13.3
Impact Factor
Theranostics 2018; 8(15):4016-4032. doi:10.7150/thno.25958 This issue Cite
Review
Aptamers in the Therapeutics and Diagnostics Pipelines
1. AstraZeneca USA.
2. Operational Technologies Corporation, 4100 NW Loop 410, Suite 100, San Antonio, Texas 78229, USA.
3. Discipline of Biosciences and Biomedical Engineering, Indian Institute of Technology Indore, Simrol, Indore, 453552, India.
4. Center for Biodesign and Diagnostics, Translational Health Science and Technology Institute (THSTI), Faridabad-121001, Haryana, India.
Received 2018-3-8; Accepted 2018-5-16; Published 2018-7-1
Abstract
Aptamers are short single-stranded DNA or RNA oligonucleotides that can selectively bind to small molecular ligands or protein targets with high affinity and specificity, by acquiring unique three-dimensional structures. Aptamers have the advantage of being highly specific, relatively small in size, non-immunogenic and can be easily stabilized by chemical modifications, thus allowing expansion of their diagnostic and therapeutic potential. Since the invention of aptamers in the early 1990s, great efforts have been made to make them clinically relevant for diseases like macular degeneration, cancer, thrombosis and inflammatory diseases. Furthermore, owing to the aforementioned advantages and unique adaptability of aptamers to point-of-care platforms, aptamer technology has created a stable niche in the field of in vitro diagnostics by enhancing the speed and accuracy of diagnoses. The aim of this review is to give an overview on aptamers, highlight the inherent therapeutic and diagnostic opportunities and challenges associated with them and present various aptamers that have reached therapeutic clinical trials, diagnostic markets or that have immediate translational potential for therapeutics and diagnostics applications.
Keywords: aptamers, therapeutics, diagnostics, clinical trials, clinical diagnostics
Introduction
Aptamers are single-stranded DNA or RNA oligonucleotides that bind with high affinity and high specificity to various targets ranging from various ions, small organic compounds to large proteins and live cells [1-8]. Possessing a specific and stable three-dimensional shape both in vitro and in vivo, aptamers are able to recognize their targets with high binding affinity and selectivity, and are thus termed “chemical antibodies” [9, 10]. Aptamers are selected from large libraries of random oligonucleotides that can contain up to 1016 unique sequences. The process of aptamer selection, termed “Systematic Evolution of Ligands by EXponential Enrichment” (SELEX), was first developed in 1990 by Tuerk and Gold [11]. SELEX is an iterative process that starts with a randomized library of oligonucleotides being incubated with the target molecule and involves iterative rounds of affinity selection and PCR amplification steps to screen for the highest affinity sequences. To date, several aptamers with high affinity and specificity have been identified for various targets with promising therapeutic and diagnostic potential using the SELEX method [12, 13]. Aptamers are considered to be strong chemical rivals of antibodies due to their inherent advantages over antibodies. Compared to antibodies: (i) aptamers can be produced using cell-free chemical synthesis and are therefore less expensive to manufacture on the mg scale, (ii) aptamers exhibit extremely low variability between batches and have better controlled post-production modification, (iii) are minimally immunogenic, and (iv) are small in size [14]. The rapidly growing aptamer industry is predicted to reach US $244.93 million by 2020 [13, 15, 16]. Presently around 40 aptamer-based companies are actively engaged in diagnostics and therapeutics research to commercialize aptamers globally [13, 15, 16]. These companies have a long and robust pipeline to address various unmet therapeutic and diagnostic needs. Some of the aptamer products are already commercially available while other products have immediate translational potential and may hit the market in the next 5-10 years. In recent years, several attempts have been made to summarize the existing information about therapeutic aptamers [17-19]. However, no attempt has been made to review the theranostic potential (combined therapeutics and diagnostics potential) of aptamers considering their current status and existing and future pipelines. Therefore, to bridge this knowledge or information gap, the current article highlights the theranostic potential of aptamers with existing therapeutics and diagnostics pipelines along with their respective bottlenecks. We herein summarize various aptamers that have already been commercialized or have the potential to be commercialized in the coming years.
Aptamers as therapeutic agents
The first SELEX experiment was carried out by Tuerk and Gold in 1990, who were first to identify an RNA aptamer against bacteriophage T4 DNA polymerase [11]. It has been nearly 3 decades since the discovery of the first aptamer and so far, only one aptamer (Macugen/Pegaptanib sodium) has received US FDA regulatory approval and is available to patients. The therapeutic use of aptamers is limited by their susceptibility to ubiquitous serum nucleases, rapid excretion by renal filtration and insufficient in vivo binding affinity for their target [21, 22]. The stability problem has been largely addressed by developing aptamers with chemical modifications to the nucleic acid backbone or sugars as well as the 3' and 5' ends. Different chemical and structural modification strategies have been adopted that are introduced during SELEX or post-SELEX to overcome these limitations and to make aptamers suitable for different biological applications [1-4, 23, 24]. Among these modifications, 2'-fluoro, 2'-amino or 2'-O-methyl-substitutions, introduction of locked nucleic acid (LNA) or phosphorothioate linkages (PS-linkages) and 3'- end capping with inverted thymidine or other blocking molecules are some common chemical modifications introduced in aptamers for resisting nuclease degradation [25-27]. Another strategy to generate biostable aptamers is represented by the Spiegelmer technology. Spiegelmers are essentially L-RNA (levorotatory RNA) built from L-ribose units (L-deoxyribose units in the case of DNA) and are non-superimposable mirror images of the natural dextrorotatory forms (D-forms) of the nucleic acid nucleotide monomers. Due to their mirror image nature, Spiegelmers display high resistance to nuclease degradation and retain their affinity for their cognate targets [28]. In addition, aptamer stem-loop structural modification strategy has been adopted to improve the binding affinity of aptamers for their targets [29]. Chemical conjugation with high molecular weight polyethylene glycol (PEG) or proteins helps to reduce the renal filtration rate and improve aptamer retention in vivo [12, 30, 31]. This conjugation strategy not only retards their renal elimination but also extends aptamer half-life up to 24-48 hours, thus improving the pharmacokinetic properties of aptamers. Some aptamers are developed with the intent of inhibiting the activity of their target proteins. However, several aptamers have been developed that use other mechanisms of action besides antagonistic interactions to exhibit their biological effects. Yunn et al. has developed an agonistic insulin receptor (IR) aptamer that binds specifically to the allosteric site of the IR and stimulates metabolic activity of the receptor for glucose uptake without proliferation of cancer cells [20,23]. In addition, aptamers have been used for targeted drug delivery applications. Sgc8 DNA aptamer, which targets protein tyrosine kinase-7 (PTK-7), has been conjugated to the chemotherapeutic drug daunorubicin (Dau) to specifically deliver and internalize Dau to leukemic T-cells resulting in less cytotoxic effects of the drug on non-cancerous healthy cells [24].
Aptamers in the therapeutic pipeline
Aptamers are promising drug candidates and are being increasingly employed in all phases of the drug discovery and development process. To date, eleven aptamers have successfully entered clinical trials for different therapeutic indications (Table 1). This section will summarize the details of aptamers in clinical studies and their therapeutic potential.
Therapeutic aptamers for macular degeneration
Pegaptanib sodium/Macugen
Vascular endothelial growth factor (VEGF) is a mitogenic protein that plays a critical role in pathological processes such as angiogenesis (the formation of new blood vessels) and increased permeability (leakage from blood vessels) that are responsible for the vision loss associated with neovascular age-related macular degeneration (AMD) [32]. Pegaptanib is a pegylated 27 nucleotide RNA aptamer that specifically binds to the predominant 165 isoform of VEGF, blocks its interaction with the cognate receptor, and ultimately stops intraocular blood vessel growth. The aptamer has been chemically modified with 2'-fluoropyrimidines, 2'-O-methyl purines, a 3′-linked deoxythymidine residue and conjugated to 40 kDa polyethylene glycol (PEG) to improve its nuclease resistance, inhibitory action and plasma half-life in vivo. Pegaptanib is the first therapeutic aptamer approved by the US FDA in 2004 as an anti-angiogenic medicine for the treatment of neovascular (wet) age-related macular degeneration (AMD). Pegaptanib is administered by intravitreal injection into the eyes in a 0.3 mg dose once every six weeks [33]. Several clinical studies have been conducted on pegaptanib for different disease conditions such as AMD, diabetic macular edema and proliferative diabetic retinopathy (Table 1). Pegaptanib is currently approved in the United States, Europe, Canada, Brazil and Australia.
Aptamers in different stages of clinical trials.
| Aptamer | Company | Target | Equilibrium dissociation constant (Kd) | Indication | References a |
|---|---|---|---|---|---|
| Pegaptanib sodium/Macugen | OSI Pharmaceuticals/ Pfizer | VEGF165 | 200 pM | - AMD (US FDA-approved for this indication) - Proliferative diabetic retinopathy - Diabetic macular edema | Ruckman, et al., 1998 [12] [14]; U.S National Library of Medicine, Clinical trials.gov Identifier: NCT00460408 (C), NCT01245387 (C), NCT00460408 (C), NCT01281098 (C), NCT00040313 (C), NCT01640171 (C) |
| E10030 | Ophthotech | PDGF | 0.1 nM | - AMD - Von Hippel-Lindau | Floege, et al., 1999 [36]]; U.S National Library of Medicine, Clinical trials.gov Identifier: NCT00569140 (C), NCT01089517 (C), NCT01944839 (T), NCT01940900 (T), NCT01940887 (T), NCT02214628 (T), NCT02591914 (U), NCT02859441 (R) |
| ARC1905 | Ophthotech | Human complement C5 | 2-5 nM | - AMD - Wet AMD - Stargardt disease 1 - Idiopathic polypoidal choroidal vasculopathy - Geographic atrophy conditions | Biesecker, et al. 1999[39] ; U.S National Library of Medicine, Clinical trials.gov Identifier: NCT00950638 (C), NCT00709527 (C), NCT03362190 (R), NCT03374670, NCT02686658 |
| AS1411 (AGRO100) | Antisoma | Nucleolin | 54.8 ± 7.3 nM (Huh7 hepatocarcinoma cells) | - Acute myeloid leukemia (AML) - Metastatic renal cell carcinoma - Advanced solid tumors - Leukemia, myeloid | U.S National Library of Medicine, Clinical trials.gov Identifier: NCT01034410 (T), CT00740441 (U), NCT00881244 (C) and NCT00512083 (C) |
| NOX-A12 | NOXXON PHARMA | CXCL12 (C-X-C Chemokine Ligand 12) | 200 pM | - Multiple myeloma - Hematopoietic stem cell transplantation - Autologous stem cell transplantation - Metastatic colorectal cancer - Metastatic pancreatic cancer - Chronic lymphocytic leukemia | Roccaro, et al., 2014 [48] ; U.S National Library of Medicine, Clinical trials.gov Identifier: NCT01521533 (C), NCT00976378 (C), NCT00976378 (C), NCT03168139 (R) and NCT01486797 (C) |
| NOX-E36 | NOXXON PHARMA | CCL2 (C-C Chemokine Ligand 2)/ MCP-1 (monocyte chemoattractant protein 1) | 1.32 nM | - Type 2 diabetes mellitus - Systematic lupus erythematosus - Albuminuria - Renal impairment | Oberthür et al., 2015 [51]; U.S National Library of Medicine, Clinical trials.gov Identifier: NCT01547897 (C), NCT01085292 (C), NCT00976729 (C) and NCT01372124 (C) |
| NOX-H94 | NOXXON PHARMA | Hepcidin | 0.65 ± 0.06 nM | - Anaemia of chronic diseases - Anaemia - End-stage renal disease - Inflammation | Schwoebel, et al., 2013 [54] U.S National Library of Medicine, Clinical trials.gov Identifier: NCT01372137 (C), NCT02079896 (C), and NCT01522794 (C) |
| ARC1779 | Archemix | A1 domain of vWF | 2 nM | - von Willebrand disease - Thrombotic thrombocytopaenic purpura - von Willebrand disease type 2b - Percutaneous coronary intervention - Thrombosis | Diener et al., 2009 [57]; U.S National Library of Medicine, Clinical trials.gov Identifier: NCT00694785 (W), NCT00632242 (C), NCT00507338 (T), NCT00432770 (C) |
| NU172 | ARCA Biopharma | Thrombin | Not published | - Heart disease | U.S National Library of Medicine, Clinical trials.gov Identifier: NCT00808964 (U) |
| REG1 system | Regado Biosciences | Coagulation factor IXa (FIXa) | 2.83 ± 0.4 nM (RB006 aptamer) | - Acute coronary syndrome - Coronary artery disease | Rusconi, et al., 2002 [63]; U.S National Library of Medicine, Clinical trials.gov Identifier: NCT00932100 (C), NCT00715455 (C), NCT01848106 (T) |
| BX499 (formerly known as ARC19499) | Baxter | Tissue Factor Pathway Inhibitor (TFPI) | 2.8 ± 0.3 nM | - Hemophilia | Waters, et al., 2011[70] ; U.S National Library of Medicine, Clinical trials.gov Identifier: NCT01191372 (T) |
a Recruiting clinical trials are represented by “R”, terminated clinical trials are represented by “T”, completed clinical trials are represented by “C”, clinical trials with unknown status are represented by “U” and withdrawn clinical trials are represented by “W”.
