13.3
Impact Factor
Theranostics 2018; 8(10):2672-2682. doi:10.7150/thno.23624 This issue Cite
Research Paper
Estrogen receptors orchestrate cell growth and differentiation to facilitate liver regeneration
1. Graduate Institution of Clinical Medical Science, and Graduate Institute of Biomedical Sciences, China Medical University, Taichung, Taiwan 40403
2. Department of Trauma and Critical Care, Changhua Christian Hospital, Changhua, Taiwan;
3. Sex Hormone Research Center, Organ Transplantation Center, and Department of OBs & GYN, China Medical University Hospital, Taichung, Taiwan 40403.
4. Departments of Urology and Pathology, University of Rochester Medical Center, Rochester, NY 14642, USA.
5. Department of OBS & GYN, BenQ Medical Center, Nanjing Medical University, Suzhou, Jiangsu Province, China 215004.
Received 2017-11-2; Accepted 2018-3-1; Published 2018-4-9
Abstract
Background and Aims: Improving liver regeneration (LR) capacity and thereby liver function reserve is a critical bridging strategy for managing liver failure patients. Since estrogen signaling may participate in LR, our aim was to characterize the roles of ERα and ERβ in LR.
Methods: LR capacity and estradiol levels following 2/3rd partial hepatectomy (PHx) were compared in ERα-KO or ERβ-KO vs. wildtype mice. The ERα- or ERβ-related transcriptome and interactome were analyzed from regenerating livers, and then bioinformatics was used for pathway discovery and analysis of interactome-transcriptome relationships. Human hepatic progenitors (HepRG cells) and mouse Hepa1-6 hepatocytes were used to elucidate molecular interactions and functions.
Results: This paper demonstrated that estrogen signals orchestrate hepatic repopulation and differentiation via distinct transcriptome patterns governed by ERα or ERβ. Cell repopulation pathway was associated with the ERα-transcriptome, but cell differentiation and metabolic function were associated with the ERβ transcriptome. Mechanistic studies linking ERs interactomes and transcriptomes discovered that ERα-Chd1 interaction promoted cell growth by upregulating Ssxb6, Crygc, and Cst1; and, ERβ-Ube3a interaction facilitated hepatic progenitor cell differentiation to hepatocytes and cholangiocytes, specifically by upregulating Ifna5.
Conclusions: ERα and ERβ orchestrate liver cell proliferation and differentiation respectively, thereby promoting LR.
Keywords: estrogen receptor, liver regeneration, liver failure
Introduction
The high mortality in patients with acute and chronic liver failure (LF) is attributable to multi-organ dysfunction, primarily liver dysfunction, caused by multiple factors, e.g., ingested toxins, hepatitis virus infection, drug misusage, etc. [1]. Although the most effective management of LF is liver transplantation, the shortage of donors is the bottleneck [2]. In the rodent LR model, particularly the early phase LR of 2/3rd PHx, the loss of liver weight can be partially recovered within the first days with hepatocyte hypertrophy and later recovered via hepatocyte proliferation [3]. In the 1st to 2nd days after 2/3rd PHx, the cell cycle in the regenerating liver is unconventional (intermittent S-M phase [4]), often yielding binucleated cells without significant apoptosis [5, 6]. Miyaoka et al. [5] found that the hepatocytes in LR become hypertrophic and undergo changes, e.g., cell size enlargement [5], lysosomal vacuolation [7], and endocytic uptake of nutrients [8]. When the liver is diseased or stressed, the lack of nutrients, e.g., hypolipids, in hepatocytes can result in the formation of microbodies [9] or large vacuoles [10]. Caveolin-1 (needed for caveolae formation) is abundantly expressed during LR [11, 12] to transport glucose and lipids [12]. The lack of coordination between cell growth, cellular hypertrophy, and increased formation of microbodies in the regenerating liver could disrupt LR or result in impaired liver function recovery [13].
The correlation between an increase of nuclear estrogen receptor (ER) with the onset of DNA synthesis in LR has been reported for decades [14-16]. It has also been reported that short-term estradiol treatment may initiate or facilitate LR after PHx [16]. However, it is argued that the additional estradiol administration could have a limited effect on enabling more hepatic regeneration due to estrogen spike in the early phase of LR in rodent models [17], hence the importance in searching for ER's mechanistic applicability in liver regeneration.
There are two classical estrogen receptors, ERα and ERβ [18], which bind estradiol, then translocate to the nucleus where they cooperate with association proteins to bind to their corresponding DNA sequences [19, 20]. ERα and ERβ have several similar characteristics including: high protein homology, high E2 affinities, and similar estrogen response element (ERE) sequences to facilitate target gene expression. Yet, they also are different, which sometimes results in opposing biological effects [21]. In a recent study, Batmunkh et al. demonstrated the co-expression of ERα and PCNA in regenerating livers of Wistar rats, and ERα was expressed in a spatial-temporal manner in hepatic zones 1 and 2 (periportal and transition zones) in male rats and all hepatic zones in female rats when homeostasis was established. The detection of ERα was weak in hepatic zone 1 before 2/3rd PHx in male Wistar rats, then gradually increased in all hepatic zones 48-168 h after 2/3rd PHx. And, ERα expression can be detected in all hepatic zones at all sampling time-points after 2/3rd PHx in female Wistar rats [22]. lvaro et al. showed low expression of ERα and no expression of ERβ in male and female rat hepatocytes [23]. Yet, ERβ was detectable in cholangiocytes under certain stress conditions [24]; therefore, the role of ERβ in LR has been neglected. Since the estrogen signal is important in LR and the interactomes-transcriptomes of ERα/ERβ in LR remain unclear, this study differentiated between the roles of ERα and ERβ in LR. Using ERKO mouse models and an unbiased bioinformatics approach, we elucidated the relationship of ERs to transcriptome diversity in regenerating livers.
Methods
Chemicals and cell line maintenance
The human hepatoma cell line Hepa1-6 was obtained from ATCC (CRL-1830), HepaRG from Invitrogen (San Diego, CA; HPRGC10), and Hs68 (#60038) and HEK293T cells (#60210) from Food Industry Research and Development Institute in Hsinchu, Taiwan. The cells were cultured in Dulbecco's modified Eagle's media containing 1 mg/mL D-glucose and supplemented with 0.3 mg/mL L-glutamine and 10% FBS (Invitrogen). All cells were maintained in a 5% CO2 humidified incubator at 37 °C. The 1,3,5-tris (4-hydroxyphenyl)-4-propyl-1H-pyrazole (PPT (an ERα agonist) dissolved in dimethyl sulfoxide (DMSO)), 2,3-bis (4-hydroxyphenyl) propionitrile (DPN (an ERβ agonist) dissolved in DMSO), and 17β-estradiol (E2 dissolved in ethanol) were purchased from Sigma-Aldrich (St. Louis, MO). Other media included hiHeps induction medium (described below) and hepatocyte maintenance media (HMM consisting of DMEM/F12 (Gibco) supplemented with 0.544 mg/L ZnCl2, 0.75 mg/L ZnSO4·7H2O, 0.2 mg/L CuSO4·5H2O, 0.025 mg/L MnSO4, 2 g/L bovine serum albumin, 2 g/L galactose, 0.1 g/L ornithine, 0.03 g/L proline, 0.61 g/L nicotinamide, 1X insulin-transferrin-sodium selenite media supplement, 40 ng/mL TGFα, 40 ng/mL EGF, 10 μM dexamethasone, and 1% fetal bovine serum (all from Sigma-Aldrich).
