13.3
Impact Factor
Theranostics 2018; 8(10):2646-2656. doi:10.7150/thno.24480 This issue Cite
Research Paper
Catalytic hairpin assembly gel assay for multiple and sensitive microRNA detection
1. Research Center for Bioengineering and Sensing Technology, Beijing Key Lab for Bioengineering and Sensing Technology, School of Chemistry and bioengineering, University of Science & Technology Beijing, Beijing 100083, P.R. China.
2. School of Petrochemical Engineering, School of Food Science and Technology, Changzhou University, Changzhou 213164, P.R. China.
3. School of Computer and Communication Engineering, University of Science & Technology Beijing, Beijing 100083, P.R. China.
Received 2017-12-21; Accepted 2018-2-21; Published 2018-4-3
Abstract
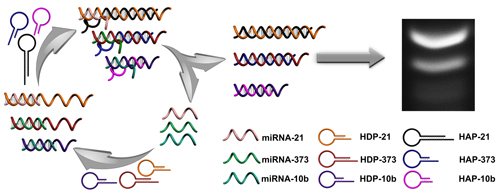
As important modulators of gene expression, microRNAs (miRNAs) have been identified as promising biomarkers with powerful predictive value in diagnosis and prognosis for several diseases, especially for cancers. Here we report a facile, multiple and sensitive miRNA detection method that uses conventional gel electrophoresis and catalytic hairpin assembly (CHA) system without any complex nanomaterials or enzymatic amplification.
Methods: In this study, three pairs of hairpin probes are rationally designed with thermodynamically and kinetically preferable feasibility for the CHA process. In the present of target miRNA, the stem of the corresponding hairpin detection probe (HDP) will be unfolded and expose the concealed domain. The corresponding hairpin assistant probe (HAP) then replaces the hybridized target miRNA to form specific HDP/HAP complexes and releases miRNA based on thermodynamically driven entropy gain process, and the released miRNA triggers the next recycle to produce tremendous corresponding HDP/HAP complexes.
Results: The results showed that the CHA gel assay can detect miRNA at fM levels and shows good capability of discriminating miRNA family members and base-mismatched miRNAs. It is able to analyze miRNAs extracted from cell lysates, which are consistent with the results of conventional polymerase chain reaction (PCR) method. Depending on the length of the designed hairpin probes, the CHA gel assay consisting of different hairpin probes effectively discriminated and simultaneously detected multiple miRNAs in homogenous solution and miRNAs extracted from cell lysates.
Conclusion: The work highlights the practical use of a conventional gel electrophoresis for sensitive interesting nucleic acid sequences detection.
Keywords: microRNA, multiple microRNA detection, catalytic hairpin assembly, gel electrophoresis, PCR
Introduction
microRNA (miRNA) are single-stranded, short (approximately 19-23 nucleotides), endogenous, noncoding regulatory RNAs that are crucial in many biological processes [1-3]. Aberrant expression levels of miRNAs are closely associated with many diseases, including cancers [4,5]. For example, miRNA-373 is validated as a potential novel oncogene for tumorigenesis, regulating tumor cell cycle and tumor metastasis [6-10]. High miRNA-21 expression levels are observed in various malignancies including breast cancer, colorectal cancer, lung cancer, stomach cancer and glioma [11,12]. In recent years, considerable efforts have been devoted to develop methods to quantify intracellular miRNA-21 [13,14]. Overexpression of miRNA-10b can promote the metastasis of cancer cells [15]. Therefore, miRNA is emerging as one of the promising biomarkers for early diagnosis and prognosis of several cancers [16,17]. However, multiple and sensitive miRNA detection is challenging because of their unique properties, such as small size, high sequence similarity, susceptibility to degradation and, especially, low abundance [18-20]. Conventional miRNA analysis methods, including real-time polymerase chain reaction (RT-PCR) [21], Northern blot [22], microarrays technology [23] have been widely used, which suffer from some defects such as low sensitivity, time-consuming methods, false positive signal and expensive equipment. Therefore, the development of a sensitive, specific and convenient strategy is urgently needed to monitor miRNA levels in early clinical diagnosis [24-28]. Especially, the multiplicity of miRNA needs a miRNA assay that detects multiple miRNA simultaneously.
Recently, a variety of amplification strategies have been established for sensitive detection of miRNA, such as nanomaterial-based [29,30], and molecular biology-involved signal amplification methods including target RNA cycling, ligase chain reaction (LCR), rolling-circle amplification (RCA), isothermal strand-displacement polymerase reaction, and strand displacement amplification (SDA) [31-33]. As a versatile SDA isothermal amplification technique, catalytic hairpin assembly (CHA) is an enzyme-free DNA circuit that relies on a thermodynamically driven entropy gain process to achieve signal amplification, possibly permitting exponential amplification of a target sequence in 15 min [34-36]. By combining CHA with other detection techniques such as colorimetry [37], fluorescence [38], electrochemistry [39,40], chemiluminescence [41], Raman spectroscopy, and surface plasmon resonance, miscellaneous CHAs platforms have been developed for miRNAs, metal ions [42] and protein molecular analysis [43]. However, these strategies are associated with complex nanoparticles and fluorescence labelling processes, time-consuming methods or expensive equipment.
Recently, numerous novel methods combining traditional gel electrophoresis and other technologies have been developed to analyze genetic material, such as microfluidic gel electrophoresis [44] and capillary gel electrophoresis [45]. Lee et al. developed a bio-barcode gel assay for miRNA detection at low concentration using a conventional gel electrophoresis platform, opening the avenue for practical application of conventional gel electrophoresis for high-sensitive analysis of miRNA without PCR amplification [46]. The final detected signal is derived from the amount of the barcode DNA in the gel electrophoresis instead of direct detection of miRNA, and the detection signal could be significantly amplified without any enzymatic amplification due to the numerous barcode DNA. The barcode gel assay provided a straightforward detection signal method on a conventional gel electrophoresis, but it also involved complicated DNA nanoparticle modification and gold nanoparticle dissolving processes.