E10030
Platelet-derived growth factor (PDGF) is a potent mitogen that plays a key role in the angiogenesis cascade, in particular for retinal vasculature. Different treatment strategies have been adopted to block PDGF activity in the angiogenesis process, thereby preventing ocular vascular diseases [34]. E10030 is a pegylated 29 nucleotide DNA aptamer obtained against platelet-derived growth factor B (PDGFB; PDGF2). E10030 was selected from a randomized DNA library using the in vitro selection method with PDGF as the target for AMD pathogenesis [35]. E100130 binds to PDGF with subnanomolar affinity and blocks the interaction of PDGF with the β subunit of its receptor. The selected aptamer sequence was chemically modified with 2'-fluoro or 2'-O-methyl-substitutions and a polyethylene glycol (PEG) moiety. In contrast to the unmodified sequence, the modified sequence exhibited no change in binding affinity and a 13-fold increase in half-life of the aptamer in plasma [36]. Upon combination with other anti-angiogenic therapies such as ranibizumab (a recombinant humanized IgG1 monoclonal antibody fragment that binds and inhibits VEGF-A), E00130 has shown promising therapeutic potential with 62% improvement in vision versus the ranibizumab alone group in a phase 2 randomized study in 449 patients with neovascular age-related macular degeneration [34] (U.S National Library of Medicine, Clinical trials.gov Identifier:NCT01089517). In a phase 3 trial, E10030 aptamer was administered in combination with Lucentis® compared to Lucentis® monotherapy in patients with acute-macular degeneration (AMD) (U.S National Library of Medicine, ClinicalTrials.gov Identifier: NCT01944839). The study was terminated with no further updates available.
ARC1905
Studies have demonstrated that the complement system (CS) plays an integral role in retinal biology and AMD pathogenesis [37, 38]. The complement system consists of proteins such as complement component 5 (C5) and the membrane attack complex (MAC) that switch on VEGF protein expression and thus promote neovascular AMD. ARC1905 is a pegylated 38 nucleotide RNA aptamer with 2'-fluoropyrimidines and 2'-O-methyl purines substitutions. ARC1905 specifically binds and inhibits the expression of the C5 component of human complement, which plays multiple roles in innate immunity and inflammatory diseases [39]. Inhibition of C5 activity further prevents the formation of other complementary proteins and the membrane attack complex (MAC), which initiates retinal cell lysis. By inhibiting these C5-mediated inflammatory and MAC activities, therapeutic benefit may be achieved in both dry and wet AMD. ARC1905 has undergone clinical studies for AMD, idiopathic polypoidal choroidal vasculopathy and geographic atrophy conditions (Table 1).
Therapeutic aptamers for oncology
AS1411
AS1411 is a 26-base guanine-rich oligonucleotide (GRO) with an unmodified (phosphodiester) DNA backbone. AS1411, previously named ARGO100, forms a G‑quadruplex structure and is the first aptamer in clinical trials for the treatment of human cancer [40-42]. The stable G-quadruplex structure imparts an unusual resistance to cellular and serum nucleases to the AS1411 aptamer. AS1411 primarily binds to nucleolin (NCL) protein, which is expressed at high levels on the surface of cancer cells, tumor-associated angiogenic endothelial cells and promotes VEGF gene expression, which is involved in blood vessel formation during tumorigenesis [43, 44]. Upon binding to cell surface nucleolin, AS1411 is internalized and may destabilize BCL2 mRNA, leading to a reduction in BCL2 protein synthesis and probably induction of apoptosis [45]. AS1411 has potential as an anticancer agent and has demonstrated anti-proliferative activity in various cancer cell lines [40]. The aptamer has been clinically evaluated for acute myeloid leukaemia (AML), metastatic renal cell carcinoma and advanced solid tumors and has shown promising results without causing major side effects. An extended Phase 1 trial in patients with renal cell carcinoma (RCC) and non-small cell lung carcinoma (NSCLC) did not show a significant tumor response with patients either sustaining the disease or showing progressive disease post-treatment [42]. No serious adverse events were observed with a 7-day infusion of AS1411 at 22 mg/kg/day, which achieved peak plasma concentrations in the range at which killing of cancer cells was observed in vitro. In a phase II single-arm study, AS1411 was administered at 40 mg/kg/day in patients with advanced renal cell carcinoma (RCC) who had failed tyrosine kinase inhibitor therapy [39]. However, no significant results were obtained from this study with low level activity and low toxicity observed in the patients.
NOX-A12
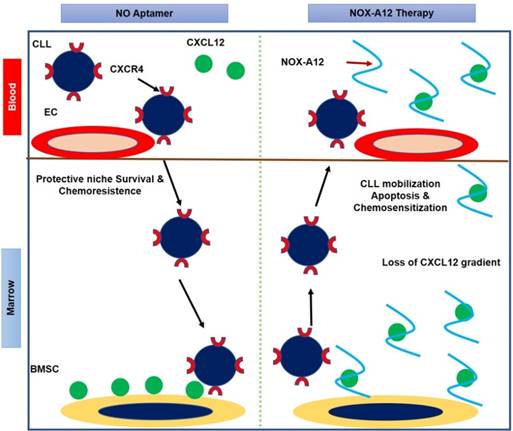
NOX‑A12 is a pegylated 45‑nucleotide L‑form RNA aptamer known as a Spiegelmer. Pegylated Spiegelmers are single-stranded RNA oligonucleotides (aptamers) in an L-configuration (i.e., the mirror image of naturally occurring RNA) containing a branched 40 kDa polyethylene glycol (PEG) [46]. NOX-A12 targets CXCL12/SDF-1 (C-X-C Chemokine Ligand 12/ Stromal Cell-Derived Factor-1), a key chemokine protein that plays an important role in tumor proliferation and promotes new blood vessel formation and metastasis. NOX-A12 disrupts CXCL12 activity and interferes with the tumor microenvironment to fight tumor proliferation and metastasis [47, 48]. In the first human clinical study, the drug was administered intravenously (i.v.) with a dose range of 0.05 mg/kg to 10.8 mg/kg in 48 healthy patients between 18 to 60 years of age. The drug was found to be safe and well-tolerated by patients and demonstrated effective and extended CXCL12 inhibition [49]. However, 27 adverse events were reported by 18 patients with no relation to the drug dose. A subsequent Phase 2a study of NOX-A12 given alone or in combination with bortezomib/ dexamethasone, was conducted in 28 patients with relapsed/refractory multiple myeloma [50]. No toxicity was observed and the drug was found to be safe and well-tolerated by patients. Additional confirmatory studies of NOX-A12 in combination with new or established drugs are required to explore its therapeutic potential. NOX-A12 is in clinical trials for multiple oncology indications including pancreatic, colorectal cancer and chronic lymphocytic leukaemia (CLL). NOX-A12 has received an orphan drug designation from the US FDA for glioblastoma treatment along with radiotherapy. Figure 1 provides a cartoon representation of the mechanism of action of the NOX-A12 aptamer.
Cartoon representation of NOX-A12 mechanism of action in chronic lymphocytic leukemia (CLL) as proposed by Marasca et al. [47]. NOX-A12 removes CXCL12 from the bone marrow stromal cell (BMSC) surface by neutralizing the chemokine, resulting in loss of CXCL12 gradient. NOX-A12 binding to CXCL12 mobilizes CLL cells from their native protective microenvironment, inducing apoptosis and chemo sensitization of leukemic cells. EC represents endothelial cell.
Therapeutic aptamer for diabetes
NOX-E36
NOX-E36 is a pegylated L-RNA aptamer (a Spiegelmer) that is 40 nucleotides long and binds with high affinity (Kd = 1.32 nM) to human chemokine CCL2, also referred to as MCP-1 (C-C Chemokine Ligand 2 / monocyte chemoattractant protein 1) [51]. CCL2/MCP-1 is one of the key chemokines that plays a crucial role in infiltration and positioning of monocytes. Overexpression of CCL2 and its receptor CCR2 is associated with a variety of inflammatory diseases, which makes CCL2 an interesting therapeutic target [52]. The strong binding of NOX-E36 to CCL2 is facilitated by a slow off rate, resulting in high complex stability that makes the blocking of CCL2 function even more efficient. NOX-E36 has successfully completed a Phase 2a clinical study in 76 patients with type 2 diabetes. The drug with a dose of 0.5 mg/kg was safe and well tolerated by patients, and results demonstrated the renoprotective potential of the drug [53]. NOX-E36 is being developed for diabetic nephropathy by NOXXON pharma.
Therapeutic aptamer for inflammation
NOX-H94
NOX-H94 is a 44-nucleotide L-RNA aptamer, referred as Spiegelmer that binds with high affinity to its target human hepcidin [54]. NOX-H94 is modified at its 5'end with a 40 kDa polyethylene glycol (PEG) tail and is the third Spiegelmer to enter clinical trials. Hepcidin is a small peptide that is the key regulator of iron hemostasis [55]. Inhibition of hepcidin overproduction could be a potential therapeutic strategy for treatment of anemia of chronic inflammation (ACI). In vitro studies have demonstrated that NOX-H94 blocked hepcidin-regulated ferroportin degradation and reversed hyperferremia activity in cynomolgus monkeys with acute systemic inflammation. NOX-H94 has been well tolerated in 64 healthy patients with minor side effects after single and repeated intravenous and subcutaneous administration at doses from 0.3 to 4.8 mg/kg in a placebo-controlled study for anemia of chronic disease [56]. A mild and transient increase in transaminase reaction was observed at doses ≥ 2.4 mg/kg, with local injection site reactions such as skin rash and hematoma.
