13.3
Impact Factor
Theranostics 2018; 8(8):2251-2263. doi:10.7150/thno.23877 This issue Cite
Research Paper
Cyclin D1-mediated microRNA expression signature predicts breast cancer outcome
1. Pennsylvania Cancer and Regenerative Medicine Research Center, Baruch S. Blumberg Institute, Pennsylvania Biotechnology Center and Lankenau Institute for Medical Research, 100 East Lancaster Avenue, Suite, 222, Wynnewood, PA. 19096
2. Research Center for Translational Medicine, East Hospital, Tongji University School of Medicine, Shanghai 200120, China
3. Department of Cancer Biology, Thomas Jefferson University, 233 South 10 th St. Philadelphia PA 19107
4. Center for Integrated Bioinformatics, Drexel University, Philadelphia, PA 19104
5. School of Biomedical Engineering, Systems and Health Sciences, Drexel University, Philadelphia, PA 19104
6. Shanghai Gongli Hospital, the Second Military Medical University, Shanghai 200120, China.
7. Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore 637551, Singapore.
# Contributed equally to this paper.
Received 2017-11-15; Accepted 2017-12-25; Published 2018-3-11
Abstract
Background: Genetic classification of breast cancer based on the coding mRNA suggests the evolution of distinct subtypes. Whether the non-coding genome is altered concordantly with the coding genome and the mechanism by which the cell cycle directly controls the non-coding genome is poorly understood.
Methods: Herein, the miRNA signature maintained by endogenous cyclin D1 in human breast cancer cells was defined. In order to determine the clinical significance of the cyclin D1-mediated miRNA signature, we defined a miRNA expression superset from 459 breast cancer samples. We compared the coding and non-coding genome of breast cancer subtypes.
Results: Hierarchical clustering of human breast cancers defined four distinct miRNA clusters (G1-G4) associated with distinguishable relapse-free survival by Kaplan-Meier analysis. The cyclin D1-regulated miRNA signature included several oncomirs, was conserved in multiple breast cancer cell lines, was associated with the G2 tumor miRNA cluster, ERα+ status, better outcome and activation of the Wnt pathway. The coding and non-coding genome were discordant within breast cancer subtypes. Seed elements for cyclin D1-regulated miRNA were identified in 63 genes of the Wnt signaling pathway including DKK. Cyclin D1 restrained DKK1 via the 3'UTR. In vivo studies using inducible transgenics confirmed cyclin D1 induces Wnt-dependent gene expression.
Conclusion: The non-coding genome defines breast cancer subtypes that are discordant with their coding genome subtype suggesting distinct evolutionary drivers within the tumors. Cyclin D1 orchestrates expression of a miRNA signature that induces Wnt/β-catenin signaling, therefore cyclin D1 serves both upstream and downstream of Wnt/β-catenin signaling.
Keywords: cyclin D1, miRNA, breast cancer
Introduction
The cyclin D1 (CCND1) gene encodes the regulatory subunit of a holoenzyme that phosphorylates and inactivates the Rb protein to promote DNA synthesis [1]. Cyclin D1 is overexpressed in up to 50% of human breast cancers. Cyclin D1 is required for the growth of cultured breast cancer cells and is necessary for the growth of oncogene-induced breast tumors in mice [2, 3]. The prognostic significance of cyclin D1 expression in human breast cancer epithelial cells has been carefully studied and correlates with ERα+ breast cancer and better patient outcome [4-6]. The common finding of cyclin D1 overexpression in the majority of human breast cancers is a consequence of gene amplification, reduced degradation or enhanced gene expression via oncogenic signals, including Ras, ErbB2 and Wnt/β-catenin signaling [7]. In addition, cyclin D1 contributes to the phosphorylation of other substrates including NRF1 in order to coordinate diverse activities within the cell, including the inhibition of mitochondrial metabolism and the phosphorylation of transcription regulatory factors [8, 9]. The DNA-bound form of cyclin D1 regulates gene expression through the recruitment of transcription factors [10, 11] and histone regulatory enzymes, in the context of local chromatin [10, 11]. The recruitment of histone deacetylase (HDAC1/HDAC3), histone acetylase (p300/CBP) and chromatin remodeling protein (SUV39, HP1α) [11] contributes to the alterations in gene expression of the coding genome [12, 13]. Both androgen- [14] and estrogen- [15] regulated growth factor gene expressions require cyclin D1 in vivo [7].