Use of experimental animals, and the generation of ERα-KO and ERβ-KO mice
All of the animal experiments followed the Guidance of the Care and Use of Laboratory Animals of the Ministry of Sciences and Technology, with approval from the China Medical University. (Approval number #103-36-N) The ERα-KO mice (ActbCre-ERαloxP/loxP) and ERβ-KO mice (ERβ-/-) used in our study were kindly provided by Prof. Shuyuan Yeh and Prof. Chawnshang Chang, respectively, University of Rochester, NY, USA [25, 26]. In principle [27], transgenic ERαloxP/loxP mice were crossed with Actb-Cre (β-actin promoter-driven Cre recombinase) transgenic mice to generate male ERα knockout (ERα-/-) mice. The control mice were ERαloxP/loxP without Actb-Cre. ERβ knockout (ERβ-/-) mice were generated by crossing heterozygote (ERβ+/-) mice and littermate wildtype (ERβ+/+) mice. PCR was used to identify mouse genotypes from DNA obtained from tail skin treated overnight with cell lysis buffer containing 0.5 mg/mL proteinase K (Sigma, P2308). All wildtype vs. ERα-/- or ERβ-/- mice used in these studies were 2-4 months old and male. All protocols related to animal use and treatment were evaluated and approved by the Animal Care and Use Committee of China Medical University, and all animals were treated in accordance with National Laboratory for Experimental Animals guidelines.
Gene expression assays
Gene expression assays [28], like RT-PCR and western blot, were performed after mining the cDNA microarray data. Samples for quantitative RT-PCR were solubilized in Trizol (Invitrogen, Carlsbad, CA) and a kit was used to isolate RNA according to the manufacturer's protocol. The BluePrint RT reagent kit (Takara, Tokyo, Japan) was used to reverse transcribe 1 µg RNA, and quantitative PCR with the CFX96 Real-time System (Bio-Rad) and SYBR Green Supermix (Bio-Rad) was used to measure cDNA levels. Fold changes in gene expression were determined by quantitation of cDNA in target (treated) samples relative to expression in a calibrator sample (vehicle).
Lentiviral-based gene expression
The cDNAs or shRNA (HNF4α, OriGene #RC217863L2; HNF1α, OriGene #RC211201L2; FOXOA3, OriGene #RC202363L2; ERα, addgene #RC213277L2; ERβ, addgene #RC218519L2; Chd1 and Ube3a [MOST-RNAi core: Chd1: TRCN0000096526; Ube3a: TRCN0000012893]) were sub-cloned into modified pLenti-puro plasmid (addgene) for over-expression and pLKO.1 for knockdown. The lentiviral gene transduction procedure was previously described [28]. In brief, the gene plasmids were then transfected into 293T cells together with packaging plasmid psPAX2 and envelope plasmid pMD2.G. After a 48-h incubation, the medium containing lentiviruses was collected, filtered through a 0.45-μm filter, and then added to cells with 4 μg/mL polybrene for 24 h. Stably transfected clones were selected by puromycin (5 μg/mL).
Two-third partial hepatectomy (2/3rd PHx)
Two-third partial hepatectomy (2/3rd PHx) was performed using a large abdominal incision and two separate ligatures [29]. In brief, the skin of 10-12-week-old isoflurane-anesthetized mice was disinfected (10% povidone-iodine), incised, and gently pulled down with a saline-moistened cotton tip to expose the median lobe of the liver with a saline-moistened cotton tip. The falciform ligament or membrane was cut with curved microsurgery scissors and 3-0 silk thread was placed at the base of the left lateral and median lobes with microdissection forceps. A cotton tip was used to rotate the left lateral or median lobe to its original position. While holding its right end with the micro-dissecting forceps, the suture thread was wrapped around the lobes as close to the base of the lobe as possible and knotted. Using the microsurgery curved scissors, the tied lobe was cut just above the suture; a 3-0 suture was used to close the peritoneum and 4-0 sutures were used to close the skin. Finally, the animal was placed on a warming pad for recovery and then housed in individual cages. Re-generated livers and blood were obtained by sacrificing mice at the 2nd and 4th days post-surgery.
cDNA microarray to investigate transcriptomic changes
RNA extraction followed a standard extraction procedure using TriZol reagent (Invitrogen, USA). The extracted RNAs were sent to Agilent Technologies (Foster City, CA) for microarray analysis. Briefly, 0.2 μg of total RNA was amplified using a Low Input Quick-Amp Labeling kit (Agilent Technologies) and labeled with Cy3 (CyDye, Agilent Technologies) during the in vitro transcription process. About 0.6 μg of Cy3-labeled cRNA was fragmented to an average size of ~50-100 nucleotides by incubation with fragmentation buffer at 60 °C for 30 min. The fragmented labeled cRNA was pooled and hybridized to an Agilent SurePrint G3 Mouse GE 8×60K Microarray (Agilent Technologies) at 65 °C for 17 h. The microarray was washed, dried with a nitrogen gun, and scanned with an Agilent microarray scanner (Agilent Technologies) at 535 nm for Cy3. Scanned images were analyzed by Feature Extraction 10.5.1.1 software (Agilent Technologies), and the signal and background intensity of each feature were quantified using image analysis and normalization software.
Proteomics for ERs interactome analysis
Immunoprecipitation (IP): The antibodies included anti-ERα (GeneTex Inc., Irvine, CA; GTX22746) and anti-ERβ (Santa Cruz Biotechnology, Santa Cruz, CA; sc-8974). The protein fraction from 1 mg of wildtype mouse liver tissue was extracted using a Crosslink Magnetic IP/Co-IP Kit according to the provided protocol (Pierce® Crosslink Magnetic IP/Co-IP Kit; Cat.#88805), precleared for 1 h with protein-A beads, and added to and allowed to react overnight with 5 μg of antibody pre-bound to protein A /G magnetic beads. The beads were washed with IP Lysis/Wash Buffer (pH 7.4, 25 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% NP40, 5% glycerol) and then with elution buffer.
Western Blot Analysis: The eluted proteins (50 μg) were denatured, subjected to 12% SDS-PAGE and transferred to a nitrocellulose membrane, which was incubated first with primary antibodies and then with secondary antibodies (HRP-conjugated Protein A; ThermoFisher Scientific, Rockford, IL). The immunoblotted protein signals were detected by enhanced chemiluminescence (ECL kit; GE Healthcare, Munich, Germany) and measured from photographs taken with a light-sensitive charge-coupled camera (ChemiDocXRS, Bio-Rad, Hercules, CA).