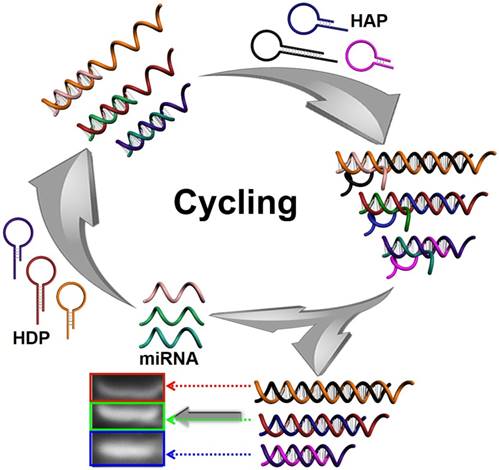
In this work, a multiple and sensitive miRNA detection strategy is designed by combing conventional gel electrophoresis with a CHA signal amplification system. As shown in Figure 1, three pairs of programmable oligonucleotide hairpin probes of hairpin detection probe (HDP) and hairpin assistant probe (HAP) are designed. In the present specific target miRNA, the stem of the corresponding HDP will be unfolded and expose the concealed domain. The corresponding HAP then replaces the hybridized target miRNA to form specific HDP/HAP complexes and releases miRNA based on thermodynamically driven entropy gain process, and the released miRNA triggers the next recycle to produce tremendous corresponding HDP/HAP complexes. In this way, one copy of target specific miRNA could trigger formation of numerous corresponding HDP/HAP complexes with designed length, resulting in significant signal amplification without enzymatic and nanoparticle-involved amplification. Depending on the length of the engineered hairpin probes, the hybridized duplex structures have different migration in the gel electrophoresis, which can be used to effectively and simultaneously detect various target miRNAs in complex biological samples.
Methods
Materials
YeaRed nucleic acid dye and 6×DNA Loading Buffer were obtained from YEASEN (Shanghai, China). The electrophoresis buffer consisted of 40 mM Tris, 20 mM acetic acid, 2 mM EDTANa2, and 12.5 mM (CH3COO)2Mg (TAE-Mg). Hybridization buffer was phosphate buffered saline (PBS) (pH 7.4, 10 mM with 137 mM NaCl), which was prepared by mixing the stock solutions of NaH2PO4 and Na2HPO4 and adjusting the pH with NaOH and H3PO4. Dimethyl sulphoxide (DMSO) was obtained from Sinopharm Chemical Reagent Co., Ltd (Beijing, China). PBS (pH 7.4, 10 mM) used in cell experiments, fetal bovine serum (FBS), Dulbecco's modified Eagle's medium (DMEM), trypsin-EDTA and penicillin-streptomycin were purchased from Gibco Life Technologies (AG, Switzerland). The ultrapure water was obtained from a Millipore water purification system (18 MΩ, Milli-Q, Millipore, USA). All of the DNA sequences were purchased from Sangon Biological Engineering Technology & Services Co., Ltd. (Shanghai, China). The RNA sequences purified using high-performance liquid chromatography were obtained from Gene Pharma (Shanghai, China) and modified with 2′-OMe to increase resistance to nucleases to improve the stability for miRNA detection [47]. Their sequences are in Table 1. All the miRNA sequences were diluted in diethyl pyrocarbonate (DEPC)-treated water for experiments, and all the experiments were performed in a laminar flow bench for a clean environment to control the influence of RNase.
Schematic illustration of CHA gel assay for multiple miRNA detection. In the presence of target miRNAs, the HDP will be unfolded to form HDP-miRNA duplex, and expose the concealed domain. Then, the corresponding HAP replaces the target miRNA to form specific HDP/HAP complexes and releases miRNA based on a thermodynamically driven entropy gain process, and the released miRNA triggers the next recycle to produce numerous corresponding HDP/HAP complexes.
Instruments
The agarose gel electrophoresis image was captured with an Alliance Ld2 (Uvitec, Cambridge, U.K.) with a UV lamp at 365 nm.
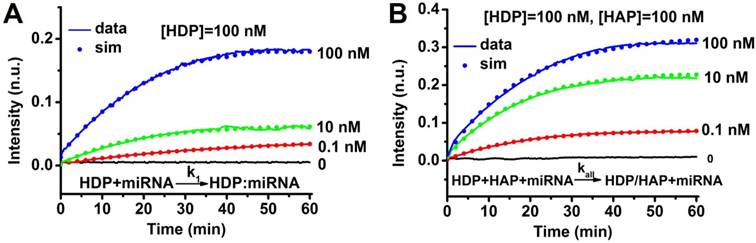
Characterization of CHA kinetics
The CHA kinetics were determined using the 'fminunc' function in Matlab. 100 nM HDP with two terminals labeled with a fluorescent dye and quencher, 100 nM HAP, and target miRNA-373 of different concentrations were mixed, and the fluorescence intensity of the solution was detected as a function of time. For comparison, the real-time fluorescence intensity of a mixture containing 100 nM HDP and target miRNA-373 of different concentrations was also measured.
DNA and miRNA oligonucleotide sequences involved in this work.
| Oligonucleotide | Sequence (5'---to---3') |
|---|---|
| HDP-373 | ACA CCC CAA AAT CGA AGC ACT TCC CAT GTG TAG AGA AGT GCT TCG ATT T |
| HAP-373 | AGC ACT TCT CTA CAC ATG GGA AGT GCT TCG ATT TCC ATG TGT AGA |
| HDP-21 | TCA ACA TCA GTC TGA TAA GCT ATG GAA CGT AGG TCT TTT GGT TAG CTT ATC AGA CTG |
| HAP-21 | TAA GCT AAC CAA AAG ACC TAC GTT CCA TAG CTT ATC AGA CTG TGG AAC GTA GGT CTT TTG GT |
| HDP-10b | CAC AAA TTC GGT TCT ACA GGG TAG TAC GCT TAC CCT GTA GAA C |
| HAP-10b | AGG GTA AGC GTA CTA CCC TGT AGA ACG TAC GCT |
| miRNA-373 | GAA GUG CUU CGA UUU UGG GGU GU |
| miRNA-21 | UAG CUU AUC AGA CUG AUG UUG A |
| miRNA-10b | UAC CCU GUA GAA CCG AAU UUG UG |
| SM miRNA-373 | GAA GUG CUU CGA UUU UGG GGU AU |
| DM miRNA-373 | GAA GUG CUU CGA UUU UGG UGU AU |
| TM miRNA-373 | GAA GUG CUU CGA UAU UGG UGU AU |
| miRNA-373* | ACU CAA AAU GGG GGC GCU UUC C |
| NC | AUU GAA UAU UCU UAU UAU AAU UA |
CHA agarose gel electrophoresis assays
Under optimized experimental conditions, HAP (1 μM), HDP (1 μM) and different concentrations of target miRNA-373 (Lane 1-11: 1 μM, 100 nM, 10 nM, 1 nM, 100 pM, 10 pM, 1 pM, 100 fM, 10 fM, 1 fM and 0) in 50 µL were reacted for 1 h at 37 °C. The resulting mixture was analyzed by gel electrophoresis. A control experiment was performed similarly without addition of HAP.