In addition to regulating the coding region, cyclin D1 has been shown to regulate non-coding miRNA [16]. The miR-17-20 locus is induced by cyclin D1, correlating with an association of cyclin D1 in the context of local chromatin within the microRNA regulatory region [16]. MicroRNAs (miRNAs) are a class of endogenous non-coding small RNAs participating in diverse biological processes through base-pairing to the complementary sequence in the 3' untranslated region (3' UTR of mRNA). Two complementary short RNA molecules are formed from stem-loop-stem pre-miRNA by Dicer cleavage, but only mature miRNA can integrate into the RNA-induced silencing complex (RISC) to regulate target mRNA expression. miRNA abundance is regulated in response to cellular signaling during cell cycle transition and cyclin D1 has been shown to govern miRNA processing through the induction of Dicer [17]. An alteration in miRNA expression is thought to contribute to the timing of cell-cycle transition, cell proliferation, stem cell self-renewal, cancer onset and progression [18]. An association of specific miRNA with initiation, progression and therapy resistance in human breast cancer has been demonstrated [19, 20]. The molecular mechanisms coordinating patterns of miRNA expression are poorly understood but are considered important for targeted therapies, in particular for therapy-resistant breast cancer [19]. In this regard, the let-7 and miR-17-20 cluster is induced by estrogen [21] and let-7f contributes to aromatase inhibitor resistance through repressing the aromatase promoter [22]. The miRNA cluster C19MC member miR-519a was identified to be the most highly upregulated miRNA in tamoxifen-resistant MCF-7 cells [23]. miR-519a overexpression increased viability and the S-phase population and decreased tamoxifen-induced apoptosis [23].
The multiple distinct mechanisms regulating the Wnt pathway are critical for the fine-tuning of signaling that occurs during physiological and biological processes. Disruption of these constraining influences occurs in human malignancies including breast cancer. Activation of the Wnt/β-catenin pathway in human breast cancer contributes to cellular proliferation, in part through the induction of cyclin D1 [24-26]. The canonical Wnt/β-catenin pathway is induced by Wnt ligands that bind to receptors including Frizzled (Fzd) [27] and the LDL receptor-related proteins 5 or 6 (LRP5, LRP6) [28]. The cytoplasmic protein Dishevelled (Dvl) is phosphorylated and thereby dissociates from a destruction complex, which includes AXIN, adenomatous polyposis coli (APC), casein kinase 1α (CK1α) and glycogen synthase kinase 3β (GSK3β). The stabilized β-catenin translocates to the cell nucleus to form a transcriptional complex with β-catenin LEF/TCF, thereby inducing downstream gene expression. Several pathways antagonize β-catenin signaling. The Dickkopf WNT signaling pathway inhibitor (DKK) proteins are antagonists of the WNT signaling pathway. DKKs function by isolating the LRP6 co-receptor so that it cannot support the WNT signaling pathway [29] and via a reduction in β-catenin abundance [30]. Although it is known that cyclin D1 is a downstream target of Wnt signaling, the notion that cyclin D1 functions upstream of Wnt signaling remains to be determined [24-26].
Given the importance of cyclin D1 in breast cancer, the current studies were conducted in order to generate a comprehensive analysis of the miRNA regulated by endogenous human cyclin D1 in breast cancer cells. The cyclin D1-regulated microRNA signature was associated with ERα positive breast tumors and activation of the Wnt pathway. The cyclin D1-mediated miRNA signature engages a network of coding genes, including DKK, that govern Wnt signaling. Thus in addition to serving as a down-stream target of Wnt/β-catenin, cyclin D1 governs an additional regulatory mechanism that activates Wnt via non-coding miRNA.