In-gel digestion and Zip-tip purification: The immunoprecipitated proteins were separated on denaturing 6% SDS-PAGE gels. After electrophoresis, the gels were stained with Coomassie blue for 3 h and washed with destaining buffer for 60 min to remove background staining. The Coomassie brilliant blue-stained protein bands were excised, washed several times, subjected to a reduction step using 50 mM dithioerythreitol (DTE; Sigma D8161) in 25 mM ammonium bicarbonate (ABC; Sigma A6141) buffer for 60 min at 37 °C, alkylated with 100 mM iodoacetamide (IAM; Sigma I6125) in 25 mM ABC for 60 min at room temperature in the dark, digested first with 1 μL of Lys-C (0.1 μg/μL in 25 mM ABC; Wako 125-05061) for 3 h at 37 °C and second with trypsin (0.1 μg/μL in 25 mM ABC; Promega V5111) overnight at 37 °C, extracted with 50 μL of 5% trifluroacetic acid (TFA; Sigma AL299537) in 50% acetonitrile (Avantor JT Baker JT-9829), and sonicated 10 times for 10 s with 10 s on ice between sonications. Eluted peptides were recovered with 0.1% formic acid (Avantor JT Baker JT-9832), further purified using Zip-tip pipette tips (Merck Millipore, Billerica, MA; ZTC18S096), diluted in 30 μL of 0.1% formic acid/50% acetonitrile, dried with Speed Vac, resuspended in 0.1% formic acid, and analyzed directly by LC-MS/MS (Thermo LTQ-Orbitrap XL, ThermoFisher Scientific, Waltham, MA).
Bioinformatics for interactome analysis
Proteins identified by mass spectrometry were subjected to extensive bioinformatics analysis, including protein data filtering, functional profiling, and pathway mapping.
Protein data filtering: mass spectrometry data were input to iProXpress (http://pir.georgetown.edu/iproxpress).
Protein annotation: The iProXpress bioinformatics system was used for protein annotation, protein function profiling, and pathway profiling. The detailed information of functional enrichment analysis for the interaction proteins is described in our previous studies [30, 31].
Data mining for known ER-associated proteins: The global ER interaction network refers to a network of genes or proteins that directly or indirectly interact or are functionally associated with ERα or ERβ. Several bioinformatics databases were used including TFcheckpoint (http://www.tfcheckpoint.org/) to identify transcription factors; AnimalTFDB (http://www.bioguo.org/AnimalTFDB/) to identify cofactors; TRANSFAC (http://www.biobase-international.com/product/transcription-factor-binding-sites) to identify eukaryotic transcription factors, consensus binding sequences (positional weight matrices), and regulated genes; and FIMO (http://meme-suite.org/doc/fimo.html?man_type=web) from MEME suite to match database sequences with ERα and ERβ motifs.
Statistics
Student's t-test or chi-square analysis were used to identify significant differences between groups or categorical variables. A p-value less than 0.05 was considered significant. All data are reported as the mean ± standard error of the mean (SEM).
Results
ERα and ERβ promotes LR via differential transcriptome reprogramming
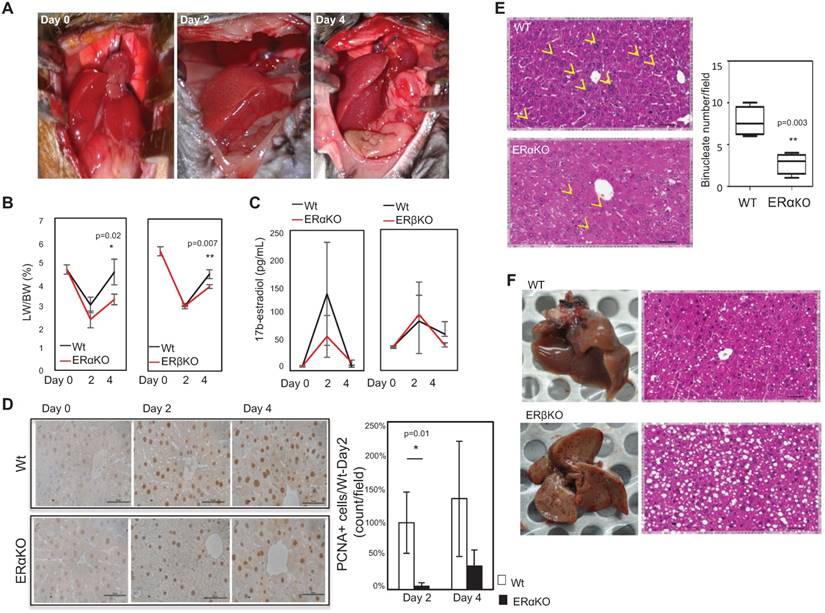
To directly examine the roles of ERα and ERβ in LR, we performed 2/3rd partial hepatectomy (PHx) surgery in mice (Figure 1A, left column). We observed LR capacity and harvested for gene expression at post-surgery day 2 (Figure 1A, middle column) and day 4 (Figure 1A, right column) in WT. vs. ERα-KO or ERβ-KO. We also measured serum estradiol levels and compared liver weight/body weight (LW/BW) (Figure 1B). The LR was reduced from 5.17±0.67% to 3.73±0.28% (p=0.02) in ERα KO mice and 4.10±0.23% to 3.45±0.1% in ERβ KO mice (p=0.007). On the other hand, the serum estradiol level surged at 2 days post-surgery and declined at 4 days post-surgery, but a comparison of serum estradiol profiles between WT vs. ERα- or ERβ-KO mice revealed no significant differences (Figure 1C). This data suggested that ERα and ERβ, but not estrogen level, play critical roles in LR. As we examined the proliferation marker PCNA in regenerating livers at post-surgery day 2 and day 4, we found PCNA-positive stains were increased at both time points. Yet, knockout of ERα decreased PCNA positivity compared to their WT littermates (Figure 1D). While examining the gross and microscopic histology of livers from ERα or ERβ-KO mice, we found that ERα-KO mice compared to their WT littermates had significantly fewer binucleated cells on day 2 post-surgery (Figure 1E) and a significantly greater vacuolated area (Figure 1F). As we compared the binucleated cell numbers, we observed a comparable degree of binucleated cell numbers in ERα-KO vs. ERβ-KO mice (Figure S1), indicating a distinct phenotype of ERα and ERβ contributing to LR.
Knockout of ERs reduced liver regeneration (LR) capacity after 2/3rd PHx surgery. (A) Photographic images showing successful 2/3rd PHx surgery (left) and liver regrowth at day 2 (middle) and day 4 (right) post-surgery. (B) LR capacity was lower in ActB Cre-ERαF/F (ERαKO) and ERβ-/- (ERβKO) mice compared to their wildtype (WT) littermates (ActB Cre- ERα+/+ and ERβ+/+, respectively). The liver weight/body weight (LW/BW; %) ratio was around 5.3% before surgery, declined significantly in ERα-KO mice compared to WT at day 2 (n=8 WT mice vs. 15 ERα-KO mice) and 4 days (n=6 WT mice vs. 9 ERα-KO mice) (p=0.02), and declined significantly in ERβKO mice compared to WT post-surgery (p=0.007; day 2, n=6 WT mice vs. 6 ERβKO mice; day 4, n=4 WT mice vs. 7 ERβKO mice. (C) Estradiol levels remained unchanged during LR. The serum estradiol was significantly increased at day 2 and decreased at day 4; it was similar between ERα- and ERβ-KO male mice vs. WT male mice. (D) The immunohistochemistry (IHC) staining of PCNA decreased in ERα-KO regenerating livers compared to their WT littermates. Left-hand side: representative photos of PCNA IHC staining (Scale bar=50 μm). Upper (WT) and lower (ERα-KO) panels show images of livers at day 0, post-surgery day 2, and day 4. Right-hand side: quantitation of PCNA+ cells/field under a microscope (400x), and normalized to post-surgery WT livers. The data were collected from 3 fields-of-view from each slide, and at least 6 mice/group were measured. (E) The number of binucleated cells was significantly decreased in ERα-KO mice compared to WT (p=0.003). The yellow arrowhead indicates the location of binucleated cells. These results are from five different slides of liver sections viewed at the same magnification (Scale bar=50 μm). (F) The vacuolation was dramatically increased in ERβ-KO mice compared to WT mice. Gross and histological morphology of the liver from a WT mouse at day 2 post-surgery (upper panel) and gross and histological morphology of a liver from an ERβ-KO mouse at day 2 post-surgery (lower panel).