Analysis of miRNA extracted from cell lysates
MCF-7 and MDA-MB-231 cells were cultured in DMEM supplemented with 10% (v/v) FBS, 1% penicillin and streptomycin (100 U/mL) at 37 °C in a humidified atmosphere containing 5% CO2. MCF-7, MDA-MB-231 and NHDF cells (~2×106 cm-3) were washed, harvested and counted with a hemocytometer. The total miRNAs were extracted using a miRcute miRNA isolation kit (TIANGEN, Beijing, China). The resulting miRNAs were redispersed in 60 μL of DEPC-treated water and stored at -80 °C for further use, and miRNA was analyzed by the same gel electrophoresis procedure as aforementioned. The negative control experiment was performed by addition of DI water instead of cell lysates.
Characterization of CHA kinetics. (A) Measurement of catalyst binding rate k1. All traces contained 100 nM HDP initially, and different amounts of target miRNA-373 were added at t ≈ 0. Dotted lines show simulation traces from modelled reactions, assuming a rate constant of k1 = 1.6 × 105 M-1s-1. (B) Measurement catalyst binding rate kall. All traces contain 100 nM HDP and 100 nM HAP initially, and different amounts of target miRNA-373 were added at t ≈ 0. Dotted lines show simulation traces from modelled reactions, assuming a rate constant of kall = 1.2×105 M-1s-1. “n.u.” denotes normalized units; all fluorescence results were normalized to 1 n.u.
Multiple miRNAs detection
Under optimized experimental conditions, a mixture containing different target miRNAs was added to a solution consisting of three types of HAP (1 μM) and HDP (1 μM) to react for 1 h at 37 °C. The resulting mixture was analyzed by gel electrophoresis. A control experiment was similarly performed, except without addition of any target miRNA.
Under optimized experimental conditions, a mixture containing three target miRNAs with different concentrations (100 nM, 100 pM, 100 fM, 0) was added to a solution consisting of three types of HDP (1 μM) and HAP (1 μM) to react for 1 h at 37 °C. The resulting mixture was analyzed by gel electrophoresis.
Results
Feasibility of the CHA gel assay
A pair of hairpin probes of HDP and HAP was rationally designed for specific miRNA detection, and the ideal structures and corresponding thermodynamic parameters (Figure S1-3) of the HDP, HAP, miRNAs, HDP-miRNA duplex and HDP/HAP complexes were analyzed by mFold under the condition of PBS buffer (pH 7.4) containing 137 mM NaCl. All the probes have lower enthalpy changes (ΔH), indicating the probes designed are stable and have few other secondary structures. For example, all the ΔH values for HDP and HAP hairpin structures are lowest in all the predicted possible structures, suggesting the good stability of the hairpin structures. The Gibbs free energy (ΔG) of the CHA reaction revealed the reaction was spontaneous, indicating the thermodynamical feasibility of the CHA process. In order to demonstrate progression of the CHA reaction, the time course of the CHA reaction over a wide range of target miRNA concentrations was measured by fluorescence spectroscopy (Figure 2 and Figure S4). The kinetic analysis of the CHA was carried out using the 'fminunc' function in Matlab [48,49]. The rate constant k1 = 1.6×105 M-1s-1 yielded the best fit to the experimental data in Figure 2A, and the kall = 1.2×105 M-1s-1 are plotted as dots in Figure 2B. Kinetic analysis of the CHA (miRNA-21 and miRNA-10b) was also carried out. The rate constants kall = 0.9×105 M-1s-1 and kall = 4.5×105 M-1s-1 yielded the best fits to the experimental data for miRNA-21 and miRNA-10b CHA processes (Figure S4). These results kinetically suggested the good feasibility of the CHA process.
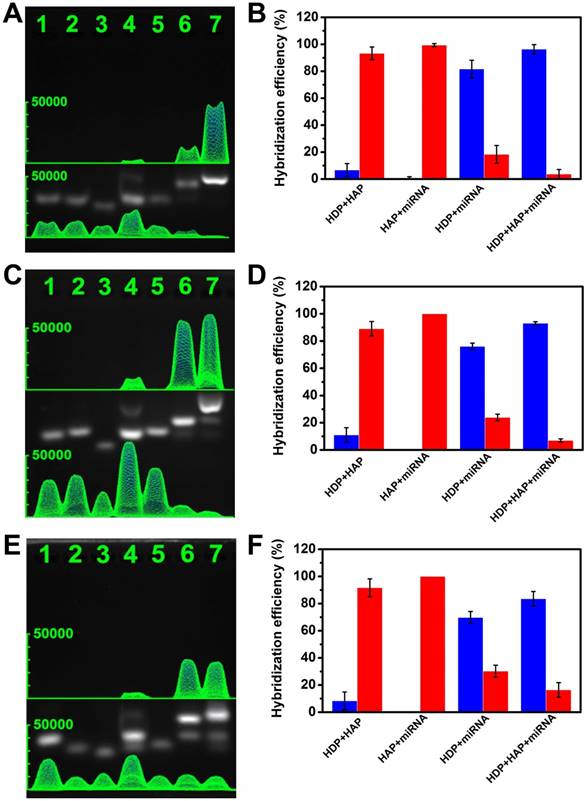
To evaluate the feasibility of CHA, agarose gel electrophoresis analysis experiments were performed. For the miRNA-373 analysis, as shown in Figure 3A-B, the HDP (lane 1), HAP (lane 2), and target miRNA (lane 3) exhibited a single obvious electrophoresis band, which indicated that secondary structures were not formed in the rationally designed hairpin probes. Although HDP and HAP contain complementary sequences, only a very weak band associated with interaction of HDP and HAP was observed (lane 4), implying the very low level of spontaneous interactions between the rationally designed hairpin probes. Similarly, there was no obvious new band assigned to hybridization of HAP and target miRNA (lane 5). However, when HDP was mixed with target miRNA, a new band with relative less migration than that of HDP appeared, which represented a good recognition of the HDP to the target miRNA (lane 6). The introduction of HAP to the mixture of HDP and target miRNA produced a more centralized band related to the HDP/HAP complex, indicating the successful dynamic assembly via cyclical toehold-induced strand displacement of target miRNA with HAP (lane 7). To further verify the signal-to-noise ratio, the gray values in Figure 3A were analyzed to investigate the corresponding hybridization efficiency. The blue column and red column represented the hybridized and free status of the mixture (Figure 3B), which indicated the excellent signal-to-noise and good feasibility of the designed CHA strategy. The designed hairpin probes of the miRNA-21 (Figure 3C-D) and miRNA-10b (Figure 3E-F) also demonstrated good feasibility for the CHA gel assay.