Results
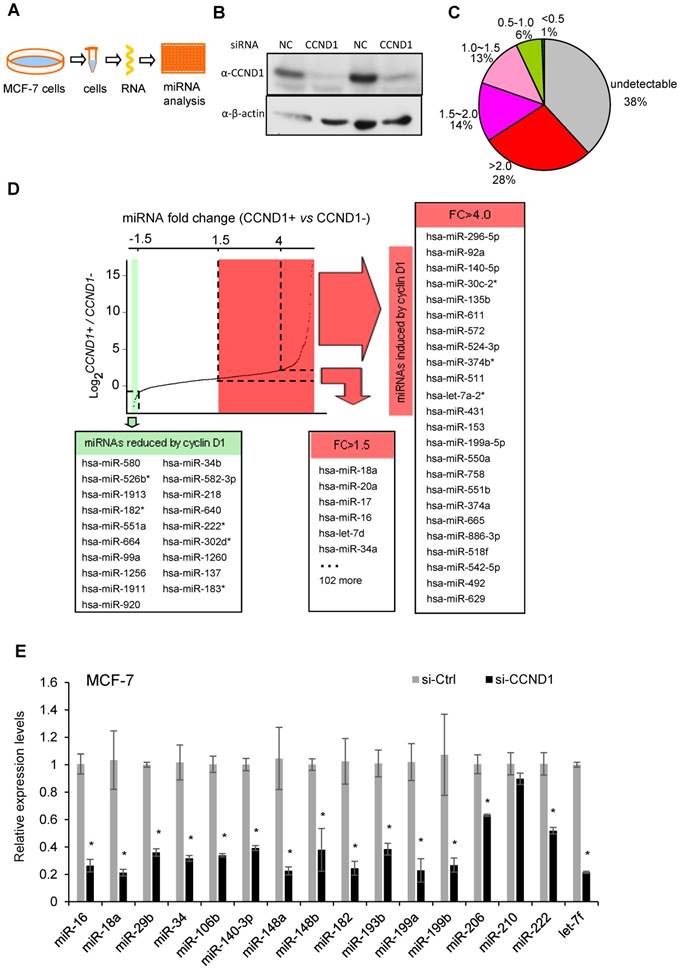
Cyclin D1 induces oncogenic miRNA abundance in breast cancer cell MCF-7. Cyclin D1 is overexpressed in up to 50% of human breast cancers and was previously shown to regulate miR-17-20 expression in breast cancer [16]. In order to define a comprehensive analysis of the miRNA regulated by cyclin D1 in human MCF-7 breast cancer cells, quantitative real-time RT-PCR-based miRNA analysis was conducted comparing the miRNA expression profiles in MCF-7 cells or MCF-7 cells treated with cyclin D1 siRNA (Figure 1A, B). Among 742 examined miRNAs, ~500 were detectable with ~30% showing >1.5-fold reduced expression in cyclin D1 knock-down cells (Figure 1C). In contrast, miR-21, the most abundant miRNA in MCF-7 cells, was not significantly regulated by endogenous cyclin D1. The fold change in expression (Cyclin D1+ vs. Cyclin D1-) of representative miRNAs is shown in Figure 1D, and further confirmed using QRT-PCR as shown in Figure 1E. The cancer-related miR-17-92 cluster (Figure S1A) and its paralogs (Figure S1B, C) were induced by endogenous cyclin D1. The Oncomir C19MC cluster [31], including miR-517c, 518d, 518c, 518f, 519, 525, was also induced by endogenous cyclin D1 (Figure S1D). In addition, members of the let-7 miRNA family were reduced by cyclin D1a siRNA (Figure S1E, F). In order to determine the miRNA signature of cyclin D1 in other breast cancer cell lines, representative miRNAs were analyzed in Hs578t cells (Figure S2). In Hs578t cells, 10 of the 15 miRNA were reduced by cyclin D1 siRNA, suggesting significant conservation of regulatory mechanisms between breast cancer cell lines.
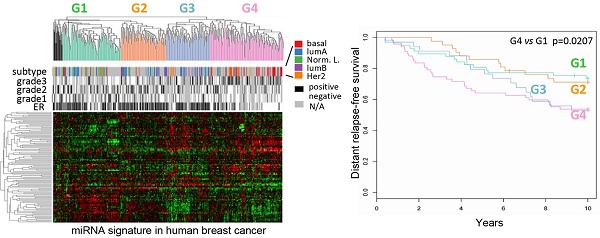
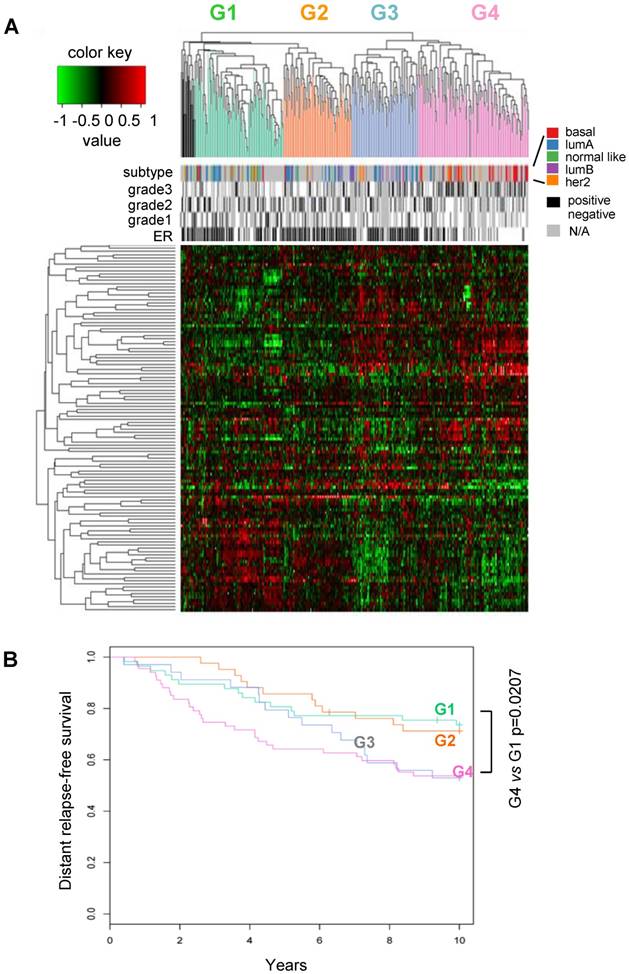
Classification of human breast cancers by their miRNA expression profiles. Emerging evidence has demonstrated miRNA involvement in human cancers including breast cancer [19, 20]. In order to determine the functional significance of the cyclin D1-regulated miRNA expression profile, a compendium of breast tumor miRNA was generated from multiple miRNA expression datasets that were associated with clinical information (Figure 2A). The compendium consisted of 114 miRNAs (Table S1) measured in 459 tumors (Figure 2A). Of these, 265 tumors were associated with estrogen receptor status, 308 tumors were associated with tumor grade, 175 tumors were classified using mRNA expression profiles of molecular subtypes and 249 tumors were linked with time to relapse data. We used this data to classify tumors using hierarchical clustering to identify four clusters of tumors (referred to as G1-4; Figure 2A) on the basis of expression of all miRNAs in the compendium. In order to determine the pathological significance of