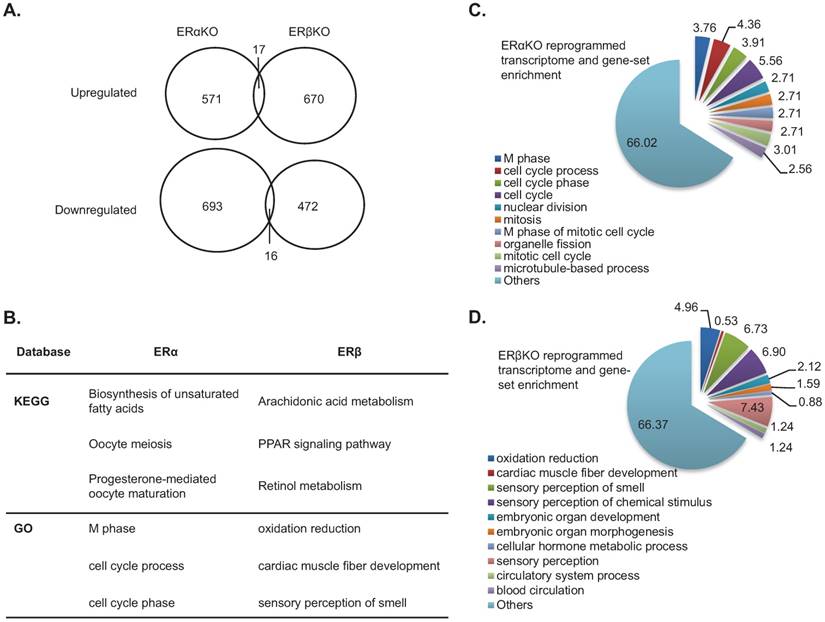
Since the phenotype contributed by ERs KO exerts significant alteration in LR capacity, we would like to dissect the mechanism on these two molecules in regenerating livers. We observed that the alteration amplitude is most dramatic at day 4 after 2/3rd PHx surgery; therefore, we would like to use this time point for measuring transcriptomes and interactomes. Comparing the transcriptomes of regenerating livers (day 4 post-surgery) between ERα- or ERβ-KO mice and their WT littermates, we showed that 588 and 687 genes were upregulated, and 709 and 488 were downregulated in ERα- or ERβ-KO mice, respectively (Figure 2A). Interestingly, ERα and ERβ is commonly thought to upregulate 17 of these genes and downregulate 16 of these genes. To interpret the transcriptome changes governed by ERα or ERβ, we used three database platforms (KEGG and GO) for pathway enrichment analysis. As shown in Figure 2B, the ERα-related pathway genes were involved in cell growth (meiosis, M phase of the cell cycle, cell cycle regulation, etc.) and the ERβ-related pathway were involved in cell differentiation and metabolism (arachidonate/retinol metabolism, oxidation reduction, etc.). The gene names of the KEGG pathway are listed in Table S1 and Table S2. Using the GSEA platform, we confirmed that around 1/3 of the genes in the ERα-related and ERβ-related pathways are involved in cell cycle/growth (Figure 2C) and cell function (Figure 2D), respectively.
Impact of ERα and ERβ knockout on the transcriptome of regenerating livers. (A) mRNA expression increased (upper circles) or decreased (lower circles) more than 2-fold in regenerating livers from ERα-KO and ERβ-KO mice compared to their wildtype littermates. (B) Pathway analyses of LR transcriptomes of ERα-KO and ERβ-KO mice compared to their wildtype littermates. Analysis using two databases (KEGG [detailed pathway analysis is shown in Table S1 and Table S2], and GO) found that ERα had major cell cycle and growth impacts, and ERβ affected cell differentiation/cell function. (C-D) The top 10 GSEA pathways enriched in regenerating liver transcriptomes of ERα- (C) and ERβ- (D) knockout mice compared to their wildtype littermates.
Distinct ERα/ERβ complexes regulate cell growth and function
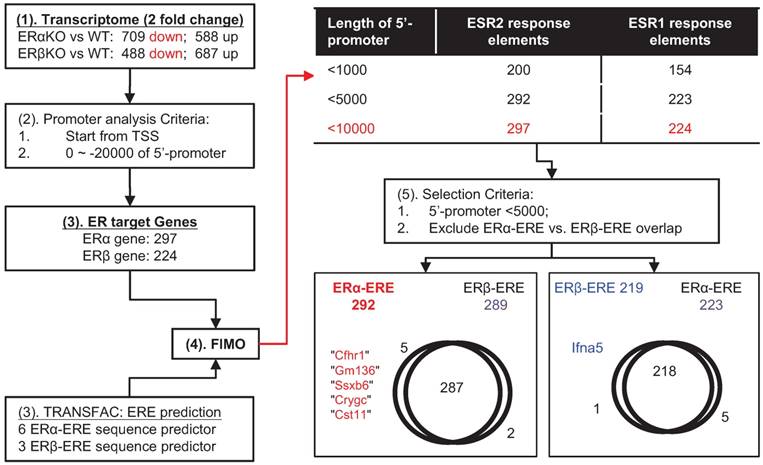
ERα and ERβ are transcription factors that interact with co-regulators and bind to similar EREs. Interestingly, the distinctness of ERα or ERβ transcriptomes (Figure 3) suggests that differences in transcriptomes of ERα or ERβ might result in functional differences. To test this idea, we identified the putative direct targets of ERα or ERβ (Figure 3 and Figure 4). First (Figure 3), we choose two-fold downregulated genes in the transcriptomes of the ERα- and ERβ-KO mice (ERα: 709 and ERβ: 488). There were 297 ERα-related and 224 ERβ-related genes identified using the following criteria: 1) the genes contain transcription start sites (TSSs); 2) the 5'-promoter sequence is located 0 to -20,000 b.p. from the TSS; and, 3) the sequences obtained using R-Software align with known sequences of ERα-ERE and ERβ-ERE (6 and 3 kinds; Figure S2) obtained using the FIMO platform. Further data analysis showed that the unique gene targets of ERα- (Cfhr1 [complement factor H related 1], Gm136 [predicted gene 136], Ssxb6 [synovial sarcoma, X member B6], Crygc [crystallin, gamma C], and Cst11 [cystatin 11]) were distinguished from the unique gene target of ERβ (Ifna5 [interferon alpha 5]) by: 1) defining EREs within -5000 bp. of the TSS as the most likely direct gene targets of ERα- and ERβ and 2) excluding the overlapping gene targets of ERα- and ERβ.
