagy. IONzymes as nanoparticles may enter the host cell by endocytosis and achieve co-localization with bacteria by fusion with autophagic vacuoles or lysosome. However, as our IONzymes are naked without surface modification, they also probably gain access by directly crossing lipid bilayers and entering cytoplasm [47]. It is still unclear that if IONzymes can cross vacuoles and lysosome to co-localize in the cytoplasm. Future study for our work is to disclose the dynamic process of IONzymes entering cell and co-localization with S. Enteritidis in autophagosomes.
Effects of IONzymes on LMH cells infected with S. Enteritidis. (A) The protein expression levels of LC3, a marker protein for autophagy, in LMH cells at 12 h. (B) The relative levels of ROS in LMH cells. (C) The antibacterial effects of NAC and IONzymes in intracellular S. Enteritidis at 12 h. (D) TEM micrographs of co-localization of IONzymes with S. Enteritidis and autophagosomes in LMH cells at 12 h. The red short triangle arrows show the IONzymes, and the yellow arrows show the S. Enteritidis. Scale bar: 0.5 μm. (E) Representative immunostaining image for IONzymes (black), S. Enteritidis (green), LMH cell nuclei (blue) and autophagosomes (LC3B, red) in LMH cells at 12 h. Scale bar: 20 μm. (F) The enlarged image of yellow delineate highlighting the co-localization of above four substances in Figure 2E. The scale bar in the figure represents 10 μm. For figure A, B and D-F, cells were treated with vehicle control (C), IONzymes alone (I), infected with S. Enteritidis in the absence (SE) or presence (SI) of IONzymes. For figure C, S. Enteritidis infected LMH cells were treated with or without NAC and IONzymes. Values represent the Mean ± SEM (n=3). Different capital letters indicate statistically significant difference (P < 0.05).
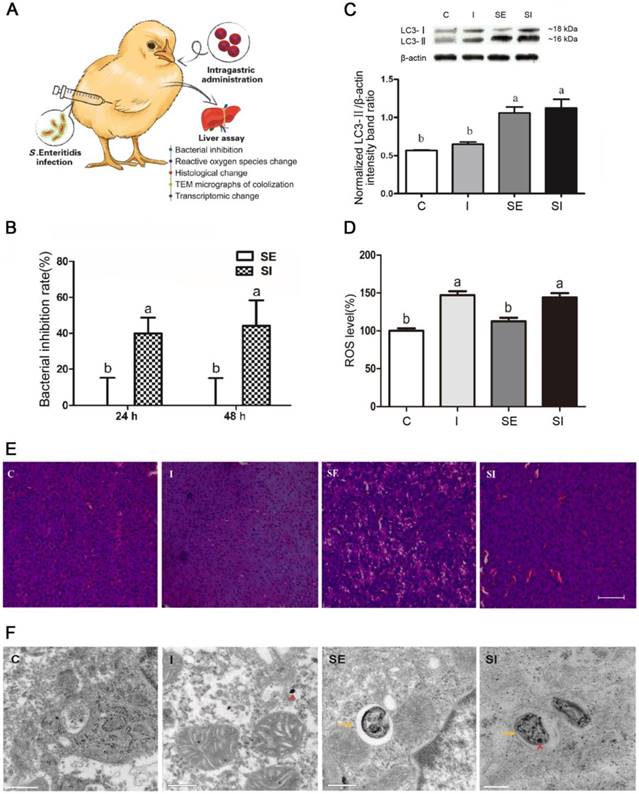
Effects of IONzymes in chickens infected with S. Enteritidis. Chickens were treated with vehicle control (C), IONzymes alone (I), infected with S. Enteritidis in the absence (SE) or presence (SI) of IONzymes. (A) Schematic illustration of the animal experiment. Chicken were orally gavaged with IONzymes and subcutaneous injected with S. Enteritidis. Livers were collected for pathological analyses. (B) Bacterial inhibition rates of IONzymes at different times in livers of S. Enteritidis infected chickens. Values represent the Mean ± SEM (n=9). Different letters indicate statistically significant difference (P < 0.05). (C) The hepatic LC3 protein expression levels in the chickens from the treatment groups. Representative images (top) and quantitative analysis (bottom) showing the effect of treatments on LC3 protein levels. The values represent the Mean ± SEM (n=3). Different letters indicate statistically significant difference (P < 0.05). (D) The relative levels of ROS in chickens. Values represent the Mean ± SEM (n=6). Different capital letters indicate statistically significant difference (P < 0.05). (E) Representative photomicrographs of the liver histological section of birds exposed to IONzymes in the absence or presence of S. Enteritidis administration. Scale bar: 50 μm. (F) TEM micrographs of co-localization of IONzymes with S. Enteritidis and autophagosomes in liver of chickens. The red short triangle arrows show the IONzymes, and the yellow arrows show the S. Enteritidis. Scale bar: 0.5 μm (in C, I, SE) and 0.2 μm (SI).