Verification of the experimental design by gel electrophoresis analysis. (A) miRNA-373, (C) miRNA-21, (E) miRNA-10b. Lane 1: HDP (1 μM); lane 2: HAP (1 μM); lane 3: miRNA (10 μM); lane 4: HDP (1 μM) and HAP (1 μM); lane 5: HAP (1 μM) and miRNA (1 μM); lane 6: HDP (1 μM) and miRNA (1 μM); lane 7: HDP (1 μM), HAP (1 μM) and miRNA (1 μM). (B, D, F) The proportion of free nucleic acid sequences (red column) and the corresponding hybridized mixture (blue column) derived from the grey value of the gel electrophoresis images.
Sensitivity of the CHA gel assay. (A) The CHA gel assay results and (B) corresponding calibration curve from different concentrations of target miRNA-373 (Lane 1-11: 1 μM, 100 nM, 10 nM, 1 nM, 100 pM, 10 pM, 1 pM, 100 fM, 10 fM, 1 fM and 0), normalized to the negative control band, and the error bars were obtained from three independent assays for each case.
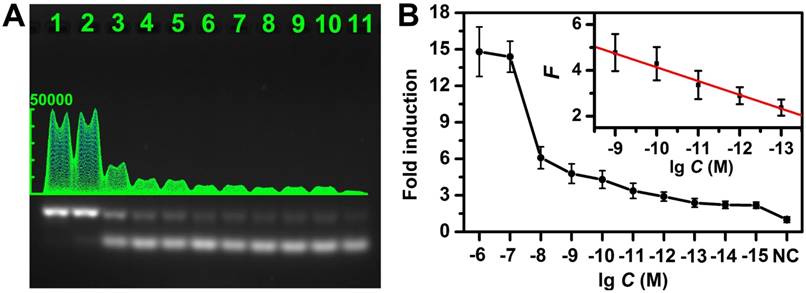
Assay performance for specific miRNA detection
The experimental conditions were first optimized (Figure S5). As shown in Figure 4A and Figure S6, the gel band intensity related to HDP/HAP complexes increased gradually with elevated concentration of target miRNA (miRNA-373), and the magnified photographs from lane 4 to lane 10 clearly showed the intensities difference. Figure 4B shows the average fold-induction values (normalized to the negative control band intensity) for the gel band intensity for each target concentration. The corresponding calibration plots showed a good linear relationship between the fold-induction values and the logarithm of target miRNA concentration in the range from 100 fM to 1 nM (inset of Figure 4B). The limit of detection (LOD) was calculated to be 10 fM by using three times of the standard deviation corresponding to the control. For comparison, we also investigated the performance of the non-amplification gel assays without adding HAP under the same experimental condition (Figure S7). It suggested the LOD of the CHA gel assay was approximately 5 orders of magnitude lower than that of the non-amplification gel assay. Moreover, the LOD of the CHA gel assays of miRNA-21 and miRNA-10b were also investigated (Figure S8), both of which could be detected at concentrations at fM levels, confirming the high sensitivity of the CHA gel assay. The LOD was also superior to previous CHA-based reports (Table S1). For example, the LOD of the CHA gel assay is about 3 orders of magnitude better than the previously reported electrochemical miRNA biosensor [39]. It is also comparable to previous electrochemiluminescence miRNA biosensors [50]. The better performance might result from the rationally designed oligonucleotide hairpin probes, higher hybridization efficiency and low background signal in the homogeneous solution of the no-label probes. The HDP was labeled with a fluorescent dye and a quencher at two terminals, respectively. The fluorescence intensity associated with the HDP/HAP complexes mixture linearly increased along with the target miRNA-373 over a wide range (Figure S9), further validating the CHA assay in quantitative analysis.
Selectivity of the CHA gel assay
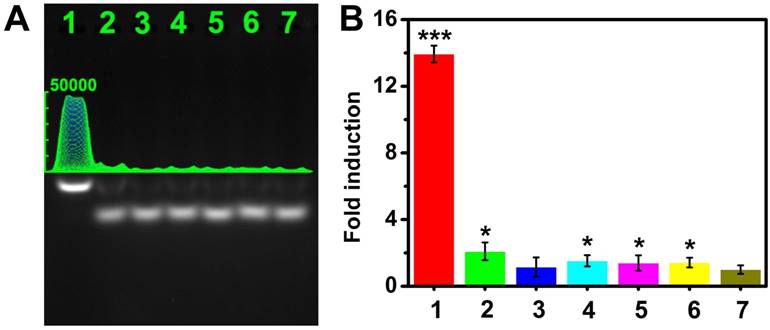
To evaluate the selectivity of the CHA gel assay, we performed a series of contrast experiments by using complementary target (CM), single-base mismatched strand (SM), double-bases mismatched strand (DM), three-bases mismatched strand (TM), miRNA-373* and non-complementary match (NC) in the same condition. The ideal structures and corresponding thermodynamic parameters of the HDP-373 with SM, DM, and TM complexes are displayed in Figure S10. The ΔG of the target miRNA CHA reaction is much lower (-5.43 kcal mol-1) than that of SM (ΔG=-3.13 kcal mol-1), DM (ΔG=-1.83 kcal mol-1) and TM (ΔG=-0.53 kcal mol-1), indicating the more preferable thermodynamic process of the target miRNA CHA reaction. Meanwhile, the HDP-miRNA duplex exhibited the lowest ΔH, which further confirmed the better stability of HDP-miRNA duplex compared to HDP-miRNA with mismatched bases. These results theoretically indicated the potential good discrimination capability of the proposed assay. As shown in Figure 5A-B, the fold-induction values of SM, DM, TM, miRNA-373* and NC were similar to that of control. However, the target miRNA-373 displayed a remarkable increase in the fold-induction values compared to control groups, which were 6.7-, 12.1-, 9.1-, 10.0- and 9.8-fold higher than that of the SM, DM, TM, miRNA-373* and NC. These results demonstrated that this CHA gel assay had high selectivity and excellent discrimination capability for base-mismatched sequences.