The target ERα- and ERβ-related genes predicted by a bioinformatics approach. (1) The non-overlapping downregulated genes (i.e., genes encoding transcription factors) in the ERα-KO or ERβ-KO mouse transcriptome were chosen for analysis. (2) The genes were subjected to promoter analysis (to locate promoter sequences within ~ -20 kb upstream from the transcription start-site). (3) Both R-software and TRANSFAC were used to analyze EREs (specific for ERα or ERβ; Figure S2) and found 297 genes for ERα and 224 genes for ERβ. (4) FIMO was used to locate the promoter sequences of ERα-ERE and ERβ-ERE, and most EREs were located within -5000 bp of their promoters. (5) The ERα-ERE vs. ERβ-ERE region of overlap was excluded to distinguish ERα-specific from ERβ-specific target genes. The final result identified Cfhr1, Gm136, Ssx6, Cryc, and Cst11 as ERα-specific and Ifna5 as ERβ-specific.
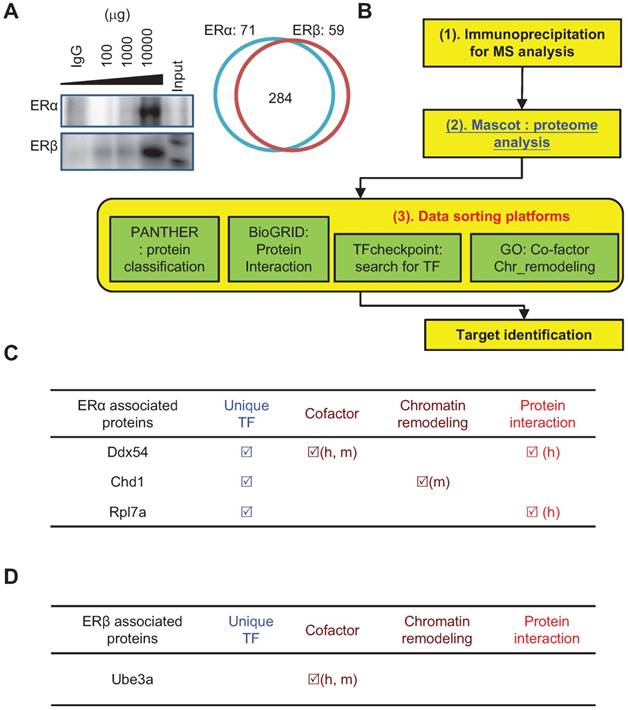
Then, ERα-specific and ERβ-specific protein interactions were identified using immunoprecipitation (Figure 4A; ERα: 71 and ERβ: 59) and mass spectrometry in tandem with MASCOT database searching (Figure 4B). ERs interactome analysis was conducted using four proteomics databases (PANTHER for protein function annotation; BioGRID to define protein interactions; TFcheckpoint for searching transcription factors; GO term to identify TF co-factor and chromatin remodeling) and identified three unique ERα-interacting proteins (Ddx54 [DEAD/H BOX 54], Chd1 [chromodomain helicase DNA-binding protein-1], and Rpl7a [ribosomal protein L7a]; Figure 4C) and one unique ERβ-interacting protein (Ube3a [ubiquitin-protein ligase E3A]; Figure 4D).
ERα and ERβ promote hepatic growth and differentiation through interaction with proteins and regulation of downstream target genes
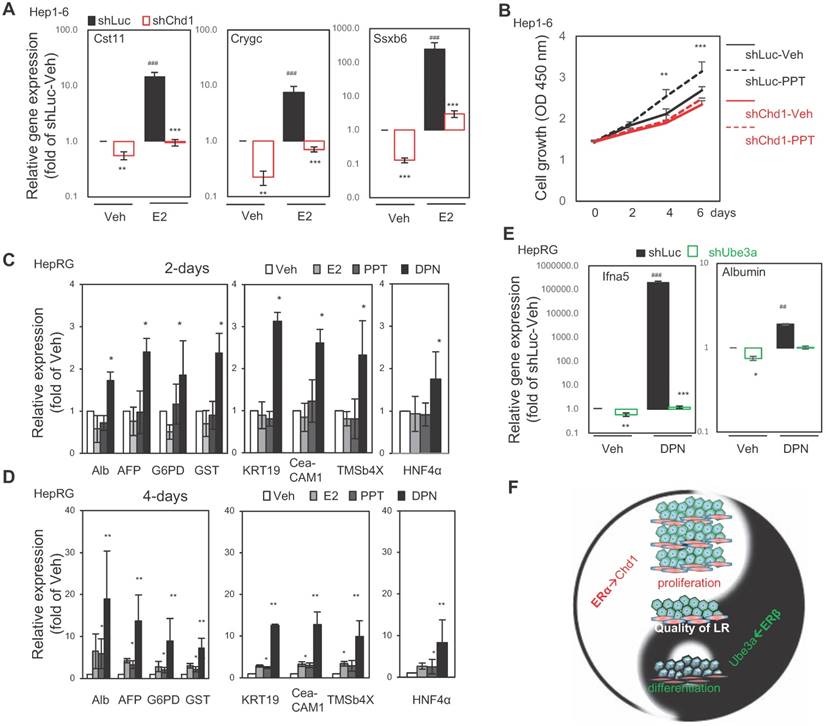
The unique direct target genes (Figure 3) and interacting proteins (Figure 4) might be involved in ERα- and ERβ-driven mechanisms underlying the LR process. Hence, we tested this hypothesis by knocking down expression of these interacting proteins in cells and measuring their target gene expression and cellular function. The expressions of Cst11, Crygc, and Ssxb6 were significantly downregulated by knockdown of Chd1 (Figure 5A, lane 1 vs. lane 2) and upregulated by treatment with E2 (Figure 5A, lane 1 vs. lane 3). Notably, E2-induced Cst11, Crygc, and Ssxb6 expression was almost completely abolished by knockdown of Chd1 (Figure 5A, lane 3 vs. lane 4). Furthermore, Hepa1-6 cell growth was significantly enhanced by treatment with PPT (ERα-specific ligand; Figure 5B, black-dashed line vs. black-solid line), and knockdown of Chd1 completely abolished this enhancement (Figure 5B, red-dash line vs. red-solid line). These data suggested a specific interaction between ERα and Chd1 to regulate genes and promote cell growth.