IONzymes suppress the growth of S. Enteritidis in vivo
Lastly, we investigated whether IONzymes could suppress the growth of S. Enteritidis in vivo (Figure 3A). In-vivo effect of IONzymes (50 mg/kg) against S. Enteritidis was tested for 24 h and 48 h (Figure 3B, S11). Bacterial loads and pathological changes in livers of infected chickens were examined. IONzyme-treated birds infected with S. Enteritidis showed a significant reduction in the viable bacterial counts at 24 h and 48 h compared to the birds infected with S. Enteritidis (Figure 3B). We also analyzed the autophagy protein (LC3-II) protein expression and ROS level in livers from chickens. We found that IONzymes did not change LC3-II protein levels compared to control groups while S. Enteritidis significantly increased the LC3-II expression (Figure 3C), and significantly increased ROS level in liver tissues (Figure 3D), which were consistent with the results in in vitro cell experiments (Figure 2A). Birds in group SE showed pathological changes with more vacuoles and widened interstitial space, IONzymes administration alleviated the liver pathological changes caused by S. Enteritidis infection (Figure 3E). IONzymes were co-localized with S. Enteritidis in autophagic vacuoles from liver cells in vivo (Figure 3F). The results from in vivo studies clearly demonstrated the potential application of IONzymes against S. Enteritidis infection in animals.
IONzymes change oxidative and autophagic gene expressions in S. Enteritidis-infected chicken livers
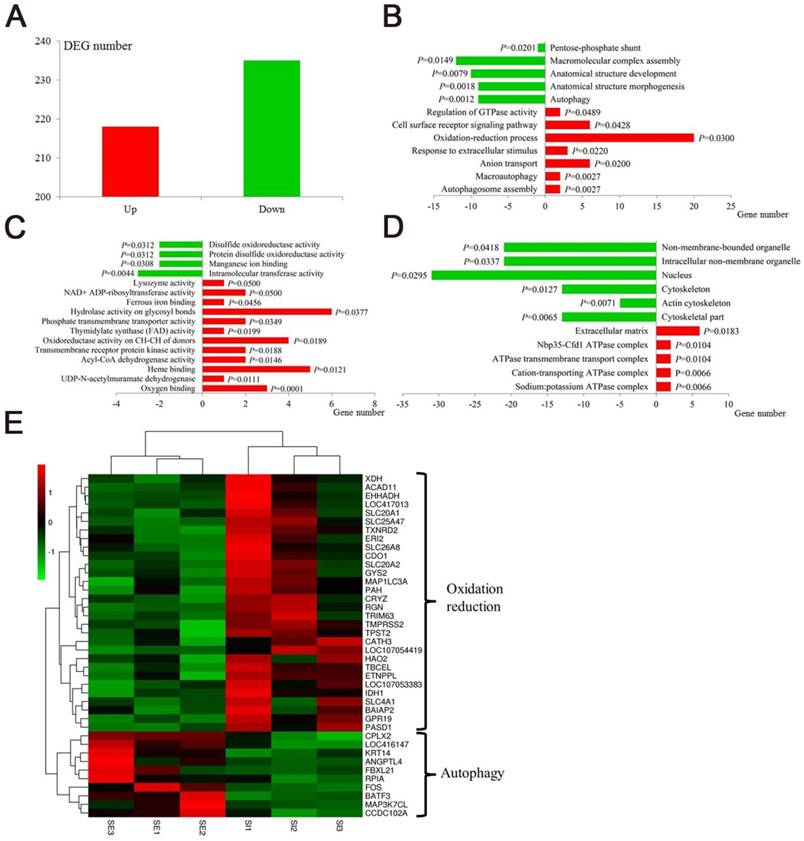
In recent years, the emerging “non-target high-throughput omics” technologies hold great promise for the discovery of the biochemical alterations and pathways that are linked to disease processes. Transcriptomic profiling can simultaneously monitor thousands of genes to provide an in-depth investigation of how external factors affect gene expression [48,49]. We selected the RNA-seq transcriptomic method to uncover the effects of IONzymes on gene expressions in S. Enteritidis-infected chicken livers. A total of 390 hepatic genes were identified as differentially expressed genes (DEGs), among which 218 genes were up-regulated and 235 genes were down-regulated in group SI compared with group SE (Figure 4A, Table S3, Table S4). DEGs were then used for Gene ontology (GO) analysis to uncover their functional enrichment. These DEGs were categorized into three main GO categories of biological process, molecular function, and cellular component. In total, there were 175, 76, and 22 significantly enriched GO terms (P < 0.05) identified in three GO categories respectively (Table S5, Table S6, Table S7). In particular, most of the significantly enriched biological process terms are associated with oxidation-reduction (GO:0055114∼regulation of oxidation-reduction process, GO:0006098∼pentose-phosphate shun) and autophagy (GO:0000045∼autophagosome assembly, GO:0016236∼macroautophagy, GO:0006914∼autophagy) (Figure 4B). Meanwhile, the same related terms were included in molecular function (Figure 4C) and cellular component (Figure 4D). The heat map of gene expressions of the DEGs enriched in oxidation-reduction and autophagy functions revealed that oxidation-reduction related genes were up-regulated in three samples of group SI, while autophagy related genes in same group SI were down-regulated conversely (Fig 4E). This non-target sequencing technology further confirmed IONzymes could alter oxidation-reduction and autophagy functions in S. Enteritidis infected chickens.