miRNA expression profiles, we evaluated the association of these miRNA subtypes with relapse-free survival (Figure 2B). Kaplan-Meier analysis indicated that tumors in groups G1 and G2 have relatively good prognosis whereas tumors in groups G3 and G4 have poor prognosis. In addition, a significant difference was detected in the risk of relapse between the four groups (p = 0.04). Cox-regression analysis verified that tumors in the G4 group have significantly higher risk of relapse than tumors in groups G1 and G2 (Figure 2B). The findings suggest that miRNA expression profiles are associated with differences in prognosis in breast cancer.
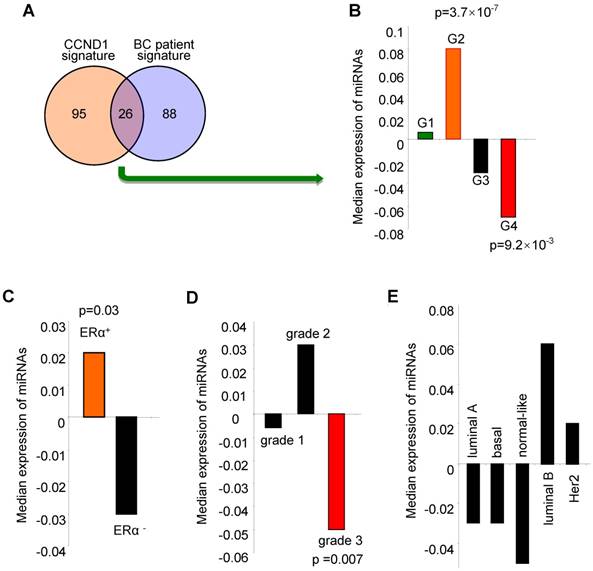
The cyclin D1-regulated miRNA signature correlated with ERα positive, low grade tumors and better outcome for breast cancer. We investigated the clinical significance of the cyclin D1-regulated miRNA signature identified in breast cancer cells by examining associations with clinical covariates, including breast cancer-associated miRNA, ERα status, tumor grade and molecular genetic subtype (coding genome), and relapse-free survival using the breast cancer miRNA compendium. The cyclin D1 miRNA signature consists of 121 miRNAs (Table S2) that are upregulated by 2-fold or more in cyclin D1+ vs. cyclin D1- cells (Figure 1). 26 of the cyclin D1-regulated miRNA displayed corresponding regulation in breast cancer patients (Figure 3A, Figure S3 and Figure S4). The cyclin D1-regulated 26 cancer-associated miRNA signature was assessed for clinical covariates. First, we calculated expression of the cyclin D1-regulated miRNA signature as the average expression of these miRNA in each tumor in the compendium. We next sorted the tumors based on these expression values. We compared the distributions of the cyclin D1 miRNA signature in sets of tumors classified by clinical covariates using non-parametric statistical testing. Notably, high expression of the cyclin D1 miRNA signature was associated with the good prognosis cluster G2 (Figure 3B; p = 3.7×10-7). In contrast, low expression of the cyclin D1 miRNA signature was associated with the poor prognosis cluster G4 (Figure 3B; p = 9.2×10-3). The cyclin D1 miRNA signature was positively associated with ERα+-status and negatively associated with poor outcome histological grade. The cyclin D1-regulated miRNA were expressed at a higher level in ERα+ positive relative to ERα- disease (Figure 3C, p = 0.03; Figure S5). Similarly, cyclin D1-regulated miRNA were downregulated in tumors with high histological grade (Figure 3D, p = 0.007; Figure S6). In contrast, the association with the molecular genetic subtype and the cyclin D1-regulated miRNA signature was not statistically significant (Figure 3E, Figure S7).