For examining the role of ERβ in hepatic differentiation, hepatocyte differentiation markers (albumin, Alb; alpha-fetoprotein, AFP; glucose-6-phosphate dehydrogenase, G6PD; and glutathione S-transferase, GST), cholangiocyte differentiation markers (keratin 19, KRT19; carcinoembryonic antigen-related cell adhesion molecule 1, CECAM1; and thymosin beta 4 X-linked, TMSb4x), and the liver progenitor cell marker HNF4α were monitored in human hepatic progenitor cells (HepRG) treated with E2, PPT, or DPN (ERβ specific ligand) for 2 and 4 days. After 2 days of treatment, the upregulation of hepatocyte and cholangiocyte markers and HNF4α expression was significant by DPN but not significant by E2 and PPT (Figure 5C). After 4 days of treatment, only DPN markedly upregulated these expressions (Figure 5D). Regarding the role of the ERβ-Ube3a axis in differentiation and gene expression, Ifna5 (ERβ-specific upregulated gene) and albumin (hepatocyte differentiation marker) were significantly reduced by knockdown of Ube3a (Figure 5E, lane 1 vs. lane 2) and dramatically upregulated (Figure 5E, lane 1 vs. lane 3) by treatment with DPN. Knockdown of Ube3a also abolished DPN-induced gene expression (Figure 5E, lane 3 vs. lane 4). These results suggested that Ube3a partially mediates ERβ-specific gene expression and hepatocyte differentiation.
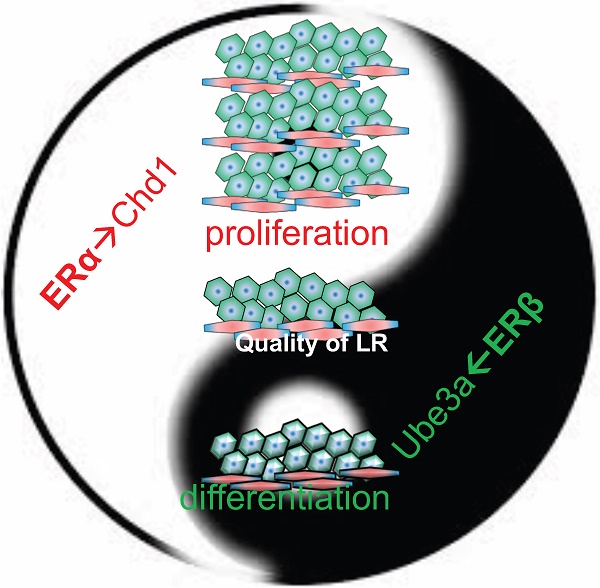
Together, the results in Figures 2-5 indicate that estrogenic signals either promote cell growth through the ERα-Chd1 axis or facilitate hepatic differentiation through the ERβ-Ube3a axis (Figure 5F). In short, ERα and ERβ expression play critical roles to ensure the quality of the tissue regenerated during LR.
Analysis of the ERα- and ERβ-specific interactomes using a bioinformatics approach. (A) Immunoprecipitation experiment demonstrated the successful pull-down of ERα or ERβ complex from regenerating livers of wildtype mice. Left panel: Immunoblots of ERα (upper) or ERβ (lower panel) complex. IgG indicates that 100 μg irrelevant primary antibody was used for IP; 100-10000 μg ERs antibody was subjected to total 10 mg of total protein extract; Input indicates that 20 μg crude extract total protein was subjected to immunoblot as immunoblot reference. Right panel: The protein numbers identified by MASCOT platform. There were 284 common ERs-associated proteins, while there were 71 ERα- and 59 ERβ-specific interacting proteins. The enrichment-based pathway ranking is listed in Figure S3. (B) ERα- and ERβ-specific interactome with a bioinformatics approach. (1) Co-IP combined with mass spectrometry revealed the ERα- and ERβ-specific proteomes. (2) Data were subjected to Mascot database searching to identify interacting proteins. (3) Data sorting with four platforms was used for function identification. These include PANTHER (for functional classification), BioGRID (for identifying interaction signal proteins), TFcheckpoint (for identifying transcription factors), and GO term analysis (for identifying transcriptional co-factor and chromatin modification proteins). (C-D) Functional annotation of the ERα (C) or ERβ (D) interactome by bioinformatics sorting discovered Dsx54, Chd1, and Rpl7a specifically interacted with ERα, and Ube3a specifically interacted with ERβ.
The ERα→Chd1 axis for cell proliferation and the ERβ→Ube3a axis for liver function/differentiation in vitro. (A) The expressions of ERα-specific target genes, Cst11, Crygc, and Ssxb6 were dramatically upregulated by E2 treatment, but abolished by the infection of Hepa1-6 hepatic cells with Chd1 shRNA-expressing lentivirus. The gene expression in shLuc (control lentivirus) infected/vehicle (Veh; ethanol)-treated cells served as baseline for expression in the experimental cells. (B) Treatment with 100 nM PPT in culture for 6 days facilitated growth of Hepa1-6 cells, but shChd1 infection totally abolished the PPT-promoted cell growth effect. (C) The expressions of hepatocyte, cholangiocyte differentiation, and hepatic progenitor marker genes were upregulated by suppressing ERβ signaling with 2-day DPN treatment in HepRG hepatic progenitor cells. Albumin (Alb), alpha-fetoprotein (AFP), glucose-6-phosphate dehydrogenase (G6PD), and glutathione S-transferase (GST) were the hepatocyte markers and keratin 19, (KRT19), carcinoembryonic antigen-related cell adhesion molecule 1 (CECAM1), and thymosin beta 4 X-linked (TMSb4x) were the cholangiocyte markers. The hepatic progenitor marker HNF4α was also measured. (D) The expressions of hepatocyte, cholangiocyte differentiation, and hepatic progenitor marker genes were upregulated by suppressing ERα signaling with 4-day E2 and PPT treatment. (E) The ERβ→Ube3a axis in hepatic gene expression (left panel) and differentiation (right panel) in HepRG cells. The expression of the target ERβ-specific gene Ifna5 was dramatically upregulated by treatment with 100 nM DPN, but abolished by infection of HepRG cells with Ube3a shRNA-expressing lentivirus. Meanwhile, the expression of hepatocyte marker albumin was increased by 2-day treatment with DPN, yet this increase was abolished by shUbe3a. The gene expression in shLuc (control lentivirus) infected/vehicle (Veh; ethanol)-treated cells served as the baseline for expression in the experimental cells. (F) A diagram showing the roles of ERα and ERβ in the process of LR. The ERα promotes LR via regulating Chd1 expression to increase hepatic cell number, whereas ERβ ensures the quality of LR via regulating Ube3a expression to facilitate hepatic progenitor cell differentiation. The cooperation between ERα and ERβ maximizes LR efficiency.
Discussion
Comparing the liver regenerating capacity in ERs-knockout and wildtype mice, we found that ERs' expressions determine estrogenic signaling during the LR process. Bioinformatic approaches revealed that ERα and ERβ orchestrate cell proliferation and differentiation, respectively, in regenerating livers. ERα and ERβ activities in LR were mediated via specific protein interactions with target genes. This report clearly demonstrated the molecular and cellular mechanisms of ERα and ERβ in LR.
ERα/ERβ expression promotes LR
It is known that resistance to hepatic damage under stressful conditions is greater in females than males [32], and that a female factor is involved in LR. Therefore, estrogen was thought to play a role in LR [33]. Interestingly, although the surge of estradiol levels after 2/3rd PHx surgery was comparable in both wildtype and ERα and ERβ knockout mice (Figure 1), the LR ability was lower in ERα and ERβ knockout mice than their wildtype littermates. Our results suggest that systemic estrogen protects against liver removal stress by possibly upregulating ER expression [34] to promote LR. In support of this, Lars Zender's group [35] reported that ERα compensates for MKK4 suppression in the hepatocytes of damaged livers.