Conclusion
In conclusion, our study demonstrated that IONzymes could enhance the autophagic eliminating efficiency of S. Enteritidis which dramatically induced autophagy once invading to LMH cells. Normally, S. Enteritidis infection induced protective autophagic response in LMH cells but the autophagic reaction could not sufficiently suppress intracellular bacteria. We found IONzymes effectively reduced the viability of intracellular S. Enteritidis. This antibacterial reaction can be ascribed to co-localization of S. Enteritidis and IONzymes in autophagic vacuoles. Specifically, the acidic microenvironment favored the peroxidase-like activity of IONzymes to potentiate ROS generation for bacterial killing. Importantly, oral administrating IONzymes to chickens effectively prevented the invasion of S. Enteritidis in livers. Comparative transcriptomic analysis also identified the effects of IONzymes on genes related to hepatic oxidation-reduction and autophagy in S. Enteritidis infected chickens. In addition, IONzymes has high biocompatibility and environmental safety and can be easily prepared at large scale with low cost. Therefore, IONzymes may be a potential antibiotic alternative in the control of S. Enteritidis infection in clinical therapy and poultry industry. Nevertheless, this method maybe more suitable for acidic environments, when in the most cases of neutral environment, this method may be invalid. As we found that ROS was the main force of controlling S. Enteritidis infection, while IONzymes showed peroxidase-like activity in acidic environments but catalase-like activity in neutral environment.
Methods
Chemicals and antibodies
FeCl3•6H2O, ethylene glycol, sodium acetate (NaAc), H2O2 (30%), 2',7'-dichlorofluorescin diacetate (H2DCFDA) , and 3,3',5,5'-tetramethylbenzidine (TMB) were purchased from Sigma-Aldrich. Dulbecco's modified Eagle medium (DMEM) was purchased from Hyclone Co. Ltd. Fetal bovine serum (FBS) was from Gibco Co., Ltd. Penicillin-Streptomycin (100×) and gentamicin were from Beijing Solarbio Science & Technology Co., Ltd. Xylose lysine deocycholate (XLD) agar was from Qingdao-Hope Bio-Technology Co., Ltd. QIAprep spin miniprep kit was from QIAGEN Co. Ltd. Triton X-100, anti-LC3B antibody, rapamycin, bisBenzimide H33342 trihydrochloride were procured from Sigma-Aldrich Co. Ltd. Inc. Cell Counting Kit-8 (CCK-8) was from Dojindo Co., Ltd. RIPA Lysis Buffer, BCA Protein Assay kit and reactive oxygen species (ROS) detection kit were from Beyotime Biotechnology. ExpressPlus™ PAGE Gels was from GenScript Co., Ltd. Goat Anti-Rabbit IgG H&L (HRP) was from Abcam. SuperSignal West Pico Trial Kit was from Thermo Fisher Scientific Inc. Bovine serum albumin (BSA) was from Cell Signaling Technology, Inc.
Transcriptomic effect of IONzymes in chickens infected with S. Enteritidis. Chickens were infected with S. Enteritidis in the absence (SE) or presence (SI) of IONzymes. (A) Number of upregulated and downregulated differentially expressed genes (DEGs) in group SI compared with group SE. Main gene ontology (GO) categories, including biological process (B), molecular function (C) and cellular component (D), of the DEGs in group SI compared with group SE respectively. Green and red columns represent downregulated and upregulated GO terms respectively. Abscissa axis represents number of DEGs in each GO term. P value beside the columns represents enrichment significance of DEGs. (E) Heatmap of DEGs in the oxidation reduction and autophagy pathways between group SI and group SE. Rows indicate gene expression of DEGs in the oxidation reduction and autophagy pathways; columns represent individual samples from two groups. Red represents upregulation, black indicates insignificant change, and green represents downregulated expression.
Culture of LMH cells
LMH cells were purchased from the North Carolina Souren Biotechnology Co., Ltd. (BNCC337930) (Suzhou, Jiangsu, China) and cultured in DMEM media containing 10% FBS and Penicillin-Streptomycin (100 units/mL penicillin and 100 µg/mL streptomycin ) at 37 ℃ in a humidified incubator containing 5% CO2.
Culture of Salmonella enterica serovar Enteritidis (S. Enteritidis)
S. Enteritidis strain SC070, an isolate collected from broilers in China, was obtained as a gift from an Associate Research Fellow, Dr. J. Gong at Poultry Institute of Chinese Academy of Agricultural Sciences [50]. A single colony was inoculated into 5 mL advanced Martin broth and incubated at 37 ℃ with shaking at 220 rpm for 6 h, and then diluted in 250 mL advanced Martin broth for overnight growth at 37 ℃ with shaking. The bacterial culture was centrifuged at 8000 rpm for 10 min to pellet cells. The bacterial pellets were washed twice with PBS, and diluted with DMEM containing 10% FBS to the desired density, which was confirmed by plating and colony counting on XLD agar.