Therapeutic aptamers for coagulation
ARC1779
ARC1779 is a 49-nucleotide DNA/RNA aptamer, which is conjugated with a 20-kDa polyethylene glycol moiety and binds specifically to the A1 domain of von Willebrand factor (VWF) [57]. VWF is a glycoprotein that plays a key role in platelet adhesion, activation and promotion of thrombosis [58, 59]. Upon binding to activated VWF, ARC1779 aptamer prevents the interaction of the VWF A1-domain with the platelet receptor GPIb, and blocks the VWF-mediated activation pathway resulting in potential inhibition of platelet activation and pathological thrombosis [60]. ARC1779 has demonstrated a unique therapeutic opportunity in the treatment of von Willebrand disease and acute coronary syndrome [60, 61]. A randomized placebo-controlled study conducted in 47 healthy patients at doses from 0.05 to 1.0 mg/kg was well-tolerated with no serious adverse events and the drug exhibited dose- and concentration-dependent inhibition of VWF activity and platelet function. In another pilot trial study, randomized in patients with type 2B von Willebrand disease (VWF type 2B), ARC1779 completely blocked VWFA1 domain activity thereby preventing the desmopressin-induced platelet drop in patients [61].
NU172
Thrombin is a serine protease that plays a key role in the blood coagulation pathway. Unlike other therapeutic aptamers with modifications, NU172 is an unmodified 26-nucleotide DNA aptamer selected to bind and interfere with thrombin function [62]. NU172 aptamer was evaluated in a Phase 2, open label clinical study in 30 patients with heart disease by ARCA biopharma. No updates are available regarding the current recruitment or completion for this study (U.S National Library of Medicine, Clinical trials.gov Identifier: NCT00808964).
REG1 system
The REG1 is a novel two-component system that consists of the aptamer-based anticoagulant, RB006, and its complementary reversal agent, RB007. RB006 is a pegylated 31-nucleotide RNA aptamer with 2-fluorpyrimidine and inverted terminal deoxythymidine residue (idT) modifications that selectively bind and impede the activity of coagulation factor IXa (FIXa), thus preventing thrombin formation [63, 64]. RB007 is a 15-nucleotide modified RNA aptamer that functions as an antidote that binds to and neutralizes RB006 anticoagulant activity rapidly and safely. Factor IXa (FIXa) plays a key role in thrombin generation, and therefore represents a potentially promising target for therapeutic development [65]. The REG1 anticoagulation system has been clinically evaluated for acute coronary syndrome and coronary artery disease (CAD) (U.S National Library of Medicine, Clinical trials.gov Identifier: NCT00932100 and NCT00715455). In a placebo-controlled study referred as Regado 1a, randomized in 85 healthy patients, both the drug RB006 (with doses of 15, 30, 60 and 90 mg) and the antidote RB007 (with doses of 30, 60, 90 and 120 mg) were administered either individually or in combination. The drugs were well tolerated with no adverse effects [64]. REG1 combined with platelet derived therapy was also well tolerated with no major bleeding and serious adverse events in a Phase 1b placebo-controlled study randomized in 50 patients with stable coronary artery disease [66]. Inhibition of factor IXa activity followed by active restoration of coagulation activity was also observed in the patients. During a RADAR Phase 2b clinical trial, severe allergic reactions observed in some patients have been linked to pre-existing anti-polyethylene glycol antibodies rather than to the aptamer component of the REG1 system [67].
BAX499/ARC19499
In vitro and animal studies have demonstrated that inhibition of tissue factor pathway inhibitor (TFPI) could be a potential mechanism for improving coagulation in hemophilia patients [68, 69]. TFPI plays an integral role in negatively regulating factor VIIa in the extrinsic pathway. BAX499 is a 32-nucleotide PEG-modified RNA aptamer that binds specifically to TFPI via interactions with multiple domains of the protein [70, 71]. BAX499 inhibits the TFPI function as a negative regulator of coagulation, thereby promoting thrombin generation and improving blood clotting times [72, 73]. The phase I randomized clinical trial for BAX499 was initiated in 2010 in hemophilia patients; however, the study was terminated with no further updates available (U.S National Library of Medicine, Clinical trials.gov Identifier: NCT01191372).
Aptamers and conjugates with therapeutic potential
In this section we summarize two major aptamer and aptamer-conjugate projects that are in advanced development, but not yet in the clinical trials pipeline. The first of these projects involves development of DNA aptamers against oxidized linoleic acid metabolites (OLAMs) generated by heat- or burn-induced tissue damage [74]. These analgesic anti-OLAM aptamers bind and block 9- and 13-hydroxy-octadecadienoic acids (HODEs), preventing their binding to the transient receptor potential vanilloid-1 (TRPV1) receptor, thereby blocking or ameliorating heat or burn pain in rat models when administered intramuscularly or intraperitoneally [75, 76]. The researchers envision a possible topical anti-OLAM analgesic aptamer cream as well. Since OLAMs or HODEs are also mediators of some types of inflammation and shock, Hargreaves and Bruno see further applications for these aptamers as potential anti-inflammatory or anti-shock drugs.
A pictorial representation of the therapeutic pipeline of aptamers showing various phases of drug development.
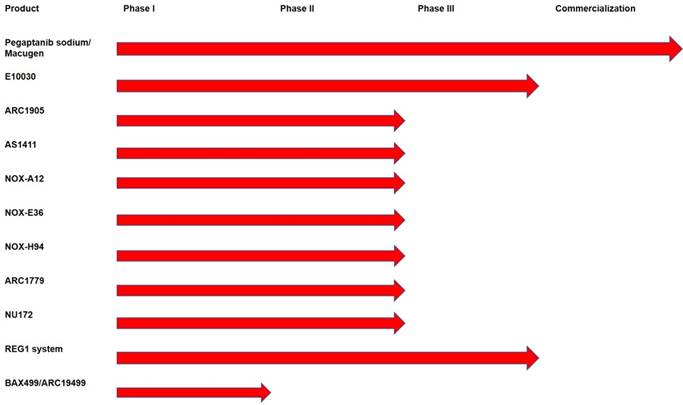
The second project was pioneered by Bruno et al., in 2008 as an alternative to conventional antibiotics to kill thinner-walled Gram-negative antibiotic-resistant bacteria using anti-lipopolysaccharide (LPS) aptamer-C1q (complement component C1q) conjugates to activate complement-mediated lysis [77]. This same mechanism was shown by Bruno and Stecker et al. to kill 50-60% of MCF-7 breast cancer cells with anti-Mucin 1 aptamer-C1q conjugates [79, 80]. Gram positive antibiotic-resistant bacteria could also be killed by enhanced phagocytosis as demonstrated by Bruno et al. using aptamer-Fc conjugates [78-80]. Subsequently, Nobel Prize laureate Kary Mullis started a company called Altermune, LLC, to develop a similar concept that he dubbed “Alphamers”. Alphamers are aptamer-alpha gal epitope conjugates that attach to the ~ 1% circulating anti-alpha gal antibodies in human serum to achieve the same general end result that Bruno et al. [78] observed previously, namely aptamer-induced killing of Group A Streptococcus and other bacteria by enhanced phagocytosis [81]. Altermune, LLC was later acquired by Centauri Therapeutics, Ltd. in the United Kingdom, which is currently pushing toward commercial Alphamer products to combat antibiotic-resistant bacteria and cancers. An overview of the existing aptamer-based therapeutics pipeline is presented in Figure 2.
One final note of potential importance to any current or future therapeutic aptamer developers is to attempt to avoid exposed cytosine-phosphate-guanine (CpG) dinucleotide sequences in aptamers, as these are well known to bind some Toll-like receptors (TLRs) and trigger innate immune responses [82, 83]. Truncation of aptamers down to their known binding sites or methylation of cytosines in CpG regions, if binding is unaffected, can ameliorate this potential toxicity problem.
Aptamers as diagnostic reagents
The aptamer market is continually growing and is expected to make great contributions to the diagnostic industry. Over the last decade, aptamers have been used in various point-of-care diagnostic platforms including Aptamer-Linked Immobilized Sorbent Assay (ALISA), Dot-blot, electrochemiluminescence (ECL) assays, fluorescence-based assays, nanoparticle-based assays, lateral flow test strips, upconverting fluorophore-based assays, electrochemical sensors and glucometer-based assays for the detection of various analytes ranging from small molecules to complex antigens to whole cell [13, 15, 16, 84-88]. Moreover, various aptamer-based diagnostic assays have exhibited the potential to follow the WHO's “ASSURED” criteria for point-of-care diagnostics: affordable, sensitive, specific, user-friendly, robust, and can always be performed outside a laboratory or hospital by unskilled labor [13]. Furthermore, point-of-care application of aptamer technology is not limited to specific diseases or sample types, rather it has wide applications, including the on-site testing of environmental pollutants like pesticides and heavy metal contaminants (e.g., arsenic, lead and mercury) [13].
Aptamers in the diagnostic pipeline
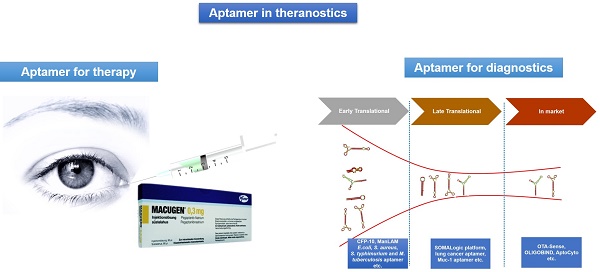
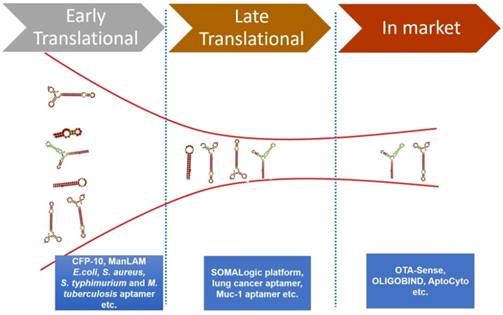
There is a vast amount of literature available that attests the success of aptamers for various diagnostic applications. However, there are few aptamer companies that have translated their research into viable commercial diagnostic products or commercial reagents. Some of these commercially successful aptamer companies are Neo Ventures Biotechnology Inc. (Ontario, Canada), SomaLogic, Inc (Boulder, CO, USA), Aptamer Sciences Inc. (AptSci, South Korea), and Base Pair (Houston, TX, USA) [89, 90]. We herein elaborate some of the important commercialized diagnostic products based on aptamer technology [16, 89, 90]. Figure 3 represents the pipeline of aptamers developed for diagnostics applications.