ERα and ERβ act as quality control gatekeepers during LR
ERα is known to promote cell growth while ERβ counteracts ERα-induced hyper-proliferation in tissues such as breast and uterus [36]. The signaling pathways involved in LR are complex and interconnected [37].
Regarding the ERα→Chd1 axis, HNF4α is a transcription factor known to regulate hepatocyte differentiation and early liver development [38]. Loss of HNF4α expression results in poorly differentiated hepatocytes in fetal liver [39], and severe hepatic derangement in adults [40]. Re-expression of HNF4α attenuates liver fibrosis [41] and suppresses hepatocarcinogenesis through repression of “stemness” gene expression and induction of dedifferentiated hepatoma cell re-differentiation [40]. Chd1 is a chromatin-remodeling enzyme that may contribute to cellular reprogramming, the efficient induction of pluripotency of mouse embryonic stem cells, and iPS cell generation [42]. Furthermore, HNF4α inhibits “stemness” gene expression by suppressing Chd1 [43], which mediates the deposition of variant histone H3.3 into de-condensing chromatin during the cell cycle [44]. In our work, the ERα→Chd1 axis was revealed by measurement of HNF4α activity in undifferentiated and differentiated HepaRG cells. E2/PPT/DPN increased HNF4α activity in undifferentiated HepaRG cells, thereby decreasing Chd1 and subsequent stemness gene expression, and vice versa in differentiated HepaRG cells.
Regarding the ERβ→Ube3a axis, Ube3a (also named as E6-AP) is an E3 ubiquitin ligase and many of its protein targets are part of the ubiquitin proteasome system. The role of Ube3a in transcriptional regulation of nuclear steroid hormone receptors may contribute to the Ube3a-associated phenotypes in humans and mouse models [45]. Its functions linked to human health and disease include co-activation of steroid hormone receptors, e.g., progesterone, estrogen, etc. [46]. Co-activation involves direct binding of the UBE3A protein to the transcription complex and is independent of the ubiquitin ligase activity [47, 48]. The coactivator function usually accompanies ubiquitination and degradation of the functional initiation complex by the UPS (ubiquitin-proteasome system) after transcriptional elongation [46],[49]. In this work, we identified another important mode of ERβ action (ERβ→Ube3a axis) to promote hepatocyte differentiation.
Conclusion
This report examined the physiological role of ERα and ERβ in liver regeneration using transgenic animal models and deciphered their molecular functions using bioinformatic and cell biological approaches. This paper demonstrated that estrogen signals orchestrate hepatic repopulation and differentiation via ERα and ERβ, respectively, during LR.
Abbreviations
Alb: albumin; AFP: alpha-fetoprotein; CECAM1: carcinoembryonic antigen-related cell adhesion molecule 1; Cfhr1: complement factor H related 1; Chd1: chromodomain helicase DNA-binding protein-1; Crygc: crystallin, gamma C; Cst11: cystatin 11; Ddx54: DEAD-box helicase 54; DPN: 2,3-bis (4-hydroxyphenyl) propionitrile; E2: 17β-estradiol; ERα: estrogen receptor α; ERβ: estrogen receptor β; ERE: estrogen response element; G6PD: glucose-6-phosphate dehydrogenase; GST: glutathione S-transferase; Gm136: predicted gene 136; HNF1α: hepatocyte necrosis factor 1α; HNF4α: hepatocyte necrosis factor 4α; Ifna5: interferon alpha 5; iPS: inducible pluripotent stem; KO: knockout; KRT19: keratin 19; LF: liver failure; LR: liver regeneration; LW/BW: liver weight/body weight; MKK4: mitogen-activated protein kinase kinase 4; PHx: partial hepatectomy; PPT: 1,3,5-tris (4-hydroxyphenyl)-4-propyl- 1H-pyrazole; Rpl7a: ribosomal protein L7a; Ssxb6: synovial sarcoma, X member B6; TMSb4x: thymosin beta 4 X-linked; Ube3a: ubiquintin-protein ligase E3A; UPS: ubiquitin-proteosome system; WT: wildtype.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
MetaCore bioinformatics analysis was performed using the system provided by the Bioinformatics Core at the National Cheng Kung University, supported by the Ministry Of Sciences and Technology (MOST), Taiwan.
Author contributions
TL Kao executed most of the experiments, and drafted the manuscript. YP Kuan performed the in vitro study, assisted with animal breeding, and edited the manuscript. WC Cheng assisted and led the bioinformatics study. S Yeh provided transgenic animals. LB Jeng supported the project and edited the manuscript. WL Ma initiated, coordinated, supported this project, and edited the final approved manuscript.
Grant Support
This work was supported by grants from the Taiwan Ministry of Sciences and Technology (MOST104-2628-B-039-001-MY4); Taiwan National Health Research Institute (NHRI-EX107-10705BI); China Medical University/Hospital (CMU102-S-15; CMU105-S-41; CMU-106-S-30); and Taiwan Ministry of Health and Welfare Clinical Trial and Research Center of Excellence (MOHW106-TDU-B-212-133019).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bernal W, Wendon J. Acute liver failure. N Engl J Med. 2013;369:2525-34
2. Stravitz RT, Kramer DJ. Management of acute liver failure. Nat Rev Gastroenterol Hepatol. 2009;6:542-53
3. Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213:286-300
4. Satyanarayana A, Wiemann SU, Buer J, Lauber J, Dittmar KE, Wustefeld T. et al. Telomere shortening impairs organ regeneration by inhibiting cell cycle re-entry of a subpopulation of cells. EMBO J. 2003;22:4003-13
5. Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, Miyajima A. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr Biol. 2012;22:1166-75
6. Ding BS, Nolan DJ, Butler JM, James D, Babazadeh AO, Rosenwaks Z. et al. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. 2010;468:310-5
7. Verity MA, Travis G, Cheung M. Lysosome-vacuolar system reactivity during early cell regeneration. Exp Mol Pathol. 1975;22:73-84
8. Mori M, Novikoff AB. Induction of pinocytosis in rat hepatocytes by partial hepatectomy. J Cell Biol. 1977;72:695-706
9. Svoboda D, Grady H, Azarnoff D. Microbodies in experimentally altered cells. J Cell Biol. 1967;35:127-52
10. Matsunaga T, Toba M, Teramoto T, Mizuya M, Aikawa K, Ohmori S. Formation of large vacuoles induced by cooperative effects of oncostatin M and dexamethasone in human fetal liver cells. Med Mol Morphol. 2008;41:53-8
11. Fernandez-Rojo MA, Ramm GA. Caveolin-1 Function in Liver Physiology and Disease. Trends Mol Med. 2016;22:889-904