Preparation and characterizations of IONzymes
IONzymes were prepared as described previously [32,41-43]. The size distribution and morphology of IONzymes were characterized by transmission electron microscopy (TEM; Philips CM100, 80 kV), scanning electron microscope (SEM; Philips XL-30 field, 15 kV), X-ray diffractometer (XRD, D8 Advance, Bruker AXS, Germany) and particle analyze (ES90 Nano, Malvern, UK). The peroxidase-like activity of IONzymes was examined in a mixture of NaAc buffer (0.1M, pH 4.5) containing IONzymes and different concentrations of H2O2 or chromogenic substrate, 3,3',5,5'-tetramethylbenzidine (TMB) for kinetics assay. The absorbance (652 nm) changes were monitored and calculated according to the molar concentration changes of TMB by using a molar absorption coefficient of 39000 M-1 cm-1 for TMB-derived oxidation products according to the Beer-Lambert law. When H2O2 concentration was variable, the reactions were conducted with variable H2O2 amounts in NaAc buffer (0.1M, pH 4.5) containing 10 µg/mL of IONzymes and 833.33 μM TMB. When TMB concentration was variable, the reactions were conducted with variable TMB amounts in NaAc buffer (0.1 M, pH 4.5) containing10 µg/mL of IONzymes and 148.46 mM H2O2. The Michaelis-Menten constant was calculated using Lineweaver-Burk plots of the double reciprocal of the Michaelis-Menten equation, ν=Vmax×[S]/(Km+[S]) by GraphPad Prism (GraphPad Prism 7, GraphPad Software, USA), where ν is the initial velocity, Vmax is the maximal reaction velocity, [S] is the substrate concentration and Km is the Michaelis-Menten constant. Kcat is calculated by Vmax/[IONzyme], where [IONzyme] is the molar concentration converted from nanoparticle number in the reaction volume. IONzymes were sterilized at 121 ℃ for 30 min and were subsequently suspended in sterile ddH2O to a concentration of 10 mg/mL as a stock solution. The sterilized IONzymes stock solution was sonicated 10 min before diluting to desired concentrations for enzymatic assays and the following antibacterial experiments.
Effect of IONzymes on planktonic S. Enteritidis
The growth phase of bacteria was monitored by measuring the optical density (OD) at 600 nm and the value of OD600 reached 1.0 ( ~ 109 CFU/mL) within 4 h. Then, 0.1M sodium acetate (NaAc, pH 4.5) was used to dilute the bacterial solution to a concentration of 106 CFU/mL for antibacterial test. The IONzyme stock solution was diluted at varied concentrations ranging from 0.25 to 0.5 mg/mL and added at equal volume to the above bacterial solution for antimicrobial test. The combination treatment was performed by adding H2O2 at the final concentration of 0.05% to IONzyme regimen. A control group with only 0.1 M NaAc (pH 4.5) was examined at the same time. After incubated for 30 min, the mixture was 10-times gradient diluted, and 100 μL of the diluted solution was cultured on an XLD agar plate at 37 ℃ for 24 h before counting the colony-forming units. For morphology characterization, the bacteria was first collected right after the IONzymes treatment and then was dehydrated with ethanol. Morphological characteristics of S. Enteritidis was determined with scanning electron microscope (SEM, Hitachi S-4800).
Construction of Green Fluorescent Protein (GFP) Labeling of S. Enteritidis
pGFPmut3.1 which contains the GFPmut3.1 variant of the Aequorea victoria GFP and anti-ampicillin protein (Ampr) in Escherichia coli was kindly provided by Dr. G. Zhu in Yangzhou University. The plasmids were extracted using a QIAprep plasmid spin miniprep kit and stored at -20 ℃. GFP-S. Enteritidis was prepared by introducing pGFPmut3.1 into the cultured S. Enteritidis using an electroporator (Eppendorf Eporator, Eppendorf AG, Germany) and selected by Luria broth (LB) agar plates containing ampicillin.
GFP-S. Enteritidis were cultivated continuously and observed under a fluorescence microscope (DM4000B, Leica, Germany). Then the GFP-S. Enteritidis were stored at -80 ℃ for the later experiments.
Exposure of IONzymes to S. Enteritidis-infected LMH cells
LMH cells were cultured at 37 ℃ in a humidified environment containing 5% CO2 until they reached about 70% confluence. LMH cells were washed twice with pre-warmed PBS and infected with approximately 5×107 CFU/mL of S. Enteritidis suspension in DMEM supplemented with 10% FBS. Each plate was then centrifuged at 1,000 × g for 5 min to promote the interaction of bacteria and host cells. Infected cells were incubated at 37 ℃ for 1 h and then treated with 200 μg/mL gentamicin for 30 min to eradicate extracellular bacteria [51]. Finally infected-LMH cells were incubated with DMEM media containing 10 μg/mL gentamicin in the presence of desired concentrations of IONzymes for the remainder of the assay.
Effect of IONzymes on the growth of intracellular S. Enteritidis
LMH cells were infected with S. Enteritidis and exposed to different concentration of IONzymes (0, 50, 100, 200, and 400 μg/mL) described as above. Cells at 12, 18, and 24 h were lysed with 500 μL 0.5% (vol/vol) Triton X-100 in PBS for 10 min at 42 ℃. CFUs were enumerated by plating aliquots of 10-fold serial dilutions of the lysates onto XLD agar plates.
Cell viability assay
LMH cell viability was assessed with CCK-8 kit according to the instruction from the manufacturer. Briefly, LMH cells were infected with S. Enteritidis and exposure to different concentration of IONzymes (0, 50, 100, 200, and 400 μg/mL) as above. CCK-8 solution (10 μL) was added to each well at 6, 12, 18, 24 h, followed by incubation at 37 ℃ for 4 h. The absorbance value (A) was measured at 450 nm with a microplate reader (Infinite M200 Pro, Tecan, Switzerland). Relative cell viability was expressed as a relative percentage of control (solvent treated cells) according to the following formula: Relative cell viability (%) = [(Asample - Ablank)/(Acontrol - Ablank)] × 100.