Selectivity of the CHA gel assay. (A) The CHA gel assay results and (B) the corresponding fold-induction values responding to 1 μM of different miRNAs (Lane 1-6: CM, SM, DM, TM, miRNA-373* and NC) and RNase-free water for control (Lane 7). The statistical significance (P < 0.05) was determined by the two-tailed Student t test (*P < 0.05, **P < 0.01, ***P < 0.001), as compared with the control.
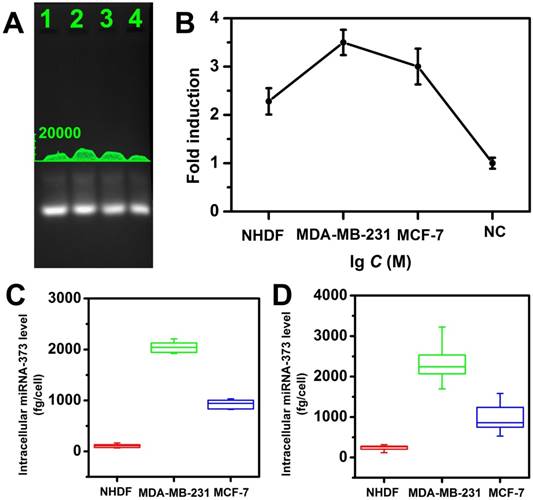
miRNA-373 detection in cell lysates
The feasibility of this strategy for miRNA detection in a real sample was also investigated by analyzing the miRNA extracted from cell lysate. Two breast cancer cell lines MDA-MB-231, MCF-7 (high expression of miRNA-373), and a control cell line normal human dermal fibroblasts (NHDF, low expression of miRNA-373) were investigated for analyzing target miRNA expression levels. Total miRNA extracted from each cell line was analyzed with the proposed CHA gel assay (Figure 6A) and the corresponding fold-induction values were recorded (Figure 6B). It demonstrated the MDA-MB-231 displayed a higher miRNA-373 expression level than that of MCF-7, and the NHDF expressed the lowest miRNA-373 among the three types of cell lines. It provided a powerful tool to analyze the miRNA expression in cell lysates.
From the linear regression equation (Figure 4B) and fold-induction values in Figure 5B, we calculated that the average quantity of miRNA-373 in a single NHDF, MDA-MB-231 and MCF-7 cell was 108.6 ± 35.9, 2082.4 ± 134.4 and 952.6 ± 104.4 fg, respectively, and exhibited no statistical differences (ANOVA and Tukey test, 99% confidence level; Figure 6C). We further performed the standard qRT-PCR experiments to confirm our results. The target miRNA-373 expression levels from the three types of cell lines were also analyzed using a standard qRT-PCR. The miRNA-373 ranging from 100 pM to 1 μM were reverse transcribed, and the PCR amplification of the cDNA was then analyzed to obtain the linear regression equation. The average quantity of miRNA-373 in a single NHDF, MDA-MB-231 and MCF-7 cell determined by qRT-PCR was 237.5 ± 77.2, 2354.9 ± 573.7 and 992.8 ± 419.2 fg, respectively (Figure 6D). Notably, the CHA gel assay showed a competitive precision for miRNA detection compared to the qRT-PCR, which indicated the good feasibility of the proposed CHA gel assay for real sample detection.
Multiple miRNA detection
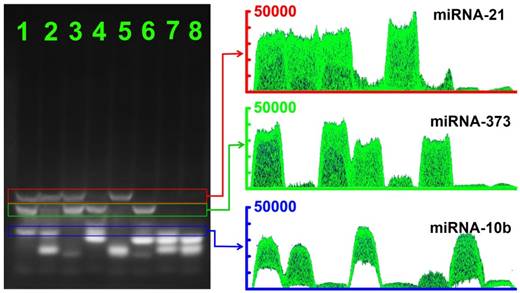
Encouraged by the initial analysis of sensitivity and selectivity, we investigated the multiplexing detection capability of the CHA gel assay for three different miRNA targets (miRNA-373, miRNA-21 and miRNA-10b). As shown in Figure 7, depending on the different lengths of the HDP/HAP complexes, the CHA gel assay could effectively discriminate and simultaneously detect the target miRNA in the solution containing a random combination of two types of miRNA targets including miRNA-21and miRNA-10b (lane 2), miRNA-373 and miRNA-21 (lane 3), and miRNA-373 and miRNA-10b (lane 4). The gel band intensity of miRNA-10b was weaker than that of the miRNA-21 and miRNA-373 gel band, which resulted from the relatively shorter sequences of HDP-10b and HAP-10b (Table 1). Notably, for the homogenous solution containing three types of miRNAs, gel bands of specific miRNAs-mediated HDP/HAP complexes were easily distinguishable by the different lengths of the designed hairpin probes (lane 1), indicating the good capability of the CHA gel assay for multiple miRNA detection. This is easily extended to more types of miRNA detection just by simply adjusting the length of the hairpin probes.