The various phases of the diagnostics pipeline of aptamers and some representative examples of early, late and commercialized aptamers
OTA-Sense
OTA-Sense is an aptamer-based sensing technology developed by Neoventures Biotechnology Inc. for the detection of Ochratoxin A (OTA), a toxin produced by fungi (Aspergillus and Penicillium species) that grow on agricultural products including, but not limited to grapes, dried fruits, cereals, coffee etc. OTA is one of the most common mycotoxins with known carcinogenic properties [91, 92]. This mycotoxin is highly toxic even at low doses, thus the maximum allowed levels of this toxin as set by the European Union are 5, 2 and 0.5 ppb for cereals, wine, and baby food respectively. Therefore, highly sensitive detection of this mycotoxin is critical. Dr. Gregory Penner of Neoventures Biotechnology Inc. has developed an aptamer-based diagnostic kit (OTA-Sense) for the accurate and highly sensitive detection of OTA [93]. The OTA-Sense kit is a fluorescence-based diagnostic kit that can detect OTA in a high-throughput manner with a high level of sensitivity [93]. This kit contains two components: an aptamer-based clean-up column and a detection solution. In order to detect OTA, one has to extract the toxin from the grain using an organic solvent like acetonitrile. The extracted toxin is then filtered through glass wool and then loaded onto the OTA-Sense affinity column, where an OTA-specific aptamer is immobilized on the resin. After washing with a copious amount of wash buffer, OTA is then eluted. The eluent is then mixed with a detection solution containing OTA-specific aptamer and terbium in solution. Terbium present in the solution acts like a cationic bridge between the aptamer and OTA. As soon as this bridge is created, fluorescence of terbium enhances several fold. This increase in fluorescence correlates well with the amount of OTA present in the grain. Application of the OTA-Sense system has been extended to detect OTA in all alcoholic beverages, including various beers and white/red wines [93]. This product is now available commercially.
AflaSense
Aflatoxins are another class of toxin produced by Aspergillus spp. Like OTA, aflatoxins are also carcinogenic agents [89, 94-96]. In addition to OTA-Sense, Neoventures has also developed an aptamer-based detection system for aflatoxins. This AflaSense system is principally similar to the OTA-Sense detection system [93]. The only difference between the kits is an aflatoxin B1-specific 5' biotinylated aptamer is immobilized on streptavidin agarose. This system can efficiently detect aflatoxin in corn and peanuts with a low-end detection limit of 1 ppb [93]. Like OTA-Sense, this product is also available commercially.
AptoCyto
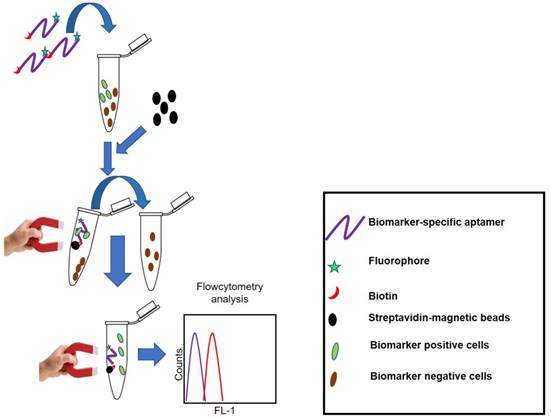
The name AptoCyto represents a range of aptamer products developed for flow cytometry applications [97]. Aptamer Science Inc. (AptSciSci Inc) is a South Korea-based aptamer company that has successfully developed aptamers against CD-31 (platelet endothelial cell adhesion molecule-1), epithelial growth factor receptor (EGFR), hepatocyte growth factor receptor (HGFR), intracellular adhesin molecule-2 (ICAM-2), vascular endothelial growth factor receptor-2 (VEGFR-2) and human epidermal growth factor receptor-2 (HER-2). Using these aptamers, they have developed several magnetic bead-based cell isolation kits that are able to efficiently isolate CD-31, EGFR, HGFR, ICAM-2, VEGFR-2 or HER-2 positive cells. Enriched cell subpopulations can easily be assessed using flow cytometry as the aptamers used in these kits are dual-labeled. On one end there exists a biotin label so as to capture the target cells by streptavidin-coated magnetic beads; on the other end the aptamer has a FITC label to monitor the captured cells using flow cytometry. This aptamer and magnetic bead-based cell separation kit provides a rapid and efficient platform to isolate the aforementioned biomarker positive cells with high yield, purity and cell viability. Figure 4 represents the principle of cell separation by the AptoCyto kit.
Principle of the AptoCyto kit to isolate and label biomarker-positive cells. This kit contains a biomarker (cell subpopulation)-specific dual-labeled aptamer having biotin on one end and a fluorophore on the other. Aptamers are incubated with a mixture of cells having both biomarker-positive and -negative subpopulations. Owing to the high specificity for a particular biomarker, aptamers will recognize and bind to the biomarker-positive cells and on addition of streptavidin-coated magnetic beads, aptamer-bound biomarker-positive cells can easily be separated from the biomarker-negative cells. These separated cells can be subjected to flow cytometry analysis in bead-bound form or after release of cells from the magnetic beads.
AptoPrep
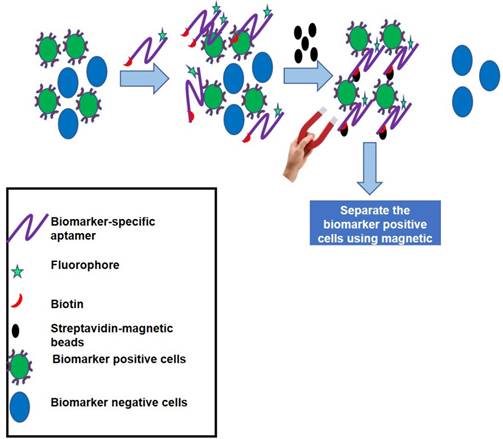
Some biomarkers are expressed at very low levels and their detection is difficult by routine diagnostic approaches like Enzyme-Linked Immunosorbent Assay (ELISA) and Western blot analysis. In such cases, immunoprecipitation yields better detection of the biomarker of choice; but, due to the cross-reactivity of some antibodies, in addition to the target biomarker, several undesired contaminating proteins may also be precipitated that hinder the downstream applications. To overcome such problems, AptSci Inc has developed a diagnostic kit called AptoPrep [98]. This kit contains conjugates of aptamers specific to CD-31, EGFR, HGFR, ICAM-2, VEGFR-2 or HER-2 and magnetic beads and can be used for “aptoprecipitation” (aptamer-based assay for protein pull down). Using the biomarker-specific aptamer, a particular biomarker can be pulled down from serum or other fluids and detected. Owing to the high affinity and specificity of aptamers, this method yields biomarker precipitates of the highest purity. Apart from the aforementioned aptamers, there are several companies including Aptamer Sciences that are offering various individual catalogue aptamers with desired modifications (linkers, fluorophores, etc.) for various analytes. Figure 5 represents the principle of biomarker-positive cell separation by aptamers and magnetic bead-based pull down assay.
SOMAscan
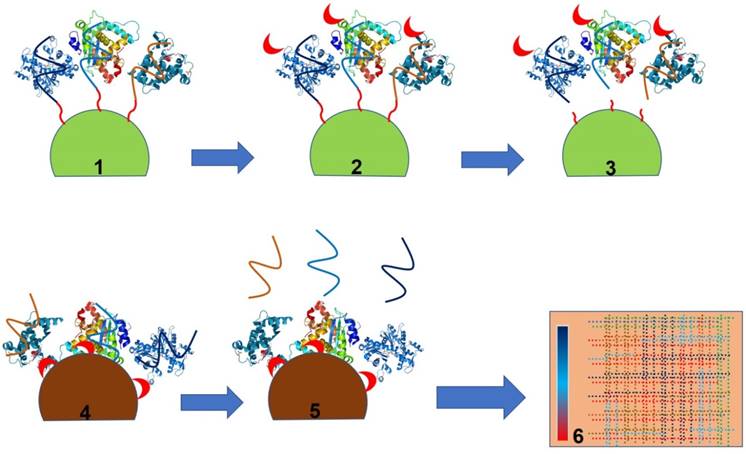
SELEX inventor Larry Gold founded a company called SomaLogic. This company has created a highly multiplexed SOMAmer (Slow Off-rate Modified Aptamer)-based biomarker discovery and clinical diagnostic platform called SOMAscan. SOMAscan is a highly versatile platform with the ability to detect and quantify >1,300 proteins simultaneously from a small volume (~50 µL) of clinical sample [99-103]. This platform is compatible with a wide variety of clinical samples ranging from serum to cerebrospinal fluid (CSF), cell and tumour extracts, synovial fluid, etc. [99-106]. Moreover, this platform is highly sensitive and can detect a protein signature down to 40 fM concentration [99-106]. This unmatched ultrasensitivity of SOMAscan has allowed the company to discover several trace biomarkers for a variety of diseases including, but not limited to, tuberculosis (TB), non-small cell lung cancer, Alzheimer's disease (AD), cardiovascular disease and inflammatory bowel disease [99-111]. Using the SOMAscan platform in addition to the previously known TB-associated serum biomarkers (IP-10, LBP, FCG3B and TSP4), several new biomarkers (serum amyloid protein A, secreted phospholipase A2 and carbonic anhydrase) were discovered that can efficiently discriminate between diseased and non-diseased states. Similarly, a recent report demonstrated the utility of SOMAscan to predict AD outcome in human subjects [111]. Using SOMAscan the authors established an efficient positive correlation between the level of prostate-specific antigen complexed to a1-antichymotrypsin (AD diagnosis), pancreatic prohormone, clusterin and fetuin B to Alzheimer's disease [111]. The application of this platform has been further extended to discover a proteomic signature for myocardial infarction subjects. Using this platform in a recent study, two novel myocardial injury markers, Dickkopf-related protein 4, a WNT pathway inhibitor, and cripto, a growth factor important in cardiac development, were identified [111]. Recently, the SOMAscan platform has also been used for the detection of non-small cell lung cancer (NSCLC) [103]. Using this highly sensitive approach, a panel of 12 proteins (cadherin-1, pleiotrophin, RGM-C, CD30 ligand, HSP90a, endostatin, LRIG3, MIP-4, sL-selectin, SCF-sR, PRKCI and YES) was also discovered. Study of archived sera of non-small cell lung cancer patients and control sera demonstrated 91% sensitivity and 84% specificity with an ability to detect early and late stage NSCLC with equal accuracy. Taken together, this technology is highly valuable to predict the disease outcome or to study the impact of treatment with wider utility in disease diagnostics [103]. Figure 6 portrays the principle of biomarker identification by the SOMAmer-based approach.
Principle of the AptoPrep kit to pull down cells having rare biomarkers on their surfaces. This kit contains a biomarker-specific dual-labeled aptamer having biotin on one end and a fluorophore on the other. Aptamers are incubated with a mixture of cells having both biomarker-positive and -negative subpopulations. Owing to the high specificity for a biomarker, aptamer will bind to the biomarker-positive cells and on addition of streptavidin-coated magnetic beads, aptamer-bound biomarker-positive cells can easily be pulled down from the mixture of cells for downstream applications.