Transmission electron microscopy (TEM) analysis
LMH cells or liver samples were fixed in 2.5% glutaraldehyde for at least 2 h followed by 1% osmium tetroxide fixation for 2 h. The samples were then gradient dehydrated in a series of ethanol washes, and were embedded in Epon 812 epoxy resin. Ultra-thin sections (about 70 nm) were cut by a ultramicrotome (EM UC6, Leica) and double stained with uranyl acetate and citromalic acid lead. Then these samples were observed using a CM100 transmission electron microscope (Philips) at 80 kV.
Western blot analysis
Cellular and liver (12h after the S. Enteritidis challenge) proteins were extracted using RIPA Lysis Buffer. After complete lysis of samples, the supernatant was collected after centrifugation at 12,000 × g for 10 min at 4 ℃. Protein concentrations were quantified using a BCA Protein Assay kit. Protein (30 μg) extract from each sample was denatured at 100 ℃ for 4 min, and subjected to SDS-PAGE electrophoresis using 12% ExpressPlus™ PAGE Gels. The separated proteins were transferred to polyvinylidene fluoride membranes, which were blocked in 5% nonfat milk in Tris-buffer-saline with Tween-20 (TBST) buffer for 1 h. The membranes were incubated at 37 ℃ either with anti-LC3B antibody at 1:850 dilution for 1 h. The membranes were washed and incubated at 37 ℃ with Goat Anti-Rabbit IgG H&L (HRP) at 1:8000 dilution for 45 min. Protein signals were detected by using a SuperSignal West Pico Trial Kit, and signals were captured with an gel imaging System (Bio-Rad ChemidocTM XRS+, USA) and then analyzed with an Image Lab software (Bio-Rad, USA).
Effect of rapamycin (RAPA) on the number of intracellular S. Enteritidis
LMH cells were pretreated with 100 nM rapamycin or vehicle media (control) for 1 h, followed by infection with S. Enteritidis and analyzed intracellular S. Enteritidis number at 12 h as above.
Immunofluorescence (IF) Staining
LMH cells growing on the glass slides in 24-well plates were infected with 5×107 CFU/mL of GFP-S. Enteritidis and exposure to 100 μg/mL IONzymes as above. Cells at 12 h were fixed with 4% paraformaldehyde, and then incubated with 0.25% Triton X-100 for 10 min. After washing with PBS, cells were incubated in a blocking buffer containing 1% BSA in PBST for 1 h at room temperature, followed by addition of a solution containing the primary antibody LC3 at a dilution of 1:200. After 1 h at 37 ℃, the plates were washed and treated with Goat Anti-Rabbit IgG H&L (Alexa Fluor® 647) at 1:600 dilution for 1 h. Then bisBenzimide H33342 trihydrochloride was used to stain nuclei of cells. The cells were visualized by a laser confocal microscopy Leica TCS SP8 (Leica Inc., Solms, Germany).
Determination of reactive oxygen species (ROS)
ROS levels were determined with a ROS detection kit by measuring the oxidative conversion of cell permeable 2',7'-dichlorofluorescein diacetate (DCFH-DA) to fluorescent dichlorofluorescein (DCF) in a microplate reader (Infinite M200 Pro, Tecan, Switzerland). For in vitro test, LMH cells in 96-well plates infected or non-infected with S. Enteritidis were incubated with DCFH-DA (10 μM) for 20 min and washed three times with PBS. Then cells were exposed to 0 or 100 μg/mL IONzymes, and each treatment set 5 replicate wells. The DCF fluorescence was continuously monitored at an excitation wave length of 488 nm and at an emission wave length of 525 nm. For in vivo test, ROS was measured as described previously [52]. Briefly, the liver tissues were diluted 1:20 times with ice-cold PBS and homogenized. After centrifugation, 0.2 mL supernatant were taken and incubated with 0.01 mL DCFH-DA (5 mM) for 30 min at 37 ℃. The conversion of DCFH-DA to the fluorescent product DCF was measured using a spectrofluorimeter with excitation at 484 nm and emission at 530 nm as above. Background fluorescence (conversion of DCFH-DA in the absence of homogenate) was corrected by the inclusion of parallel blanks.
Animals and management
Three hundred and fifty of fertilized specific pathogen free (SPF) eggs were obtained from Beijing Merial Vital Laboratory Animal Technology Co., Ltd. After 21 days hatchery, 260 healthy SPF chickens were obtained and randomly divided into four groups, denoted as groups C, I, SE, and SI. At the age of 2, 4 and 6 days-old, birds in group I and group SI were orally gavaged IONzymes with a dose of 50 mg/kg, while those birds in group C and group SE received with same volume of sterile PBS. At day 7, birds in group SE and group SI were subcutaneous injected with 0.1 mL of S. Enteritidis (1×108 CFU/mL), while birds in group C and group I received the same volume of sterile PBS. The birds were reared in cages with a wire screen floor. Water and feed were provided ad libitum, with the photoperiod set at 24L throughout the study. The temperature in the broiler house during the first week was 32 to 35 ℃. This experiment was repeated twice independently. The study was conducted according to the Regulations of the Experimental Animal Administration issued by the State Committee of Science and Technology of the People's Republic of China. The animal use protocol was approved by the Animal Care and Use Committee of the Poultry Institute, Chinese Academy of Agriculture Science. The diet of the birds was formulated to meet or slightly exceed all nutrient requirements without antibiotics or anticoccidial drugs (NRC, 1994), which were prepared at the Poultry Institute, Chinese Academy of Agriculture Science and were negative for S. Enteritidis.