Quantitative measurement of miRNA-373 in cell Lysates. (A) The CHA gel assay results and (B) corresponding fold induction values responding to different cell lysates (Lane 1-4: NHDF, MDA-MB-231, MCF-7 cells and RNase free water ). (C) Quantification of miRNA-373 by CHA gel assay and (D) qRT-PCR.
The CHA gel assay results and corresponding gray curves for multiple miRNA detection. Lane 1: miRNA-21 (1 μM), miRNA-373 (1 μM) and miRNA-10b (1 μM); Lane 2: miRNA-21 (1 μM) and miRNA-10b (1 μM); Lane 3: miRNA-21 (1 μM) and miRNA-373 (1 μM); Lane 4: miRNA-373 (1 μM) and miRNA-10b (1 μM); Lane 5: miRNA-21 (1 μM); Lane 6: miRNA-373 (1 μM); Lane 7: miRNA-10b (1 μM) and RNase-free water for control.
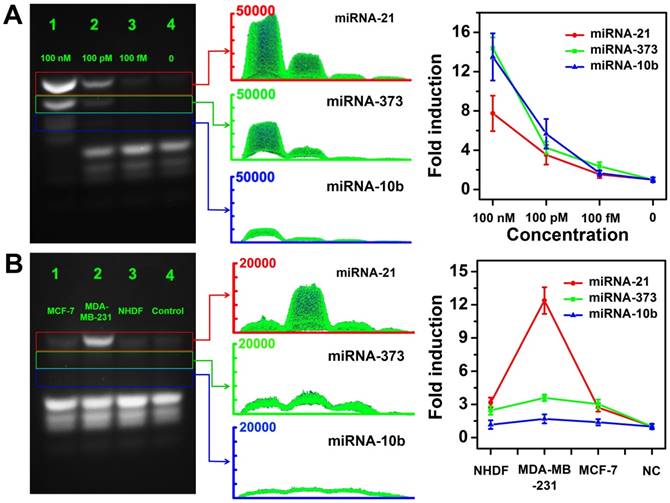
We further investigated the feasibilities of the CHA gel assay for multiple miRNA detection at different concentrations. As shown in Figure 8, three types of target miRNAs were effectively discriminated and simultaneously detected even at concentrations down to 100 fM without any enzymatic amplification. The results revealed the good sensitivity and selectivity of the multiplexed CHA gel assay. The expression levels of three miRNAs in three different cell lines were also evaluated by the proposed CHA gel assay. Much higher expression levels of miRNA-21, miR-373 and miRNA-10b in MDA-MB-231 cells [17,51], aberrantly higher expression of miRNA-21, miR-373 but normal expression level of miRNA-10b were observed in comparison with the control NHDF cells. The results indicated that the proposed strategy holds great potential in biomedical research and early clinical diagnostics.
Multiple miRNA detection in homogeneous solutions and cell lysates. (A) The CHA gel assay results and corresponding gray curves for multiple miRNA detection at different concentrations (Lane 1: 100 nM, Lane 2: 100 pM; Lane 3:100 fM.) and RNase-free water for control (Lane 4). (B) The CHA gel assay results and corresponding gray curves for multiple miRNA detection extracted from cell lysates (Lane 1-4: MCF-7 cells, MDA-MB-231 cells, NHDF cells and RNase-free water).
Discussion
In this study, we demonstrated that a facile CHA gel assay could effectively and simultaneously detect three miRNA species even at concentrations down to 100 fM without any enzymatic amplification in homogeneous solutions and cell lysates. Specifically, we showed that the CHA gel assay was effective for quantifying different miRNAs over a wide spectral range (0.1-1000 pM) with good capability of discriminating miRNA family members and base-mismatched miRNAs. This assay has the potential to be a powerful tool for quantifying miRNA expression in clinical specimens without involving complex conventional PCR and nanomaterials-based amplification, and shows great potential for nucleic acid detection applications.
CHA is an enzyme-free DNA circuit that relies on a thermodynamically driven entropy gain process to achieve signal amplification, possibly permitting exponential amplification of a target sequence in 15 min. Since traditional gel electrophoresis could distinguish nucleic acids according to their length, we developed three pairs of programmable oligonucleotide hairpin probes to simultaneously detect three miRNA species, depending on the length of the designed hairpin probes (Figure 1). First, the ideal structures (Figure S1-3) of the HDP, HAP, miRNAs, HDP-miRNA duplex and HDP/HAP complexes were thermodynamically optimized to ensure the good feasibility of the CHA process. The designed probes possessed a kinetically preferable reaction process with a fast kinetic rate.
To experimentally evaluate the feasibility of CHA, fluorescence spectrometry and agarose gel electrophoresis experiments were performed (Figure 2-3), and the results were consistent with the theoretical prediction. It demonstrated that the miRNA target unfolded the HDP with a fast rate to form an exposed HDP-miRNA duplex and expose the concealed domain. The exposed toehold terminal could be recognized by HAP, which displaced the target miRNA to form specific HDP/HAP complexes and released miRNA based on a thermodynamically driven entropy gain process. The released miRNA triggered the next recycle to produce numerous corresponding HDP/HAP complexes structures. The CHA gel assay analysis also confirmed that all the designed probes displayed few self-assembly secondary structures, indicating low false-signal production and reliability of the CHA gel assay. These findings impelled us to explore the CHA gel assay for multiple and sensitive miRNA quantitative detection in homogeneous solutions and cell lysates.
The very low level of spontaneous interactions between the rationally designed hairpin probes resulted in low background (Figure 3), which is significant for the sensitivity of the CHA gel assay. After optimization of the experimental conditions, the sensitivity of the CHA gel assay was measured. The CHA gel assay exhibited significant signal amplification with five orders of magnitude sensitivity enhancement for target miRNA detection compared to non-amplification gel assays without adding HAP under the same experimental condition, leading to a limit of detection down to 10 fM without enzymatic amplification (Figure 4). Meanwhile, we validated the selectivity of the CHA gel assay (Figure 5), and the results verified high selectivity for discriminating the target sequences from single-base mismatched miRNA and the family members.
Generally, quick detection methods, such as fluorescence methods, require complex probe labels, possess high background signals (due to incomplete quenching) and high LOD, and are inefficient for multiple detection due to overlap of the dye fluorescence spectra. The proposed CHA gel assay is cost-effective, avoiding laborious probe labelling and complex instruments. Especially, it is favorable for multiplex miRNA simultaneous detection by simply adjusting the length of the probe. The low background signal and CHA signal amplification of the assay can lead to highly sensitive detection performance.