CibusDx, a food pathogen diagnostics platform
The World Health Organization, in 2015, reported that one in ten people worldwide fall ill due to ingestion of contaminated food. Thus, detection of foodborne pathogens or toxins produced by them is critical to ensure food safety. To address this task, a USA-based start-up (Pronucleotein, Inc.) developed a range of aptamers against foodborne [112] and waterborne pathogens, bacterial and fungal toxins, lipopolysaccharides and bacterial capsule polysaccharides. Unlike antibodies these aptamers can undergo target-induced structural changes [112]. This attribute makes them ideal candidates to design an electrochemical sensor where a target-specific aptamer is modified with a redox label (methylene blue or ferrocene) that has the ability to convert aptamer-target binding events into measurable electrochemical outputs. On interaction with the target, conformational changes in aptamers place redox labels closer or further apart from the gold-coated working electrode where the thiolated aptamer is immobilized. Accordingly, electrical current either increases or decreases as a function of the binding event and target concentration. Moreover, such an electrochemical sensor is quite handy and can be operated through a portable smart mobile phone, which makes these devices real point-of-care biosensors. This sensor is highly sensitive, rapid and can give sample-to-answer results within 5-10 minutes. Using the foodborne pathogen-specific aptamers developed by Pronucleotein, CibusDx (USA), an electrochemical sensor company, has fully developed a handheld, smart phone-enabled electrochemical sensing platform for the detection of various foodborne and waterborne pathogens. CibusDx is planning to launch this product for food safety in 2018. Owing to the rapid turnaround time, this product can create a huge impact in the food safety testing industry by replacing the traditional bacterial culture-based central laboratory paradigm that takes 24-72 hours to provide test results.
Principle of the SOMAscan platform as demonstrated by Kraemer et al. [101]. Step 1 shows bead-bound SOMAmers binding to their cognate target proteins followed by biotinylation (red crescent) of the SOMAmer-bound protein (Step 2). Step 3 is release of the SOMAmer (aptamer) and biotinylated protein complex by the photocleavage of UV-sensitive linkers. The released aptamer and biotinylated protein complex is captured on a streptavidin bead (Step 4) followed by release of SOMAmers from the protein (Step 5) and detection of SOMAmers by a standard hybridization-based DNA quantification method (Step 6).
Aptamer technology for the detection of non-small cell lung cancer
Recently Jung et al. (2016) have developed an aptamer-based proteomics technology for the ultrasensitive detection of non-small cell lung cancer [108]. Using this approach, they have identified seven protein biomarkers (EGFR, CA6, MMP7, CRP, KIT, C9, SERPINA3). Using aptamer-based proteomics technology, they were able to detect differential expression of these biomarkers and differentiate non-small cell lung cancer patients from healthy subjects with a sensitivity of 75% and ~ 92% specificity [108]. More recently, in March 2017 Aptamer Sciences (South Korea) introduced this technology for commercial use [113].
OLIGOBIND
This product was developed by Sekisui diagnostics, GmbH. Thrombin comes under the category of essential serine proteases and is known to play a vital role in the blood coagulation pathway. Therefore, estimation of active thrombin levels is critical to identifying patients predisposed to bleeding or thromboembolic complications. The limitation of the current diagnostic modalities is that they cannot measure the true in vivo levels of active thrombin, rather they either measure prothrombin fragment F1.2 released after proteolytic activation of thrombin, or the inactivated thrombin-antithrombin-complexes (TAT) [114]. To address this unmet need, Müller et al. (2011) discovered a novel oligonucleotide-based enzyme capture assay that can accurately measure the true in vivo levels of thrombin in blood [115]. This product is available in the market as OLIGOBIND® Thrombin activity assay by Sekisui Diagnostics (https://www.sekisuidiagnostics.com/products/722-thrombin-activity-assay). This kit accurately measures true in vivo levels of thrombin through an aptamer-based enzyme-capture fluorescent assay [115]. In order to measure the levels of active thrombin using the aforementioned assay, plasma samples are first added to microwell plates pre-coated with thrombin-specific aptamers followed by incubation and washes to remove unbound material. Finally, aptamer-bound thrombin is measured by the addition of a fluorogenic substrate (excitation, 360 nm; emission, 460 nm). This kit can detect thrombin down to picomolar concentrations in blood samples. Further, this kit is highly selective and evinces ~100-fold higher selectivity for thrombin than prothrombin [117]. The only limitation of this kit is that platelet contamination in the plasma samples can interfere with the assay.
Taq DNA polymerases from New England Biolabs (NEB)
NEB, a leading manufacturer of enzymes and other molecular biology reagents, has commercialized Hot Start Taq DNA polymerase (Cat No. M0496S and M0490S). Both of these products principally rely on an aptamer-based hot start technology to avoid nonspecific amplification, thereby allowing users to perform room temperature reaction setup [118]. These aforementioned Taq DNA polymerase products are supplied as a mixture of Taq DNA polymerase and an aptamer-based inhibitor. An aptamer-based inhibitor binds reversibly to the polymerase, thereby inhibiting the polymerase activity at below 45 °C, but releases the enzyme during normal thermal cycling conditions. Unlike traditional polymerases, the main advantage of this aptamer-based hot start polymerase is that it does not require a separate incubation step at high temperature to activate the enzyme, thus making hot start polymerase chain reaction (PCR) a facile task. This product can be used for various PCR-based diagnostics applications. A list of aptamer-based diagnostic products along with their principles of detection, applications and limitations is tabulated in Table 2.
Aptamers with immediate translational potential
Apart from the commercially available aptamers, there are several aptamers reported in literature that may have immediate translational potential, if adapted to an appropriate diagnostic platform. These aptamers have high translational value in cancer, bacterial and small molecule diagnostics.
Aptamers for cancer diagnostics
Cancer cells are different from normal cells in many ways including the expression of cell surface proteins. The early and accurate detection of such cancer-specific biomarkers is critical to improvement of survival rates and treatment options for cancer patients. In recent years, a large number of aptamers have been developed against cancer-specific protein biomarkers or whole cells with the aim of early cancer detection. Some of these aptamers are able to detect very difficult-to-treat forms of lung cancer. Usually lung cancer is not diagnosed until it has progressed to a late stage with poor patient survival. Recently, Li et al. [113] have discovered a panel of six aptamers by using a modified SELEX approach where aptamer selection was done by incubating the aptamer library with serum of lung cancer patients and counterselecting with serum from healthy individuals. Using this approach, they discovered a panel of aptamers that can efficiently discriminate between lung cancer patients and healthy individuals in an aptamer-based PCR assay [113]. This aptamer-based approach appears to be much more sensitive than the contemporary detection methods and has the potential for early lung cancer diagnosis. Though preliminary findings are encouraging, the translation of these aptamers into a diagnostic product will require validation of the aptamers at multiple sites on a large number of clinical samples.
Like lung cancer, early detection of ovarian cancer is also highly challenging. To address this challenge, Tsai et al. [119] have developed an aptamer-based microfluidic system that can detect rarely found circulating tumour cells (CTCs) present in body fluid. This aptamer-based approach yields rapid and highly sensitive detection of ovarian cancer with low false positives when compared with antibody-based detection of such cancer cells [119].
In addition to the detection of a specific type of cancer, a generic diagnostic tool than can detect and discriminate between cancer cells and normal healthy cells could be highly valuable. In order to achieve this diagnostic tool, one must target a biomarker that is abundantly expressed on cancer cells. One such biomarker is Mucin-1 (MUC1), a glycoprotein that is present in ample amounts on the surface of most malignant epithelial cancer cells including, but not limited to, lung, breast, prostate, bladder, pancreas, colorectal and ovarian cancers [79, 113]. Owing to the importance of this biomarker in cancer diagnosis, a number of research groups have developed aptamer-based diagnostic platforms for the detection of MUC1 including fluorescent and electrochemical aptasensors. These sensors can detect as few as 100 cancer cells/mL, thus showing a high value for clinical diagnostics [113]. Considering such performance of these sensors, there will be no surprise that after careful clinical validation this technology may hit the market in coming years.
Commercially available aptamer-based diagnostic products and their principles of detection, applications and limitations.
| S.N. | Product | Company | Mode of detection | Application | Limitation |
|---|---|---|---|---|---|
| 1 | OTA-Sense and AflaSense | Neoventures Biotechnology Inc. | Fluorescence- based assay | Detection of Mycotoxins (produced by Aspergillus and Penicillium species) in food | Require extraction of toxin from the sample |
| 2 | AptoCyto | AptSci Inc | Aptamer-based flowcytometry | Aptamer and magnetic bead-based isolation of biomarker positive cell | Dependence on expensive instrument (flow cytometer) to visualize the outcome |
| 3 | AptoPrep | AptSci Inc | Fluorescence-based assay and polyacrylamide gel electrophoresis (PAGE) | Aptamer-magnetic-bead-based pulldown of biomarker-positive cells and protein isolation | 4 °C Storage is recommended for long term storage |
| 4 | SOMAscan | SomaLogic | SOMAmer-based detection and quantification of biomarkers | A highly efficient platform with multiplex capability for Biomarkers discovery and diagnostics | Multistep process |
| 5 | CibusDx | CibusDx | Electrochemical sensing | Detection of foodborne pathogens | Instrument-based technology |
| 6 | OLIGOBIND | Sekisui diagnostics | Fluorogenic activity assay | Detection of active thrombin | Platelet contamination in plasma sample may interfere with assay |
| 7 | Hot Start Taq DNA polymerase | NEB | Aptamer-based reversible inhibition of Taq DNA polymerase | Hot start PCR | In comparison to normal polymerase this is relatively expensive |
Aptamers for bacterial pathogens
Bacterial pathogens are amongst the most notorious microorganisms and they are responsible for a variety of infectious food, water and nosocomial diseases. Therefore, their detection in food, water or patient specimens is critical. To address this challenge, a number of research groups have developed aptamers to detect various bacterial pathogens including, but not limited to, pathogenic Escherichia coli, Staphylococcus aureus, Pseudomonas aeruginosa, Salmonella enterica and Mycobacterium tuberculosis [110, 120-128]. Broadly speaking two strategies are used: 1) aptamers are developed against whole bacterial cells and 2) aptamers are developed against extracted bacterial antigens or toxins produced by them. In either case, aptamers have been adapted to various diagnostic platforms to achieve highly sensitive and specific detection. Aptamers developed against Salmonella enterica can detect as low as 10 CFU/gram of biological samples in a fluorescence-based assay [125]. Similarly, aptasensors designed for the detection of S. aureus can detect ~800 CFU/mL from skin swab samples based on the redox response [129]. A more sensitive potentiostatic aptasensor is also reported for pathogenic serotypes of E.coli that can detect as low as 6 and 26 CFU/mL in milk and apple juice, respectively [130, 131]. More recently, the utility of aptamer technology has been demonstrated in several independent reports for the detection of both pulmonary and extra-pulmonary tuberculosis. All these studies have shown detection of Mycobacterium tuberculosis (Mtb)-specific antigens (CFP-10, ESAT-6 and ManLAM) in clinical samples including serum and sputum. Tang et al. (2014) developed aptamers against the heterodimer of CFP-10 and ESAT-6 using SELEX [132]. After a series of experiments, they identified two aptamer candidates CE24 and CE15. In an Enzyme-Linked Oligonucleotide Assay (ELONA), CE24 yielded 100% sensitivity and 94.1% specificity when evaluated in serum samples of TB subjects and healthy volunteers, while the CE15 aptamer had relatively lower sensitivity (89.6%) and comparable specificity (94.1%) [132]. These aptamers are highly promising and can be used as a valuable clinical diagnostic tool to detect the presence of TB. More recently, Sypabekova et al. [127] have identified an aptamer specific to MPT-64, a secretory antigen specific to M.tb, and the clinical evaluation of this aptamer in sputum samples has shown a very high sensitivity (91.3%) and specificity (90 %) in an ELONA format. In addition to the aforementioned antigen, there is a report of a ManLAM-specific DNA aptamer that has shown equal utility for pulmonary and extra-pulmonary TB detection. In a study performed by Tang et al. [128], ManLAM-specific DNA aptamer, when tested in serum samples, yielded specificity and sensitivity of 95.31% and 83.00% respectively in pulmonary TB samples and 94.44% and 88.71% specificity and sensitivity respectively in extra-pulmonary TB samples. When the same aptamer was tested on sputum samples, it displayed ~93% specificity and ~95% sensitivity [128]. Taken together, these aptamers have very high translational value and may be adapted to various point-of-care diagnostics platforms for the detection of tuberculosis. Further, in recent years the WHO has issued an advisory against the Sero-diagnostic assay used for TB (https://doi.org/10.1016/S0140-6736(11)60005-6), which opens up a new vista for aptamers to demonstrate real diagnostic potential against a disease that affects a large population in the developing countries.