Collection of liver samples
At 24 and 48 h after the S. Enteritidis challenge, 15 birds of each group were randomly selected and killed by jugular bleeding. Livers in the left were removed, minced and snap-frozen in liquid nitrogen, and then stored at -80 ℃ until analysis. Part of the right livers was immediately fixed in a 10% formaldehyde solution for histopathological examination and 2.5% glutaraldehyde for TEM examination. The rest of livers were collected for intracellular salmonella counts and ROS level determination.
Enumeration of bacterial liver load from infected chicken
For bacterial enumeration in liver, pre-weighed and labelled 5-mL tubes were used to collect samples during necropsy, stored on ice. The samples were weighed, and homogenized in 3 mL of sterile PBS using SCIENTZ-48 homogeniser (NingBo Scientz Biotechnology Co., Ltd.). Viable bacterial counts in homogenates were determined by enumerating viable bacteria culture on XLD agar plates after incubating plates for overnight at 37 ℃.
Histopathology Examination
The histopathology of livers was examined according to the method reported previously [53].
RNA extraction, cDNA library construction and RNA-sequencing
Three biological replicates of liver samples (1.5 g each) of chickens in group SE and group SI at 24h after the S. Enteritidis challenge were selected for further transcriptomic analysis. Total RNA samples were extracted using the TRIzol reagent (Invitrogen) and then treated by RNase-free DNase I (Takara) to remove genomic DNA contamination. The quality of mRNA including purity, quantity and integrity was evaluated using Nanodrop, Qubit, and Agilent 2100 bioanalyzer. cDNA libraries were constructed according to the standard protocols provided by Illumina. Briefly, mRNA was fragmented using fragmentation buffer and first strand cDNA was synthesized using Dynabeads oligo (dT) (Dynal; Invitrogen). Second-strand cDNA was synthesized using reverse transcriptase (Superscript II; Invitrogen) and random hexamer primers, and further purified using AMPure XP beads (Beckman-Coulter). Finally, the purified double-stranded cDNA samples were further enriched by PCR to construct the final cDNA libraries that were sequenced using Hiseq 2500 (150 bp paired ends) by Novogene (China).
RNA-Seq Data Analysis
Adaptor sequences and low-quality sequences were removed from the raw reads (Q < 20). The clean reads obtained after data filtering were mapped to the chicken reference genome (ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF_000002315.4_Gallus_gallus-5.0/GCF_000002315.4_Gallus_gallus-5.0_genomic.fna.gz) using TopHat software. The mapped reads were used for further transcript annotation and for calculating the expression level using the FPKM (fragments per kilobase per million reads) method [54]. Differential expression analysis of group SE and group SI was performed using the DESeq R package (1.18.0). Genes with an adjusted P-value <0.05 and fold change > 1.5 found by DESeq were assigned as differentially expressed genes (DEGs). Gene Ontology (GO) enrichment analysis of differentially expressed genes was implemented by the GOseq R package, in which gene length bias was corrected. GO terms with corrected P-value less than 0.05 were considered significantly enriched by DEGs.
Statistical Analysis
All experiments were performed at least three times and results are expressed as the Mean ± standard error of the mean (SEM). Statistical significance was evaluated using an independent-sample T-test or One-way ANOVA test with SPSS 19.0. P < 0.05 was considered statistically significant in all analyses (95% confidence level).
Abbreviations
S. Enteritidis: Salmonella Enteritidis; IONzymes: iron oxide nanozymes; SPF: specific pathogen free; ROS: reactive oxygen species; TMB: 3,3',5,5'-tetramethyl benzidine; H2O2: hydrogen peroxide; XRD: X-ray diffraction; EDS: energy dispersive spectrometry; NaAc: sodium acetate; DLS: dynamic light scattering; CFU: colony forming units; GFP: green fluorescent protein; RAPA: rapamycin; TEM: transmission electron microscopy; IF: immunofluorescence; DMEM: Dulbecco's modified Eagle medium; FBS: fetal bovine serum; XLD: xylose lysine deocycholate; CCK-8: cell counting Kit-8; BSA: bovine serum albumin; SEM: scanning electron microscope; Ampr: anti-ampicillin protein; DCFH-DA: 2',7'-dichlorofluorescein diacetate; DCF: fluorescent dichlorofluorescein; DEGs: differentially expressed genes; GO: Gene Ontology.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (31702132), Innovation Capacity Building Program of Jiangsu Province (BM2018026), “Three Renovations” Agricultural Project of Jiangsu Province (SXGC [2017]253, SXGC[2017]254), the Special Fund for Agro- scientific Research in the Public Interest (No.201403047), National key R & D Program of China (2017YFD0500506). The authors acknowledge Dr. Chunhong Zhu (Poultry Institute, Chinese Academy of Agricultural Sciences) for her help in animal S. Enteritidis infection and providing green fluorescent protein (pGFPmut3.1), Dr. Ruilong Song (Yangzhou University) for his help in immunofluorescence determination. Dr. Yan Lu (Yangzhou University) for her help in transmission electron microscopy determination. All animal experiments were approved by the Animal Care and Use Committee of the Poultry Institute, Chinese Academy of Agriculture Science, according to the Regulations of the Experimental Animal Administration issued by the State Committee of Science and Technology of the People's Republic of China.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hannemann S, Galán JE. Salmonella enterica serovar-specific transcriptional reprogramming of infected cells. PLoS Pathog. 2017;13:e1006532