Furthermore, using oncogenic miRNA-373 as a model, the feasibility to the assay for miRNA detection in real samples was investigated (Figure 6). Total miRNA extracted from three cell lines was analyzed with the proposed CHA gel assay using a linear regression equation. A low background signal was observed due to the spontaneous interaction between HDP and HAP. The average quantity of miRNA-373 in a single cell showed a competitive precision compared to qRT-PCR, which indicated the good feasibility of the proposed CHA gel assay for real sample detection. Notably, depending on the different lengths of the HDP/HAP complexes, the CHA gel assay could simultaneously discriminate and detect the target miRNA in homogeneous solutions (Figure 7) and cell lysates (Figure 8). The results indicated that the proposed CHA gel assay holds great potential in nucleic acid detection-based biomedical research and early clinical diagnostics.
Overall, we developed a CHA-based gel assay for multiple and sensitive miRNA detection using a conventional gel electrophoresis platform and rationally designed CHA hairpin probes. Although relatively complex and time-consuming (1 h running time) operation steps for running gel electrophoresis are involved, the CHA gel assay exhibited significant signal amplification for target miRNA detection and could detect the miRNA at fM levels without enzymatic amplification. It showed high selectivity for discriminating the target sequences from single-base mismatched miRNA and the family members. This method had good feasibility to detect miRNA extracted from different cell lysates, and the results were consistent with the conventional qRT-PCR-based results. Notably, the CHA gel assay consisting of different hairpin probes could effectively discriminate and simultaneously detect three types of miRNAs in homogenous solution and miRNAs extracted from cell lysates. It is easy to extend to simultaneous detection of more types of miRNA just by rationally designing the hairpin probes, which provides a powerful tool for the sensitive sequence detection of miRNAs of interest in practical applications without enzymatic amplification.
Abbreviations
miRNAs: microRNAs; CHA: catalytic hairpin assembly; HDP: hairpin detection probe; HAP: hairpin assistant probe; PCR: polymerase chain reaction; RT-PCR: real-time polymerase chain reaction; LCR: ligase chain reaction; RCA: rolling-circle amplification; SDA: strand displacement amplification; ΔH: enthalpy changes; ΔG: gibbs free energy; LOD: the limit of detection; CM: complementary target; SM: single-base mismatched strand; DM: double-bases mismatched strand; TM: three-bases mismatched strand; NC: non-complementary match.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
The work was supported by National Key R&D Program of China (Grant Nos. 2016YFC0106602 and 2016YFC0106601); National Natural Science Foundation of China (Grant No. 21645005, 21475008); the Open Research Fund Program of Beijing Key Lab of Plant Resource Research and Development, Beijing Technology and Business University (PRRD-2016-YB2).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
2. Ambros V. The functions of animal microRNAs. Nature. 2004;431:350-5
3. Dong HF, Lei JP, Ding L, Wen YQ, Ju HX, Zhang XJ. MicroRNA: function, detection, and bioanalysis. Chem Rev. 2013;113:6207-33
4. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-66
5. Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-69
6. Wei FR, Cao CH, Xu XQ, Wang JF. Diverse functions of miR-373 in cancer. J Transl Med. 2015;13:8
7. Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY. et al. Human embryonic stem cells express a unique set of microRNAs. Dev Bio. 2004;270:488-98
8. Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221-9
9. Wu NN, Liu XY, Xu XM, Fan XX, Liu M, Li X. et al. MicroRNA-373, a new regulator of protein phosphatase 6, functions as an oncogene in hepatocellular carcinoma. Febs J. 2011;278:2044-54
10. Hafez MM, Hassan ZK, Zekri ARN, Gaber AA, Al Rejaie SS, Sayed-Ahmed MM. et al. MicroRNAs and metastasis-related gene expression in egyptian breast cancer patients. Asian Pac J Cancer P. 2012;13:591-8
11. Volinia S, Calin GA, Liu C-G, Ambs S, Cimmino A, Petrocca F. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257-61
12. Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029-33
13. Li S, Xu L, Ma W, Wu X, Sun M, Kuang H. et al. Dual-mode ultrasensitive quantification of microRNA in living cells by chiroplasmonic nanopyramids self-assembled from gold and upconversion nanoparticles. J Am Chem Soc. 2016;138:306-12
14. Zhao X, Xu L, Sun M, Ma W, Wu X, Kuang H. et al. Gold-quantum dot core-satellite assemblies for lighting up microRNA in vitro and in vivo. Small. 2016;12:4662-8
15. Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682-8
16. Keklikoglou I, Koerner C, Schmidt C, Zhang JD, Heckmann D, Shavinskaya A. et al. MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-κB and TGF-β signaling pathways. Oncogene. 2012;31:4150-63
17. Chen W, Cai F, Zhang B, Barekati Z, Zhong XY. The level of circulating miRNA-10b and miRNA-373 in detecting lymph node metastasis of breast cancer: potential biomarkers. Tumor Bio. 2013;34:455-62
18. Chen CF, Ridzon DA, Broomer AJ, Zhou ZH, Lee DH, Nguyen JT. et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:9
19. Stenvang J, Silahtaroglu AN, Lindow M, Elmen J, Kauppinen S. The utility of LNA in microRNA-based cancer diagnostics and therapeutics. Semin Cancer Biol. 2008;18:89-102
20. Becker C, Hammerle-Fickinger A, Riedmaier I, Pfaffl MW. mRNA and microRNA quality control for RT-qPCR analysis. Methods. 2010;50:237-43