Aptamers for small molecules
Apart from macromolecules, a large number of aptamers have been generated against a range of small molecules including heavy metals, antibiotics, pesticides, steroid hormones, explosives, drugs and other contaminants. Some of these aptamers are available commercially (https://www.basepairbio.com/store/). Recently, DeRosa et al. [89] and Mayer et al. [90] have reviewed the generation and application of small molecule aptamers; therefore, we are not describing them in detail here. However, adaption of these aptamers on electrochemical sensing platforms or conjugation with nanomaterials yields subnanomolar or ppb level detection in water and food matrices. Overall, all these aptamers have strong potential to be used as environmental monitoring and food quality assurance reagents.
Concluding remarks
Aptamers are single-stranded oligonucleotides that bind with high affinity and high specificity to their cognate targets. In this review, we have discussed aptamers that have been subjected to clinical trials for the treatment of diverse medical conditions such as AMD, acute myeloid leukaemia (AML), metastatic renal cell carcinoma, type 2 diabetes mellitus, von Willebrand disease (VMD), thrombosis, heart disease, acute coronary syndrome and hemophilia. While discussing each of the therapeutic aptamers, we have explained their individual protective chemical modifications, their mechanisms of action and therapeutic potential. In addition, this review has also discussed aptamers in the current diagnostic pipeline while addressing challenges associated with the diagnostic application of aptamer technology.
We believe the bottleneck of commercialized aptamers for diagnostic and therapeutic applications is not due to any deficiencies in aptamer technology, except possibly overlooked CpG toxicity in some cases, but rather the general public ignorance about aptamer technology and the entrenched nature of antibody use in disease treatments and existing immunoassays. Another main reason for the aptamer bottleneck in diagnostics and therapeutics is the huge biotech industry's financial investment in the well-established antibody and humanized monoclonal antibody research, development and manufacturing processes, making these industries relatively reluctant to invest capital in new technology such as aptamers. Despite the aforementioned challenges, the advantages of aptamers over antibodies has created some market penetration in the agricultural industry (mycotoxin detection), food safety testing industry and cancer therapeutics. This penetration of aptamer technology to niche markets may create confidence in aptamer developers and users and may also create broader markets and expanded roles for aptamer-based therapeutic development and aptamer assays in the general diagnostics arena. While there is clearly an FDA approval bottleneck for therapeutic aptamers and other drugs in their respective pipelines, the recently signed federal “right to try” law for terminally ill patients, may bypass some of this bottleneck in areas such as cancer or other life-threatening infectious and genetic diseases that aptamers may treat. Acquisition of patient data in such terminal outlook cases could be invaluable for determining aptamer efficacy and safety and accelerating FDA approval of some aptamers. Thus, taken together, aptamer technology has a bright future in therapeutics and diagnostics and in the next 5-10 years, we foresee many aptamer-based therapeutics and diagnostics products building their own niches in the vast biopharmaceutical and diagnostics worlds.
Abbreviations
ACI: anemia of chronic inflammation; AD: alzheimer's disease; ALISA: aptamer-linked immobilized sorbent assay; AMD: age-related macular degeneration; AML: acute myeloid leukaemia; AptSci Inc : aptamer science inc.; CS: complement system; C5: complement component 5; CAD: coronary artery disease; CCL2: C-C chemokine ligand 2; CLL: chronic lymphocytic leukaemia; CpG: cytosine-phosphate-guanine; CSF: cerebrospinal fluid; CTCs: circulating tumour cells; CXCL12: C-X-C chemokine ligand 12; D-forms: dextrorotatory forms; Dau: daunorubicin; ECL: electrochemiluminescence; EGFR: epithelial growth factor receptor; ELISA: enzyme-linked immunosorbent assay; ELONA: enzyme-linked oligonucleotide assay; GRO: guanine-rich oligonucleotide; HER-2: human epidermal growth factor receptor-2; HGFR: hepatocyte growth factor receptor; HODEs: hydroxy-octadecadienoic acids; ICAM-2: intracellular adhesin molecule-2; idT: inverted terminal deoxythymidine residue; IR: insulin receptor; LNA: locked nucleic acid; L-RNA: levorotatory RNA; L-ribose units: L-deoxyribose units; LPS: lipopolysaccharide; MAC: membrane attack complex; MCP-1: monocyte chemoattractant protein 1; MUC1: mucin-1; Mtb: mycobacterium tuberculosis; NCL: nucleolin; NEB: new england biolabs; NSCLC: non-small cell lung cancer; OLAMs: oxidized linoleic acid metabolites; OTA: ochratoxin A; PCR: polymerase chain reaction; PEG: polyethylene glycol; PDGF: platelet-derived growth factor; PS-linkages: phosphorothioate linkages; PTK-7: protein tyrosine kinase-7; RCC: renal cell carcinoma; SDF-1: stromal cell-derived factor-1; SOMAmer: slow off-rate modified aptamer; TAT: thrombin-antithrombin-complexes; TB: tuberculosis; TFPI: tissue factor pathway inhibitor; TLRs: toll-like receptors; TRPV1: transient receptor potential vanilloid-1; VEGF: vascular endothelial growth factor; VEGFR-2: vascular endothelial growth factor receptor-2; VWF: von willebrand factor.
Competing Interests
T.K.S. Own 87% Stakes in AptaBharat Innovation Pvt. Ltd. and J.G.B. own 8% stakes in Pronucleotin and OTC Biotech.
Acknowledgements
T.K.S. wishes to thank the Department of Biotechnology, Govt. of India for DBT-Innovative Young Biotechnologist Award. T.K.S. also thanks Translational Health Science and Technology Institute (THSTI) for providing core grant funds to support aptamer research. We also thank Dr. Priya Kalra for her valuable input and proofreading of this manuscript.
References
1. Nakamura Y. Aptamers as therapeutic middle molecules. Biochimie. 2018;145:22-33
2. Morita Y, Leslie M, Kameyama H, Volk DE, Tanaka T. Aptamer therapeutics in cancer: Current and future. Cancers (Basel). 2018;10:80
3. Zhuo Z, Yu Y, Wang M, Li J, Zhang Z, Liu J. et al. Recent advances in SELEX technology and aptamer applications in biomedicine. Int J Mol Sci. 2017;18:2142
4. Wu YX, Kwon YJ. Aptamers: The evolution of SELEX. Methods. 2016;106:21-8
5. Wrzesinski J, Ciesiolka J. Characterization of structure and metal ions specificity of Co2+-binding RNA aptamers. Biochemistry. 2005;44:6257-68
6. Famulok M. Oligonucleotide aptamers that recognize small molecules. Curr Opin Struct Biol. 1999;9:324-9
7. Thomas M, Chedin S, Carles C, Riva M, Famulok M, Sentenac A. Selective targeting and inhibition of yeast RNA polymerase II by RNA aptamers. J Biol Chem. 1997;272:27980-6
8. Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P. et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci U S A. 2006;103:11838-43
9. Darfeuille F, Reigadas S, Hansen JB, Orum H, Di Primo C, Toulme JJ. Aptamers targeted to an RNA hairpin show improved specificity compared to that of complementary oligonucleotides. Biochemistry. 2006;45:12076-82
10. Kalra P, Dhiman A, Cho WC, Bruno JG, Sharma TK. Simple methods and rational design for enhancing aptamer sensitivity and specificity. Front Mol Biosci. 2018;5:1-16
11. Tuerk C. Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505-10
12. Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD. et al. 2'-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J Biol Chem. 1998;273:20556-67
13. Dhiman A, Kalra P, Bansal V, Bruno JG, Sharma TK. Aptamer-based point-of-care diagnostic platforms. Sens Actuators B Chem. 2017;246:535-53
14. Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9:537-50
15. Sharma TK, Bruno JG, Dhiman A. ABCs of DNA aptamer and related assay development. Biotechnol Adv. 2017;35:275-301
16. Sharma TK, Bruno JG, Cho WC. The point behind translation of aptamers for point of care diagnostics. Aptamers and Synthetic Antibodies. 2016;3:36-42
17. Lee JW, Kim HJ, Heo K. Therapeutic aptamers: developmental potential as anticancer drugs. BMB Rep. 2015;48:234-7
18. Parashar A. Aptamers in Therapeutics. J Clin Diagn Res. 2016;10:BE01-6
19. Zhou J, Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov. 2017;16:181-202
20. Yunn NO. et al. Agonistic aptamer to the insulin receptor leads to biased signaling and functional selectivity through allosteric modulation. Nucleic Acids Res. 2015Sep18;43(16):7688-701
21. Healy JM, Lewis SD, Kurz M, Boomer RM, Thompson KM, Wilson C. et al. Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm Res. 2004;21:2234-46
22. Abeydeera ND, Egli M, Cox N, Mercier K, Conde JN, Pallan PS. et al. Evoking picomolar binding in RNA by a single phosphorodithioate linkage. Nucleic Acids Res. 2016;44:8052-64
23. Darmostuk M, Rimpelova S, Gbelcova H, Ruml T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol Adv. 2015;33:1141-61
24. Taghdisi SM. et al. Targeted delivery of daunorubicin to T-cell acute lymphoblastic leukemia by aptamer. J Drug Target. 2010May;18(4):277-81
25. Burmeister PE, Lewis SD, Silva RF, Preiss JR, Horwitz LR, Pendergrast PS. et al. Direct in vitro selection of a 2'-O-methyl aptamer to VEGF. Chemistry & biology. 2005;12:25-33
26. Pieken WA, Olsen DB, Benseler F, Aurup H, Eckstein F. Kinetic characterization of ribonuclease-resistant 2'-modified hammerhead ribozymes. Science. 1991;253:314-7
27. Kaur H, Li JJ, Bay BH, Yung LY. Investigating the antiproliferative activity of high affinity DNA aptamer on cancer cells. PLoS One. 2013;8:e50964
28. Vater A, Klussmann S. Toward third-generation aptamers: Spiegelmers and their therapeutic prospects. Curr Opin Drug Discov Devel. 2003;6:253-61