2. WHO. Salmonella (non-typhoidal). http://www.who.int/mediacentre/factsheets/fs139/en/.
3. Martínez-Moya M, de Pedro MA, Schwarz H, Garcfa-del Portillo F. Inhibition of Salmonella intracellular proliferation by non-phagocytic eucaryotic cells. Res Microbiol. 1998;149:309-18
4. Niedergang F, Sirard JC, Blanc CT, Kraehenbuhl JP. Entry and survival of Salmonella typhimurium in dendritic cells and presentation of recombinant antigens do not require macrophage-specific virulence factors. Proc Natl Acad Sci U S A. 2000;97:14650-5
5. Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008;6:53-66
6. Panzarini E, Inguscio V, Tenuzzo BA, Carata E, Dini L. Nanomaterials and autophagy: new insights in cancer treatment. Cancers (Basel). 2013;5:296-319
7. Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323-35
8. Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722-37
9. Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T. et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037-40
10. Birmingham CL, Cacadien V, Gouin E, Troy EB, Yoshimori T, Cossart P. et al. Listeria monocytogenes evades killing by autophagy during colonization of host cells. Autophagy. 2007;3:442-51
11. Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374-83
12. Fujita N, Saitoh T, Kageyama S, Akira S, Noda T, Yoshimori T. Differential involvement of Atg16L1 in Crohn disease and canonical autophagy: analysis of the organization of the Atg16L1 complex in fibroblasts. J Biol Chem. 2009;284:32602-9
13. Conway KL, Kuballa P, Song JH, Patel KK, Castoreno AB, Yilmaz OH. et al. Atg16L1 is required for autophagy in intestinal epithelial cells and protection of mice from Salmonella infection. Gastroenterology. 2013;145:1347-57
14. Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS. et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512-6
15. Wei H, Wang E. Nanomaterials with enzyme-like characteristics (nanozymes): next-generation artificial enzymes. Chem Soc Rev. 2013;42:6060-93
16. He W, Wamer W, Xia Q, Yin JJ, Fu PP. Enzyme-like activity of nanomaterials. J Environ Sci Heal C. 2014;32:186-211
17. Ragg R, Tahir MN, Tremel W. Solids Go Bio: inorganic nanoparticles as enzyme mimics. Eur J Inorg Chem. 2016;13:1896-1896
18. Gao L, Yan X. Nanozymes: an emerging field bridging nanotechnology and biology. Sci China Life Sci. 2016;59:400-2
19. Wang XY, Guo WJ, Hu YH, Wu JJ, Wei H. Nanozymes: next wave of artificial enzymes. Berlin, Germany: Springer-Verlag Berlin and Heidelberg GmbH & Co. KG. 2016
20. Wang XY, Hu YH, Wei H. Nanozymes in bionanotechnology: from sensing to therapeutics and beyond. Inorg Chem Front. 2016;3:41-60
21. Zhou Y, Liu B, Yang R, Liu J. Filling in the gaps between nanozymes and enzymes: challenges and opportunities. Bioconjug Chem. 2017;28:2903-9
22. Chen Z, Yin JJ, Zhou YT, Zhang Y, Song L, Song M. et al. Dual enzyme-like activities of iron oxide nanoparticles and their implication for diminishing cytotoxicity. ACS Nano. 2012;6:4001-12
23. Wei H, Wang E. Fe3O4 magnetic nanoparticles as peroxidase mimetics and their applications in H2O2 and glucose detection. Anal Chem. 2008;80:2250-4
24. Wei H, Wang E. Nanomaterials with enzyme-like characteristics (nanozymes): next-generation artificial enzymes. Chem Soc Rev. 2013;42:6060-93
25. Huang DM, Hsiao JK, Chen YC, Chien LY, Yao M, Chen YK. et al. The promotion of human mesenchymal stem cell proliferation by superparamagnetic iron oxide nanoparticles. Biomaterials. 2009;30:3645-51
26. Wang XQ, Tu Q, Zhao B, An YF, Wang JC, Liu WM. et al. Effects of poly (L-lysine)-modified Fe3O4 nanoparticles on endogenous reactive oxygen species in cancer stem cells. Biomaterials. 2013;34:1155-69
27. Zhang L, Wang XQ, Miao YM, Chen ZQ, Qiang PF, Cui LQ. et al. Magnetic ferroferric oxide nanoparticles induce vascular endothelial cell dysfunction and inflammation by disturbing autophagy. J Hazard Mater. 2016;304:186-95
28. Shi M, Cheng L, Zhang Z, Liu Z, Mao XL. Ferroferric oxide nanoparticles induce prosurvival autophagy in human blood cells by modulating the Beclin 1/Bcl-2/VPS34 complex. Int J Nanomed. 2015;10:207-16
29. Zhang D, Zhao YX, Gao YJ, Gao FP, Fan YS, Li XJ. et al. Anti-bacterial and in vivo tumor treatment by reactive oxygen species generated by magnetic nanoparticles. J Mater Chem B. 2013;1:5100-7
30. Pan WY, Huang CC, Lin TT, Hu HY, Lin WC, Li MJ. et al. Synergistic antibacterial effects of localized heat and oxidative stress caused by hydroxyl radicals mediated by graphene/iron oxide-based nanocomposites. Nanomedicine. 2016;12:431-8