21. Mei Q, Li X, Meng Y, Wu Z, Guo M, Zhao Y. et al. A facile and specific assay for quantifying microRNA by an optimized RT-qPCR approach. PLoS One. 2012;7:e46890
22. Varallyay E, Burgyan J, Havelda Z. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat Protoc. 2008;3:190-6
23. Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD. et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101:11755-60
24. Wang Y, Zheng D, Tan Q, Wang M, Gu L. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat Nanotechnol. 2011;6:668-74
25. Xie T, Li M, Long Y. Dual-channel signals for intracellular mRNA detection via a PRET nanosensor. Chem Commun. 2017;53:7768-71
26. Zhengli Hu, Jihui Du, Yilun Ying, Yueyi Peng, Chan Cao, Yi-Tao Long. Single-molecule analysis of colorectal cancer-associated microRNAs via a biological nanopore. Acta Chim Sinica. 2017;75:1087-90
27. Xu F, Zhou W, Cao J, Xu Q, Jiang D, Chen Y. A combination of DNA-peptide probes and liquid chromatography-tandem mass spectrometry (LC-MS/MS): a quasi-targeted proteomics approach for multiplexed microRNA quantification. Theranostics. 2017;7:2849-62
28. James A, Baker M, Bao G, Searles C. MicroRNA detection using a double molecular beacon approach: distinguishing between miRNA and pre-miRNA. Theranostics. 2017;7:634-46
29. Xu F, Shi H, He X, Wang K, He D, Guo Q. et al. Concatemeric dsDNA-templated copper nanoparticles strategy with improved sensitivity and stability based on rolling circle replication and its application in microRNA detection. Anal Chem. 2014;86:6976-82
30. Castañeda AD, Brenes NJ, Kondajji A, Crooks RM. Detection of microRNA by electrocatalytic amplification: a general approach for single-particle biosensing. J Am Chem Soc. 2017;139:7657-64
31. Zhang P, Zhang J, Wang C, Liu C, Wang H, Li Z. Highly sensitive and specific multiplexed microRNA quantification using size-coded ligation chain reaction. Anal Chem. 2014;86:1076-82
32. Zhang Z, Wang Y, Zhang N, Zhang S. Self-assembly of nucleic acid molecular aggregates catalyzed by a triple-helix probe for miRNA detection and single cell imaging. Chem Sci. 2016;7:4184-9
33. Dai W, Dong H, Guo K, Zhang X. Near-infrared triggered strand displacement amplification for MicroRNA quantitative detection in single living cells. Chem Sci. 2018, 9. 1753 -9
34. Walker GT, Fraiser MS, Schram JL, Little MC, Nadeau JG, Malinowski DP. Strand displacement amplification-an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 1992;20:1691-6
35. Walker GT, Little MC, Nadeau JG, Shank DD. Isothermal in vitro amplification of DNA by a restriction enzyme/DNA polymerase system. Proc Natl Acad Sci U S A. 1992;89:392-6
36. Hellyer TJ, Nadeau JG. Strand displacement amplification: a versatile tool for molecular diagnostics. Expert Rev Mol Diagn. 2004;4:251-61
37. Wen J, Chen J, Zhuang L, Zhou S. Designed diblock hairpin probes for the nonenzymatic and label-free detection of nucleic acid. Biosens Bioelectron. 2016;79:656-60
38. Zhang C-H, Tang Y, Sheng Y-Y, Wang H, Wu Z, Jiang J-H. Ultrasensitive detection of microRNAs using catalytic hairpin assembly coupled with enzymatic repairing amplification. Chem Commun. 2016;52:13584-7
39. Zhang Y, Yan Y, Chen W, Cheng W, Li S, Ding X. et al. A simple electrochemical biosensor for highly sensitive and specific detection of microRNA based on mismatched catalytic hairpin assembly. Biosens Bioelectron. 2015;68:343-9
40. Yao J, Zhang Z, Deng Z, Wang Y, Guo Y. An enzyme free electrochemical biosensor for sensitive detection of miRNA with a high discrimination factor by coupling the strand displacement reaction and catalytic hairpin assembly recycling. Analyst. 2017;142:4116-23
41. Li D, Cheng W, Li Y, Xu Y, Li X, Yin Y. et al. Catalytic hairpin assembly actuated DNA nanotweezer for logic gate building and sensitive enzyme-free biosensing of microRNAs. Anal Chem. 2016;88:7500-6
42. Wu ZK, Fan HH, Satyavolu NSR, Wang WJ, Lake R, Jiang JH. et al. Imaging endogenous metal ions in living cells using a DNAzyme-catalytic hairpin assembly probe. Angew Chem Int Ed. 2017;56:8721-5
43. Shi K, Dou B, Yang J, Yuan R, Xiang Y. Target-triggered catalytic hairpin assembly and TdT-catalyzed DNA polymerization for amplified electronic detection of thrombin in human serums. Biosens Bioelectron. 2017;87:495-500
44. Walczak R, Kubicki W, Dziuban J. Low cost fluorescence detection using a CCD array and image processing for on-chip gel electrophoresis. Sensor Actuat B-Chem. 2017;240:46-54
45. Danish A, Lee SY, Muller CE. Quantification of green fluorescent protein-(GFP-) tagged membrane proteins by capillary gel electrophoresis. Analyst. 2017;142:3648-55
46. Lee H, Park JE, Nam JM. Bio-barcode gel assay for microRNA. Nature Commun. 2014;5:7
47. Liu ZX, Chen TJ, Romesberg FE. Evolved polymerases facilitate selection of fully 2 '-OMe-modified aptamers. Chem Sci. 2017;8:8179-82
48. Zhang DY, Winfree E. Control of DNA strand displacement kinetics using toehold exchange. J Am Chem Soc. 2009;131:17303-14
49. Zhang DY, Turberfield AJ, Yurke B, Winfree E. Engineering entropy-driven reactions and networks catalyzed by DNA. Science. 2007;318:1121-5
50. Liao Y, Huang R, Ma Z, Wu Y, Zhou X, Xing D. Target-triggered enzyme-free amplification strategy for sensitive detection of microRNA in tumor cells and tissues. Anal Chem. 2014;86:4596-604
51. He X, Zeng T, Li Z, Wang G, Ma N. Catalytic molecular imaging of microRNA in living cells by DNA-programmed nanoparticle disassembly. Angew Chem Int Ed. 2016;55:3073-6
Author contact
![]() Corresponding author: hfdongedu.cn; zhangxuejiedu.cn. Phone/Fax: +86 010 82376576.
Corresponding author: hfdongedu.cn; zhangxuejiedu.cn. Phone/Fax: +86 010 82376576.
 Global reach, higher impact
Global reach, higher impact