29. Kaur H, Yung LY. Probing high affinity sequences of DNA aptamer against VEGF165. PLoS One. 2012;7:e31196
30. Dougan H, Lyster DM, Vo CV, Stafford A, Weitz JI, Hobbs JB. Extending the lifetime of anticoagulant oligodeoxynucleotide aptamers in blood. Nucl Med Biol. 2000;27:289-97
31. Watson SR, Chang YF, O'Connell D, Weigand L, Ringquist S, Parma DH. Anti-L-selectin aptamers: Binding characteristics, pharmacokinetic parameters, and activity against an intravascular target in vivo. Antisense Nucleic Acid Drug Dev. 2000;10:63-75
32. Ferrara N. Vascular endothelial growth factor: Basic science and clinical progress. Endocr Rev. 2004;25:581-611
33. Macugen. http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Summary_for_the_public/human/000620/WC500026216.pdf
34. Sadiq MA, Hanout M, Sarwar S, Hassan M, Agarwal A, Sepah YJ. et al. Platelet-derived growth factor inhibitors: A potential therapeutic approach for ocular neovascularization. Dev Ophthalmol. 2016;55:310-6
35. Green LS, Jellinek D, Jenison R, Ostman A, Heldin CH, Janjic N. Inhibitory DNA ligands to platelet-derived growth factor B-chain. Biochemistry. 1996;35:14413-24
36. Floege J, Ostendorf T, Janssen U, Burg M, Radeke HH, Vargeese C. et al. Novel approach to specific growth factor inhibition in vivo: Antagonism of platelet-derived growth factor in glomerulonephritis by aptamers. Am J Pathol. 1999;154:169-79
37. Kawa MP, Machalinska A, Roginska D, Machalinski B. Complement system in pathogenesis of AMD: Dual player in degeneration and protection of retinal tissue. J Immunol Res. 2014;2014:483960
38. Sparrow JR, Ueda K, Zhou J. Complement dysregulation in AMD: RPE-Bruch's membrane-choroid. Mol Aspects Med. 2012;33:436-45
39. Biesecker G, Dihel L, Enney K, Bendele RA. Derivation of RNA aptamer inhibitors of human complement C5. Immunopharmacology. 1999;42:219-30
40. Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp Mol Pathol. 2009;86:151-64
41. Choi EW, Nayak LV, Bates PJ. Cancer-selective antiproliferative activity is a general property of some G-rich oligodeoxynucleotides. Nucleic Acids Res. 2010;38:1623-35
42. Miller DM, Laber DA, Bates PJ, Trent JO, Taft BS, Kloecker GH. et al. Extended phase I study of AS1411 in renal and non-small cell lung cancers- Meeting Abstract. Ann Oncol. 2006;17:147-8
43. Semenkovich CF, Ostlund RE Jr, Olson MO, Yang JW. A protein partially expressed on the surface of HepG2 cells that binds lipoproteins specifically is nucleolin. Biochemistry. 1990;29:9708-13
44. Christian S, Pilch J, Akerman ME, Porkka K, Laakkonen P, Ruoslahti E. Nucleolin expressed at the cell surface is a marker of endothelial cells in angiogenic blood vessels. J Cell Biol. 2003;163:871-8
45. Soundararajan S, Chen W, Spicer EK, Courtenay-Luck N, Fernandes DJ. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008;68:2358-65
46. Hoellenriegel J, Zboralski D, Maasch C, Rosin NY, Wierda WG, Keating MJ. et al. The Spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization. Blood. 2014;123:1032-9
47. Marasca R, Maffei R. NOX-A12: Mobilizing CLL away from home. Blood. 2014;123:952-3
48. Roccaro AM, Sacco A, Purschke WG, Moschetta M, Buchner K, Maasch C. et al. SDF-1 inhibition targets the bone marrow niche for cancer therapy. Cell Rep. 2014;9:118-28
49. Vater A, Sahlmann J, Kroger N, Zollner S, Lioznov M, Maasch C. et al. Hematopoietic stem and progenitor cell mobilization in mice and humans by a first-in-class mirror-image oligonucleotide inhibitor of CXCL12. Clin Pharmacol Ther. 2013;94:150-7
50. Ludwig H, Weisel K, Petrucci MT, Leleu X, Cafro AM, Garderet L. et al. Olaptesed pegol, an anti-CXCL12/SDF-1 Spiegelmer, alone and with bortezomib-dexamethasone in relapsed/refractory multiple myeloma: A Phase IIa Study. Leukemia. 2017;31:997-1000
51. Oberthur D, Achenbach J, Gabdulkhakov A, Buchner K, Maasch C, Falke S. et al. Crystal structure of a mirror-image L-RNA aptamer (Spiegelmer) in complex with the natural L-protein target CCL2. Nat Commun. 2015;6:6923
52. Dawson J, Miltz W, Mir AK, Wiessner C. Targeting monocyte chemoattractant protein-1 signalling in disease. Expert Opin Ther Targets. 2003;7:35-48
53. Menne J, Eulberg D, Beyer D, Baumann M, Saudek F, Valkusz Z. et al. C-C motif-ligand 2 inhibition with emapticap pegol (NOX-E36) in type 2 diabetic patients with albuminuria. Nephrol Dial Transplant. 2017;32:307-15
54. Schwoebel F, van Eijk LT, Zboralski D, Sell S, Buchner K, Maasch C. et al. The effects of the anti-hepcidin Spiegelmer NOX-H94 on inflammation-induced anemia in cynomolgus monkeys. Blood. 2013;121:2311-5
55. Ganz T. Hepcidin, a key regulator of iron metabolism and mediator of anemia of inflammation. Blood. 2003;102:783-8
56. Boyce M, Warrington S, Cortezi B, Zollner S, Vauleon S, Swinkels DW. et al. Safety, pharmacokinetics and pharmacodynamics of the anti-hepcidin Spiegelmer lexaptepid pegol in healthy subjects. Br J Pharmacol. 2016;173:1580-8
57. Diener JL, Daniel Lagasse HA, Duerschmied D, Merhi Y, Tanguay JF, Hutabarat R. et al. Inhibition of von Willebrand factor-mediated platelet activation and thrombosis by the anti-von Willebrand factor A1-domain aptamer ARC1779. J Thromb Haemost. 2009;7:1155-62
58. Blann AD. Plasma von Willebrand factor, thrombosis, and the endothelium: the first 30 years. Thromb Haemost. 2006;95:49-55
59. Ruggeri ZM. Von Willebrand factor, platelets and endothelial cell interactions. J Thromb Haemost. 2003;1:1335-42
60. Gilbert JC, DeFeo-Fraulini T, Hutabarat RM, Horvath CJ, Merlino PG, Marsh HN. et al. First-in-human evaluation of anti von Willebrand factor therapeutic aptamer ARC1779 in healthy volunteers. Circulation. 2007;116:2678-86
61. Jilma B, Paulinska P, Jilma-Stohlawetz P, Gilbert JC, Hutabarat R, Knobl P. A randomised pilot trial of the anti-von Willebrand factor aptamer ARC1779 in patients with type 2b von Willebrand disease. Thromb Haemost. 2010;104:563-70
62. Waters E, Richardson J, Schaub R, Kurz J. Effect of NU172 and bivalirudin on ecarin clotting time in human plasma and whole blood. J Thromb Haemost (Supplement 2) Abstract PPWE-168. 2009:7
63. Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D. et al. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature. 2002;419:90-4
64. Dyke CK, Steinhubl SR, Kleiman NS, Cannon RO, Aberle LG, Lin M. et al. First-in-human experience of an antidote-controlled anticoagulant using RNA aptamer technology: a phase 1a pharmacodynamic evaluation of a drug-antidote pair for the controlled regulation of factor IXa activity. Circulation. 2006;114:2490-7
65. Smiley DA, Becker RC. Factor IXa as a target for anticoagulation in thrombotic disorders and conditions. Drug Discov Today. 2014;19:1445-53
66. Chan MY, Cohen MG, Dyke CK, Myles SK, Aberle LG, Lin M. et al. Phase 1b randomized study of antidote-controlled modulation of factor IXa activity in patients with stable coronary artery disease. Circulation. 2008;117:2865-74
67. Ganson NJ, Povsic TJ, Sullenger BA, Alexander JH, Zelenkofske SL, Sailstad JM. et al. Pre-existing anti-polyethylene glycol antibody linked to first-exposure allergic reactions to pegnivacogin, a PEGylated RNA aptamer. J Allergy Clin Immunol. 2016;137:1610-3 e7
68. Nordfang O, Valentin S, Beck TC, Hedner U. Inhibition of extrinsic pathway inhibitor shortens the coagulation time of normal plasma and of hemophilia plasma. Thromb Haemost. 1991;66:464-7
69. Erhardtsen E, Ezban M, Madsen MT, Diness V, Glazer S, Hedner U. et al. Blocking of tissue factor pathway inhibitor (TFPI) shortens the bleeding time in rabbits with antibody induced haemophilia A. Blood Coagul Fibrinolysis. 1995;6:388-94
70. Waters EK, Genga RM, Schwartz MC, Nelson JA, Schaub RG, Olson KA. et al. Aptamer ARC19499 mediates a procoagulant hemostatic effect by inhibiting tissue factor pathway inhibitor. Blood. 2011;117:5514-22
71. Waters EK, Genga RM, Thomson HA, Kurz JC, Schaub RG, Scheiflinger F. et al. Aptamer BAX 499 mediates inhibition of tissue factor pathway inhibitor via interaction with multiple domains of the protein. J Thromb Haemost. 2013;11:1137-45
72. Chang JY, Chantrathammachart P, Monroe DM, Key NS. Studies on the mechanism of action of the aptamer BAX499, an inhibitor of tissue factor pathway inhibitor. Thromb Res. 2012;130:e151-7
73. Gorczyca ME, Nair SC, Jilma B, Priya S, Male C, Reitter S. et al. Inhibition of tissue factor pathway inhibitor by the aptamer BAX499 improves clotting of hemophilic blood and plasma. J Thromb Haemost. 2012;10:1581-90
74. Hargreaves KM, Bruno JG. Composition of matter that reduces pain, shock, and inflammation by blocking linoleic acid metabolites and uses thereof. US Patent No 9,592,301. 2017
75. Bruno JG. Aptamers: scope, limitations and future prospects. In: (ed.) Veedu RN. Aptamers: Tools for targeted nanotherapy and molecular imaging. Oxford, UK: Taylor & Francis. 2016
76. Bruno JG. Do it yourself 3-dimensional aptamer-ligand molecular modeling. J Bionanosci. 2017;11:183-8
77. Bruno JG, Carrillo MP, Phillips T. In vitro antibacterial effects of anti-lipopolysaccharide DNA aptamer-C1qrs complexes. Folia Microbiol. 2008;53:295-302
78. Bruno JG. Aptamer-biotin-streptavidin-C1q complexes can trigger the classical complement pathway to kill cancer cells. In Vitro Cell Dev Biol Anim. 2010;46:107-13
79. Stecker JR, Savage AA, Bruno JG, Garcia DM, Koke JR. Dynamics and visualization of MCF7 adenocarcinoma cell death by aptamer-C1q-mediated membrane attack. Nucleic Acid Ther. 2012;22:275-82
80. Bruno JG, Carrillo MP, Crowell R. Preliminary development of DNA aptamer-Fc conjugate opsonins. J Biomed Mater Res A. 2009;90:1152-61
81. Kristian SA, Hwang JH, Hall B, Leire E, Iacomini J, Old R. et al. Retargeting pre-existing human antibodies to a bacterial pathogen with an alpha-Gal conjugated aptamer. J Mol Med. 2015;93:619-31