Cyclin D1 induces a miRNA signature. (A) Schematic representation of the procedure in which MCF-7 cells were treated with control or cyclin D1 siRNA knockdown. (B) Western blot analyses showing the decreased level of cyclin D1 by siRNA. (C) The pie chart shows the percentage of miRNA with expressions altered by cyclin D1 siRNA and their fold change in abundance. (D) Graphical representation showing the distribution of the log2 (control vs. cyclin D1 siRNA) of miRNA expression in MCF-7 cells. Shadowing represents the miRNAs reduced (green) and induced (red) more than 1.5-fold by endogenous cyclin D1. (E) Validation of representative miRNAs' expression in MCF-7 cells treated with negative control or cyclin D1 siRNA; the results are shown as mean ± SEM (n=3).
Together, these studies demonstrate that the cyclin D1-regulated miRNA signature correlates with ERα positive tumors, better tumor grade and better prognosis. These findings are consistent with observed associations between expression of cyclin D1 and clinical covariates in breast cancer.
Classification of breast tumors by miRNA expression profiling distinguishes good and poor prognosis tumors. (A) miRNA expression profiles and clinical information from the breast cancer compendium. Rows indicate expression of 114 miRNA. Columns indicate expression of miRNA in 459 breast tumors compiled from four datasets. Green points represent down-regulation, red points represent up-regulation. Rows and columns were sorted using hierarchical clustering with Pearson correlation and average linkage. Hierarchical clustering was used to classify tumors into four groups according to their miRNA expression profiles (G1 = green, G2 = orange, G3 = blue, G4 = pink, outliers = black). Column color bars indicate the clinical data associated with each sample. (B) Kaplan-Meier plot of relapse-free survival in breast cancer for miRNA subtypes.
Expression of the cyclin D1-regulated miRNA signature is associated with biological features and clinical outcomes in breast cancer. (A) Venn diagram indicating the number of miRNA in the 'cyclin D1 signature” (miRNA regulated by cyclin D1), the number of miRNA measured in the breast cancer compendium and the intersection of these sets. (B) Distribution of cyclin D1 miRNA signature in breast tumors classified by miRNA subtype. Hashmarks below the plot indicate the expression of the cyclin D1-regulated miRNA for each tumor. Color indicates the miRNA subtype of the associated set of tumors. High expression of the cyclin D1 miRNA signature is associated with G2 (p = 3.7×10-7). Low expression of the cyclin D1 miRNA signature is associated with G4 (p = 9.2×10-3). (C) Cyclin D1-regulated miRNA signature in tumors classified by estrogen receptor (ERα) status. Hashmarks below the plot indicate the expression of the cyclin D1 miRNA signature for each tumor. Color indicates the estrogen receptor status of the associated set of tumors. High expression of the cyclin D1 miRNA signature is associated with ERα-positive disease (p = 0.03). (D) Cyclin D1-regulated miRNA signature in tumors classified by histological grade. Low expression of the cyclin D1 miRNA signature is associated with grade 3 (p = 0.007). (E). Cyclin D1 miRNA signature in tumors classified by molecular genetic (coding genome) subtype.