12. Fernandez-Rojo MA, Restall C, Ferguson C, Martel N, Martin S, Bosch M. et al. Caveolin-1 orchestrates the balance between glucose and lipid-dependent energy metabolism: implications for liver regeneration. Hepatology. 2012;55:1574-84
13. Stanger BZ. Cellular homeostasis and repair in the mammalian liver. Annu Rev Physiol. 2015;77:179-200
14. Francavilla A, di Leo A, Eagon PK, Wu SQ, Ove P, van Thiel DH. et al. Regenerating rat liver: correlations between estrogen receptor localization and deoxyribonucleic acid synthesis. Gastroenterology. 1984;86:552-7
15. Fisher B, Gunduz N, Saffer EA, Zheng S. Relation of estrogen and its receptor to rat liver growth and regeneration. Cancer research. 1984;44:2410-5
16. Chiu EJ, Lin HL, Chi CW, Liu TY, Lui WY. Estrogen therapy for hepatectomy patients with poor liver function? Medical hypotheses. 2002;58:516-8
17. Liddle C, Farrell GC. Role of the oestrogen receptor in liver regeneration in the male rat. J Gastroenterol Hepatol. 1993;8:524-9
18. O'Malley BW. Molecular biology. Little molecules with big goals. Science. 2006;313:1749-50
19. Auboeuf D, Honig A, Berget SM, O'Malley BW. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 2002;298:416-9
20. McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465-74
21. Couse JF, Hewitt SC, Bunch DO, Sar M, Walker VR, Davis BJ. et al. Postnatal sex reversal of the ovaries in mice lacking estrogen receptors alpha and beta. Science. 1999;286:2328-31
22. Batmunkh B, Choijookhuu N, Srisowanna N, Byambatsogt U, Synn Oo P, Noor Ali M. et al. Estrogen Accelerates Cell Proliferation through Estrogen Receptor alpha during Rat Liver Regeneration after Partial Hepatectomy. Acta histochemica et cytochemica. 2017;50:39-48
23. Alvaro D, Alpini G, Onori P, Franchitto A, Glaser SS, Le Sage G. et al. Alfa and beta estrogen receptors and the biliary tree. Mol Cell Endocrinol. 2002;193:105-8
24. Meyer SK, Probert PM, Lakey AK, Leitch AC, Blake LI, Jowsey PA. et al. Environmental Xenoestrogens Super-Activate a Variant Murine ER Beta in Cholangiocytes. Toxicol Sci. 2017;156:54-71
25. Chen M, Yeh CR, Chang HC, Vitkus S, Wen XQ, Bhowmick NA. et al. Loss of epithelial oestrogen receptor alpha inhibits oestrogen-stimulated prostate proliferation and squamous metaplasia via in vivo tissue selective knockout models. J Pathol. 2012;226:17-27
26. Hsu I, Chuang KL, Slavin S, Da J, Lim WX, Pang ST. et al. Suppression of ERbeta signaling via ERbeta knockout or antagonist protects against bladder cancer development. Carcinogenesis. 2014;35:651-61
27. Ma WL, Hsu CL, Wu MH, Wu CT, Wu CC, Lai JJ. et al. Androgen receptor is a new potential therapeutic target for the treatment of hepatocellular carcinoma. Gastroenterology. 2008;135:947-55 55 e1-5
28. Ma WL, Hsu CL, Yeh CC, Wu MH, Huang CK, Jeng LB. et al. Hepatic androgen receptor suppresses hepatocellular carcinoma metastasis through modulation of cell migration and anoikis. Hepatology. 2012;56:176-85
29. Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3:1167-70
30. Chung IF, Chen CY, Su SC, Li CY, Wu KJ, Wang HW. et al. DriverDBv2: a database for human cancer driver gene research. Nucleic Acids Res. 2016;44:D975-9
31. Cheng WC, Chung IF, Chen CY, Sun HJ, Fen JJ, Tang WC. et al. DriverDB: an exome sequencing database for cancer driver gene identification. Nucleic Acids Res. 2014;42:D1048-54
32. Yokoyama Y, Nagino M, Nimura Y. Which gender is better positioned in the process of liver surgery? Male or female? Surg Today. 2007;37:823-30
33. Umeda M, Hiramoto M, Imai T. Partial hepatectomy induces delayed hepatocyte proliferation and normal liver regeneration in ovariectomized mice. Clin Exp Gastroenterol. 2015;8:175-82
34. Uebi T, Umeda M, Imai T. Estrogen induces estrogen receptor alpha expression and hepatocyte proliferation in the livers of male mice. Genes Cells. 2015;20:217-23
35. Wuestefeld T, Pesic M, Rudalska R, Dauch D, Longerich T, Kang TW. et al. A Direct in vivo RNAi screen identifies MKK4 as a key regulator of liver regeneration. Cell. 2013;153:389-401
36. Paterni I, Granchi C, Katzenellenbogen JA, Minutolo F. Estrogen receptors alpha (ERalpha) and beta (ERbeta): subtype-selective ligands and clinical potential. Steroids. 2014;90:13-29
37. Mao SA, Glorioso JM, Nyberg SL. Liver regeneration. Transl Res. 2014;163:352-62
38. Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev. 2006;20:2293-305
39. Jixuan Li GN, Stephen A. Duncan. Mammalian hepatocyte differentiation requires the transcription factor HNF-4α. Genes Dev. 2000;14:464-74
40. Lu H. Crosstalk of HNF4alpha with extracellular and intracellular signaling pathways in the regulation of hepatic metabolism of drugs and lipids. Acta Pharm Sin B. 2016;6:393-408
41. Yue HY, Yin C, Hou JL, Zeng X, Chen YX, Zhong W. et al. Hepatocyte nuclear factor 4alpha attenuates hepatic fibrosis in rats. Gut. 2010;59:236-46
42. Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A. et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863-8
43. Lu H, Lei X, Liu J, Klaassen C. Regulation of hepatic microRNA expression by hepatocyte nuclear factor 4 alpha. World J Hepatol. 2017;9:191-208
44. Konev AY, Tribus M, Park SY. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007;317:1087-90
45. LaSalle JM, Reiter LT, Chamberlain SJ. Epigenetic regulation of UBE3A and roles in human neurodevelopmental disorders. Epigenomics. 2015;7:1213-28
46. Ramamoorthy S, Nawaz Z. E6-associated protein (E6-AP) is a dual function coactivator of steroid hormone receptors. Nucl Recept Signal. 2008;6:e006
47. Nawaz Z, Lonard DM, Smith CL, Lev-Lehman E, Tsai SY, Tsai MJ. et al. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol Cell Biol. 1999;19:1182-9
48. Nawaz Z, Lonard DM, Dennis AP, Smith CL, O'Malley BW. Proteasome-dependent degradation of the human estrogen receptor. Proc Natl Acad Sci USA. 1999;96:1858-62
49. Catoe HW, Nawaz Z. E6-AP facilitates efficient transcription at estrogen responsive promoters through recruitment of chromatin modifiers. Steroids. 2011;76:897-902
Author contact
![]() Corresponding author: Wen-Lung Ma, PhD. Tel: +886-975681032; email: maverickcmu.edu.tw; Address: 91 Xueshi Rd., North District, Taichung, Taiwan 40403. Shuyuan Yeh, PhD. email: shuyuan_yehrochester.edu
Corresponding author: Wen-Lung Ma, PhD. Tel: +886-975681032; email: maverickcmu.edu.tw; Address: 91 Xueshi Rd., North District, Taichung, Taiwan 40403. Shuyuan Yeh, PhD. email: shuyuan_yehrochester.edu
 Global reach, higher impact
Global reach, higher impact