82. Avci-Adali M, Steinle H, Michel T, Schlensak C, Wendel HP. Potential capacity of aptamers to trigger immune activation in human blood. PLoS One. 2013;8:e68810
83. Mena A, Nichani AK, Popowych Y, Godson DL, Dent D, Townsend HG. et al. Innate immune responses induced by CpG oligodeoxyribonucleotide stimulation of ovine blood mononuclear cells. Immunology. 2003;110:250-7
84. Sharma S, Zapatero-Rodriguez J, Estrela P, O'Kennedy R. Point-of-care diagnostics in low resource settings: Present status and future role of microfluidics. Biosensors. 2015;5:577-601
85. Sharma T.K. RR, Weerathunge P, Mohammadtaheri M, Daima H. K, Shukla R, Bansal V. Aptamer-mediated 'turn-off/turn-on' nanozyme activity of gold nanoparticles for kanamycin detection. Chem Commun. 2014;50:15856-9
86. Sharma TK, Shukla R. Nucleic acid aptamers as an emerging diagnostic tool for animal pathogens. Adv Animal Vet Sci. 2014;2:50-5
87. Weerathunge P, Ramanathan R, Shukla R, Sharma TK, Bansal V. Aptamer-controlled reversible inhibition of gold nanozyme activity for pesticide sensing. Anal Chem. 2014;86:11937-41
88. Musumeci D, Platella C, Riccardi C, Moccia F, Montesarchio D. Fluorescence sensing using DNA aptamers in cancer research and clinical diagnostics. Cancers. 2017;9:174
89. Ruscito A, DeRosa MC. Small-molecule binding aptamers: Selection strategies, characterization, and applications. Front Chem. 2016;4:14
90. Pfeiffer F, Mayer G. Selection and biosensor application of aptamers for small molecules. Front Chem. 2016;4:25
91. J. R. Clarke RRM, A. Oosterveld, A. A. Frohlich, F. J. Madrid, M. Dawood. Development of a quantitative and sensitive enzyme-linked immunosorbent assay for ochratoxin A using antibodies from the yolk of the laying hen. J Agric Food Chem. 1993;41:1784-9
92. Soh JH, Lin Y, Rana S, Ying JY, Stevens MM. Colorimetric detection of small molecules in complex matrixes via target-mediated growth of aptamer-functionalized gold nanoparticles. Anal Chem. 2015;87:7644-52
93. Penner G. Commercialization of an aptamer-based diagnostic test. IVD Technology. 2012:31-7
94. Bai W, Zhu C, Liu J, Yan M, Yang S, Chen A. Gold nanoparticle-based colorimetric aptasensor for rapid detection of six organophosphorous pesticides. Environ Toxicol Chem. 2015;34:2244-9
95. Liu A, Zhang Y, Chen W, Wang X, Chen F. Gold nanoparticle-based colorimetric detection of staphylococcal enterotoxin B using ssDNA aptamers. Eur Food Res Techol. 2013;237:323-9
96. Shim WB, Kim MJ, Mun H, Kim MG. An aptamer-based dipstick assay for the rapid and simple detection of aflatoxin B1. Biosens Bioelectron. 2014;62:288-94
97. Cell separation kit (AptoCytoTM). http://www.aptsci.com/product/product_1.html
98. Protein isolation kit (AptoPrepTM). http://www.aptsci.com/product/product_2.html
99. Candia J, Cheung F, Kotliarov Y, Fantoni G, Sellers B, Griesman T. et al. Assessment of variability in the SOMAscan assay. Sci Rep. 2017;7:14248
100. De Groote MA, Nahid P, Jarlsberg L, Johnson JL, Weiner M, Muzanyi G. et al. Elucidating novel serum biomarkers associated with pulmonary tuberculosis treatment. PLoS One. 2013;8:e61002
101. Kraemer S, Vaught JD, Bock C, Gold L, Katilius E, Keeney TR. et al. From SOMAmer-based biomarker discovery to diagnostic and clinical applications: A SOMAmer-based, streamlined multiplex proteomic assay. PLoS One. 2011;6:e26332
102. Lollo B, Steele F, Gold L. Beyond antibodies: New affinity reagents to unlock the proteome. Proteomics. 2014;14:638-44
103. Mehan MR, Ayers D, Thirstrup D, Xiong W, Ostroff RM, Brody EN. et al. Protein signature of lung cancer tissues. PLoS One. 2012;7:e35157
104. Ashley SL, Xia M, Murray S, O'Dwyer DN, Grant E, White ES. et al. Six-SOMAmer index relating to immune, protease and angiogenic functions predicts progression in IPF. PLoS One. 2016;11:e0159878
105. Eid C, Palko JW, Katilius E, Santiago JG. Rapid slow off-rate modified aptamer (SOMAmer)-based detection of C-reactive protein using isotachophoresis and an ionic spacer. Anal Chem. 2015;87:6736-43
106. Hathout Y, Brody E, Clemens PR, Cripe L, DeLisle RK, Furlong P. et al. Large-scale serum protein biomarker discovery in Duchenne muscular dystrophy. Proc Natl Acad Sci U S A. 2015;112:7153-8
107. De Groote MA, Sterling DG, Hraha T, Russell TM, Green LS, Wall K. et al. Discovery and validation of a six-marker serum protein signature for the diagnosis of active pulmonary tuberculosis. J Clin Microbiol. 2017;55:3057-71
108. Jung YJ, Katilius E, Ostroff RM, Kim Y, Seok M, Lee S. et al. Development of a protein biomarker panel to detect non-small-cell lung cancer in Korea. Clin Lung Cancer. 2017;18:e99-e107
109. Ostroff RM, Bigbee WL, Franklin W, Gold L, Mehan M, Miller YE. et al. Unlocking biomarker discovery: Large scale application of aptamer proteomic technology for early detection of lung cancer. PLoS One. 2010;5:e15003
110. Russell TM, Green LS, Rice T, Kruh-Garcia NA, Dobos K, De Groote MA. et al. Potential of high-affinity, slow off-rate modified aptamer reagents for Mycobacterium tuberculosis proteins as tools for infection models and diagnostic applications. J Clin Microbiol. 2017;55:3072-88
111. Sattlecker M, Kiddle SJ, Newhouse S, Proitsi P, Nelson S, Williams S. et al. Alzheimer's disease biomarker discovery using SOMAscan multiplexed protein technology. Alzheimers Dement. 2014;10:724-34
112. Bruno JG, Phillips T, Montez T, Garcia A, Sivils JC, Mayo MW. et al. Development of a fluorescent enzyme-linked DNA aptamer-magnetic bead sandwich assay and portable fluorometer for ultrasensitive and rapid Listeria detection. J Fluorescence. 2015:25 173-83
113. Hori SI, Herrera A, Rossi JJ, Zhou J. Current advances in aptamers for cancer diagnosis and therapy. Cancers. 2018;10:9
114. Merlini PA, Ardissino D. Laboratory measurement of thrombin activity-What every clinician scientist needs to know. J Thromb Thrombolysis. 1995;2:85-92
115. Muller J, Friedrich M, Becher T, Braunstein J, Kupper T, Berdel P. et al. Monitoring of plasma levels of activated protein C using a clinically applicable oligonucleotide-based enzyme capture assay. J Thromb Haemost. 2012;10:390-8
116. Konigsbrugge O, Koder S, Riedl J, Panzer S, Pabinger I, Ay C. A new measure for in vivo thrombin activity in comparison with in vitro thrombin generation potential in patients with hyper- and hypocoagulability. Clin Exp Med. 2017;17:251-6
117. Muller J, Becher T, Braunstein J, Berdel P, Gravius S, Rohrbach F. et al. Profiling of active thrombin in human blood by supramolecular complexes. Angew Chem Int Ed Engl. 2011;50:6075-8
118. EpiMark® Hot Start Taq DNA Polymerase. https://www.neb.com/products/m0490-epimark-hot-start-taq-dna-polymerase#Product%20Information
119. Tsai SC, Hung LY, Lee GB. An integrated microfluidic system for the isolation and detection of ovarian circulating tumor cells using cell selection and enrichment methods. Biomicrofluidics. 2017;11:034122
120. JG Bruno MPC, T Phillips, CJ Andrews. A novel screening method for competitive FRET-aptamers applied to E. coli assay development. J Fluorescence. 2010 20: 1211-23
121. Cao X, Li S, Chen L, Ding H, Xu H, Huang Y. et al. Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus. Nucleic Acids Res. 2009;37(14):4621-8
122. Kim HJ, Kim HS, Lee JM, Yoon SS, Yong D. Rapid detection of Pseudomonas aeruginosa and Acinetobacter baumannii Harboring bla(VIM-2), bla(IMP-1) and bla(OXA-23) genes by using loop-mediated isothermal amplification methods. Ann Lab Med. 2016;36:15-22
123. Kim LH, Yu HW, Kim YH, Kim SI, Jang A. Potential of fluorophore labeled aptamers for Pseudomonas aeruginosa detection in drinking water. J Korean Soc Appl Biol Chem. 2013;56:165-71
124. Lavu PS, Mondal B, Ramlal S, Murali HS, Batra HV. Selection and characterization of aptamers using a modified whole cell bacterium SELEX for the detection of Salmonella enterica Serovar typhimurium. ACS Comb Sci. 2016;18:292-301
125. Park HC, Baig IA, Lee SC, Moon JY, Yoon MY. Development of ssDNA aptamers for the sensitive detection of Salmonella typhimurium and Salmonella enteritidis. Appl Biochem Biotechnol. 2014;174:793-802
126. Qin L ZR, Ma Z, Feng Y, Liu Z, Yang H, Wang J, Jin R, Lu J, Ding Y, Hu Z. The selection and application of ssDNA aptamers against MPT64 protein in Mycobacterium tuberculosis. Clin Chem Lab Med. 2009;47:405-11
127. Sypabekova M, Bekmurzayeva A, Wang R, Li Y, Nogues C, Kanayeva D. Selection, characterization, and application of DNA aptamers for detection of Mycobacterium tuberculosis secreted protein MPT64. Tuberculosis. 2017;104:70-8
128. Tang XL, Wu SM, Xie Y, Song N, Guan Q, Yuan C. et al. Generation and application of ssDNA aptamers against glycolipid antigen ManLAM of Mycobacterium tuberculosis for TB diagnosis. J Infect. 2016;72:573-86
129. Thet NT, Jenkins ATA. An electrochemical sensor concept for the detection of bacterial virulence factors from Staphylococcus aureus and Pseudomonas aeruginosa. Electrochemistry Communications. 2015;59:104-8
130. Wen he Wu, Min Li1, Yue Wang, Hou-xian Ouyang, Lin Wang C-xL, Yu-chen Cao. et al. Aptasensors for rapid detection of Escherichia coli O157:H7 and Salmonella typhimurium. Nanoscale Research Letters. 2012;7:658-65
131. Wu W, Zhao S, Mao Y, Fang Z, Lu X, Zeng L. A sensitive lateral flow biosensor for Escherichia coli O157:H7 detection based on aptamer mediated strand displacement amplification. Anal Chim Acta. 2015;861:62-8
132. Tang XL, Zhou YX, Wu SM, Pan Q, Xia B, Zhang XL. CFP10 and ESAT6 aptamers as effective Mycobacterial antigen diagnostic reagents. J Infect. 2014;69:569-80
Author contact
![]() Corresponding author: Tarun K. Sharma, tarunres.in; Telephone: +91 8800931999.
Corresponding author: Tarun K. Sharma, tarunres.in; Telephone: +91 8800931999.
 Global reach, higher impact
Global reach, higher impact