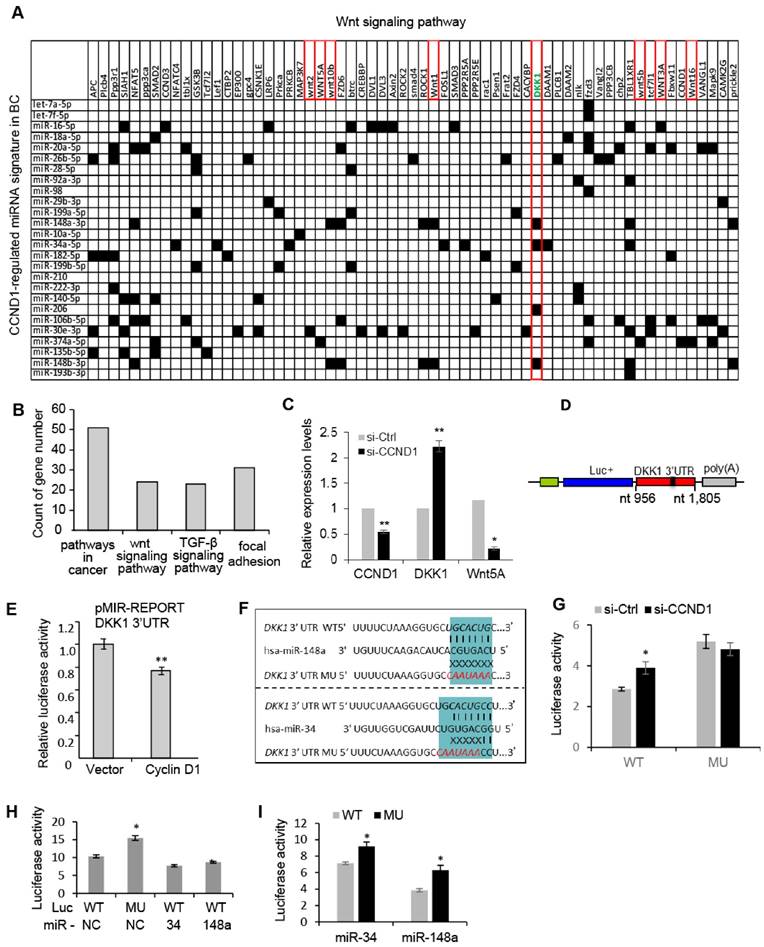
The cyclin D1 miRNA signature intersects Wnt signaling. In order to investigate further the biological significance of the cyclin D1-mediated miRNA signature in breast cancer, we performed a target gene screening and pathway analysis to the 26 miRNA signature regulated by endogenous cyclin D1 in breast cancer. Alignment of the cyclin D1-regulated breast cancer-associated miRNA signature with gene sequences identified 63 genes in the Wnt pathway potentially targeted by the miRNA signature (Figure 4A). KEGG pathway analysis identified enrichment for signaling pathways involved in breast cancer regulation including Wnt signaling pathway (p = 0.048, Figure 4B). The increased DKK1 and decreased Wnt5A expression were associated with knockdown of cyclin D1 in MCF-7 cells (Figure 4C). Because the Wnt signaling inhibitor DKK1 was found to be targeted by multiple cyclin D1-regulated miRNAs (Figure 4A), a reporter plasmid carrying the 3'UTR of DKK1 was applied to cyclin D1-overexpressing cells, showing ~20% inhibition of DKK1 by cyclin D1 in MCF-7 cells (Figure 4D, E). We hypothesized that cyclin D1-regulated miRNAs, including miR-34a and miR-148, suppressed DKK1 expression. A comparison was therefore made between the DKK1 3'UTR and a DKK1 3'UTR reporter with point mutations in the miR-34 and miR-148 binding sites, linked to a luciferase reporter vector (Figure 4F). Cyclin D1 siRNA upregulated the DKK1 WT 3'UTR, but did not regulate the DKK1 3'UTR that was mutated in the miR-34 and miR-148 binding site (Figure 4G). In addition, enforced overexpression of miR-34 or miR-148a suppressed the DKK1 WT 3'UTR reporter, but failed to repress the DKK1 mutated 3'UTR (Figure 4H, I and Figure S8). Together these studies provide further support for a model in which the cyclin D1-regulated miRNAs contribute to Wnt signaling regulation by cyclin D1 in MCF-7 cells.