31. Gao LZ, Liu Y, Kim D, Li Y, Hwang G, Naha PC. et al. Nanocatalysts promote Streptococcus mutans biofilm matrix degradation and enhance bacterial killing to suppress dental caries in vivo. Biomaterials. 2016;101:272-84
32. Gao LZ, Zhuang J, Nie L, Zhang JB, Zhang Y, Gu N. et al. Intrinsic peroxidase-like activity of ferromagnetic nanoparticles. Nat Nanotechnol. 2007;2:577-83
33. Gao LZ, Giglio KM, Nelson JL, Sondermann H, Travis AJ. Ferromagnetic nanoparticles with peroxidase-like activity enhance the cleavage of biological macromolecules for biofilm elimination. Nanoscale. 2014;6:2588-93
34. Gao LZ, Fan KL, Yan XY. Iron Oxide Nanozyme: a multifunctional enzyme mimetic for biomedical applications. Theranostics. 2017;7:3207-27
35. Kuehn BM. Salmonella cases traced to egg producers: findings trigger recall of more than 500 million eggs. JAMA. 2010;304:1316
36. Ong SY, Ng FL, Badai SS, Yuryev A, Alam M. Analysis and construction of pathogenicity island regulatory pathways in Salmonella enterica serovar Typhi. J Integr Bioinform. 2010;7:145
37. Chai SJ, White PL, Lathrop SL, Solghan SM, Medus C, McGlinchey BM. et al. Salmonella enterica serotype Enteritidis: increasing incidence of domestically acquired infections. Clin Infect Dis. 2012;54:S488-497
38. Jackson BR, Griffin PM, Cole D, Walsh KA, Chai SJ. Outbreak-associated Salmonella enterica serotypes and food commodities, United States, 1998-2008. Emerg Infect Dis. 2013;19:1239-44
39. Shah DH, Zhou XH, Kim HY, Call DR, Guar J. Transposon mutagenesis of Salmonella enterica serovar Enteritidis identifies genes that contribute to invasiveness in human and chicken cells and survival in egg albumen. Infect Immun. 2012;80:4203-15
40. Wang CL, Fan YC, Wang CL, Tsai HJ, Chou CH. The impact of Salmonella Enteritidis on lipid accumulation in chicken hepatocytes. Avian Pathol. 2016;45:450-457
41. Zeng Y, Hao R, Xing B, Hou YL, Xu ZC. One-pot synthesis of Fe3O4 nanoprisms with controlled electrochemical properties. Chem Commun (Camb). 2010;46:3920-2
42. Yang C, Wu J, Hou Y. Fe3O4 nanostructures: synthesis, growth mechanism, properties and applications. Chem Commun (Camb). 2011;47:5130-41
43. Sun SH, Zeng H, Robinson DB, Raoux S, Rice PM, Wang SX. et al. Monodisperse MFe2O4 (M = Fe, Co, Mn) nanoparticles. J Am Chem Soc. 2004;126:273-9
44. Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313-26
45. Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720-8
46. Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, Magalhaes MA. et al. Activation of antibacterial autophagy by NADPH oxidases. Proceedings of the national academy of sciences. 2009;106:6226-6231
47. Zanella D, Bossi E, Gornati R, Bastos C, Faria N, Bernardini G. Iron oxide nanoparticles can cross plasma membranes. Sci Rep. 2017;7:11413-23
48. Hendriksen PJM, Freidig AP, Jonker D, Thissen U, Bogaards JJP, Mumtaz MM. et al. Transcriptomics analysis of interactive effects of benzene, trichloroethylene and methyl mercury within binary and ternary mixtures on the liver and kidney following subchronic exposure in the rat. Toxicol Appl Pharmacol. 2007;225:171-88
49. Shi SR, Shen YR, Zhao ZH, Hou ZC, Yang Y, Zhou HJ. et al. Mol. Integrative analysis of transcriptomic and metabolomic profiling of ascites syndrome in broiler chickens induced by low temperature. Mol BioSyst. 2014;10:2984-93
50. Gong JS, Wang CM, Shi SR, Bao HD, Zhu CH, Kelly P. et al. Highly drug-resistant Salmonella enterica Serovar Indiana clinical isolates recovered from broilers and poultry workers with diarrhea in China. Antimicrob Agents Chemother. 2016;60:1943-47
51. Liu CM, Ma JQ, Sun YZ. Puerarin protects the rat liver against oxidative stress-mediated DNA damage and apoptosis induced by lead. Exp Toxicol Pathol. 2012;64:575-82
52. Shah DH, Zhou XH, Kim HY, Call DR, Guar J. Transposon mutagenesis of Salmonella enterica serovar Enteritidis identifies genes that contribute to invasiveness in human and chicken cells and survival in egg albumen. Infect Immun. 2012;80:4203-15
53. Desmidt M, Ducatelle R, Haesebrouck F. Pathogenesis of Salmonella enteritidis phage type four after experimental infection of young chickens. Vet Microbiol. 1997;56:99-109
54. Anders S, Huber W. Differential expression of RNA-Seq data at the gene level-the DESeq package. Heidelberg, Germany: European Molecular Biology Laboratory. 2012
Author contact
![]() Corresponding authors: ssr236com (Shourong Shi), Email: lzgaoedu.cn (Lizeng Gao)
Corresponding authors: ssr236com (Shourong Shi), Email: lzgaoedu.cn (Lizeng Gao)
 Global reach, higher impact
Global reach, higher impact