Cyclin D1-regulated miRNAs govern the Wnt signaling pathway. (A) Interaction between cyclin D1-regulated miRNAs and target mRNAs in the Wnt signaling pathway. The y-axis shows Wnt signaling pathway genes that are predicted targets of the cyclin D1-regulated miRNA. The x-axis shows cyclin D1-regulated miRNAs in MCF-7 cells. Black boxes indicate predicted interactions between the gene and miRNA. Both axes were ordered using (pseudo-) hierarchical clustering. (B) Signaling pathways potentially targeted by cyclin D1-regulated miRNAs (p < 0.05) in MCF-7 cells. (C) Upregulation of DKK1 and downregulation of Wnt 5A were associated with cyclin D1 knockdown by siRNA in MCF-7 cells. (D) Schematic representation of the Luciferase reporter carrying DKK1 3'UTR. (E) The DKK1 3'UTR activity is suppressed by cyclin D1 in MCF-7 cells. (F) Sequencing alignment indicated that DKK1 3'UTR has binding sites to multiple cyclin D1-regulated miRNAs including miR-34 and miR-148a. Point mutations to the binding sites were applied to demonstrate the direct interaction between the 3'UTR of DKK1 and miR-34/miR-148a. (G) Luciferase reporter assays indicated that WT DKK1 3'UTR activity was inhibited by endogenous cyclin D1 in MCF-7 cells, but not the mutated DKK1 3'UTR. (H, I) Luciferase reporter assays are shown as mean ± SEM (n=3). miR-34 or miR-148a overexpression inhibited WT DKK1 3'UTR activity, but not mutated DKK1 3'UTR.
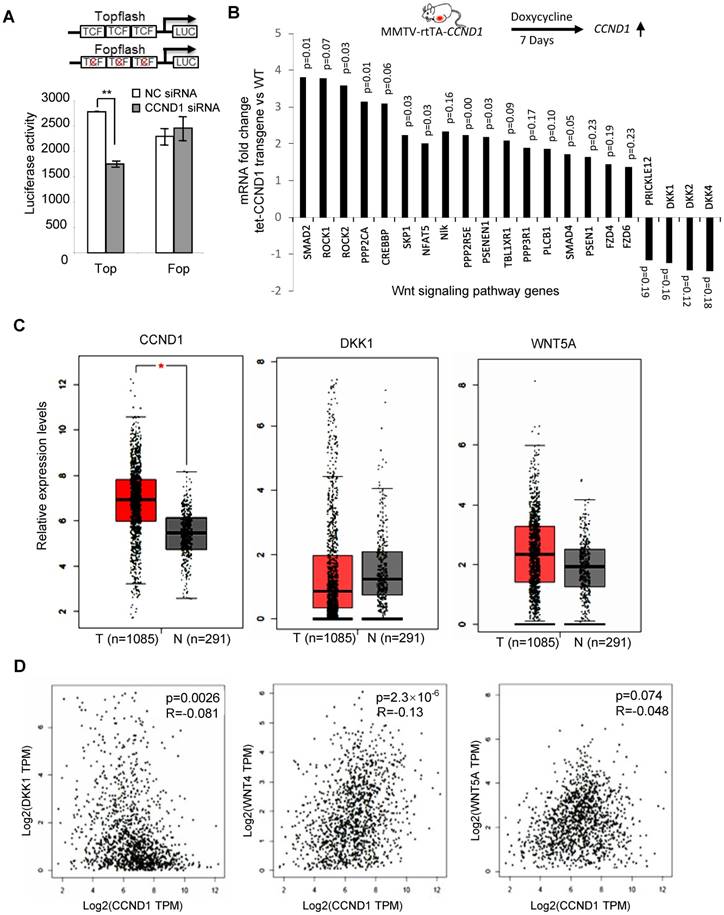
Cyclin D1-mediated regulation of the Wnt signal transduction pathway was assessed in MCF-7 cells using cyclin D1 siRNA, and the surrogate TCF-responsive luciferase reporter (TOP-LUC). TOP-LUC activity, compared with the TCF-mutant reporter (FOP-LUC), was increased 80% by endogenous cyclin D1 (Figure 5A). Consistent with the repression of the DKK1 3' UTR by cyclin D1, and the identification of binding sites for miR-34 and miR-148 in the DKK1 3' UTR, cyclin D1 was shown to induce these miRNAs in MCF-7 cells (Figure 1E and Figure S9). In order to determine whether acute induction of cyclin D1 was sufficient to induce Wnt-mediated gene expression in vivo, transgenic mice were employed. Tetracycline was used to induce the cyclin D1 transgene and after 1 week the mammary glands were harvested. The induction of the mammary epithelial cell (MEC)-targeted cyclin D1 transgene for one week was sufficient to induce Wnt signaling pathway genes including Smad, Rock, Fzd and others (Figure 5B). Consistent with the DKK reporter studies, induction of the cyclin D1 transgene reduced expression of the Wnt signaling pathway inhibitors DKKs and Prickle (Figure 5B).
In order to examine the possibility that the abundance of cyclin D1 may be negatively correlated with DKK1, we examined mRNA expression from the TCGA database. Analysis was conducted of 1,085 breast cancer patients and 291 normal control samples. As shown in Figure 5C, upregulation of cyclin D1 and Wnt5A and downregulation of DKK1 in breast cancer samples were observed compared to normal control. In addition, the expression of cyclin D1 was positively correlated with Wnt4 and Wnt5A but negatively correlated with DKK1 expression in breast cancer patients (Figure 5D).
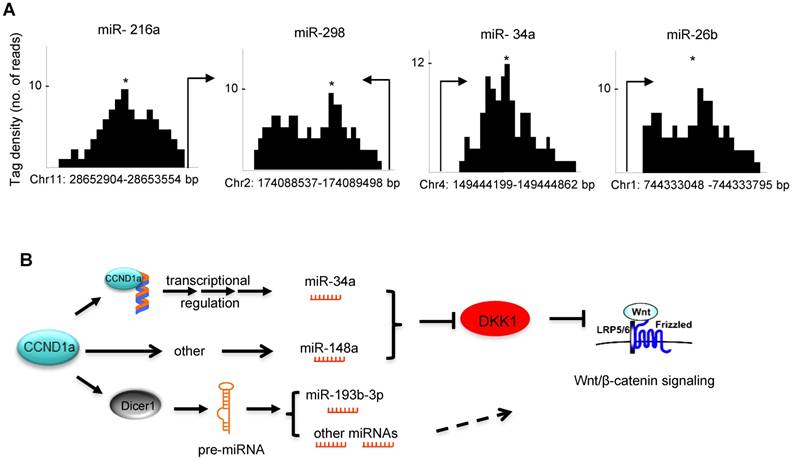
In order to examine potential mechanisms by which cyclin D1 regulated miRNA expression herein, we considered two previously characterized processes. Firstly, in this regard, cyclin D1 has been shown to regulate gene transcription via binding in the context of chromatin to either repress or induce gene expression [7, 32]. Cyclin D1 has also been shown to selectively regulate miRNA expression at the transcriptional level through binding to the promoter region of miRNAs [14]. ChIP-sequence analysis demonstrated that within the cyclin D1-regulated miRNA signature, cyclin D1 was recruited to the regulatory region of miR-216a, 26b, 298 and 34a (Figure 6A). A second mechanism by which cyclin D1 regulates miRNA involves Dicer. Our previous work has demonstrated the cyclin D1 induction of Dicer promotes miRNA biogenesis [17]. In addition to miRNA biogenesis, Dicer itself contributes to the regulation of miRNA expression, as the Dicer-/- cells have altered expression of specific miRNA [33]. We therefore compared the miRNA expression profile regulated by Dicer, and the miRNA expression profile regulated by cyclin D1. They showed similar miRNA expression patterns. The miRNA miR-10a, let-7d, miR-34c, miR-193* and miR-202, were regulated both by Dicer and by cyclin D1 (Figure S10, Table S2).
Taken together these data show that cyclin D1 induces expression of miRNA (miR-34 and miR-148a) that restrains DKK1 via its 3' UTR through seed elements for miR-34 and miR-148a. Cyclin D1 regulates miRNA expression either through Dicer or through regulating miRNA promoter expression (Figure 6B).
Discussion
The current studies identified several novel findings illustrating an important role of cyclin D1 in maintaining the abundance of miRNA that govern Wnt signaling. Firstly, we show that cyclin D1 maintains a specific cluster of miRNA in human breast cancer cells. Secondly, we showed that the cyclin D1-mediated miRNA expression profile was concordant with the direction of expression of ~25% of the breast cancer-associated miRNA. Thirdly, this miRNA signature had specific biological correlates: correlated with ERα+ breast cancer, anti-correlated with poor outcome and highly correlated with Wnt signaling. Fourthly, the cyclin D1-mediated miRNA expression profile correlated highly with a matrix of cyclin D1-mediated miRNA identified target 3' UTR sequences in the known Wnt target genes. Evidencing the significance of cyclin D1 in Wnt signaling, DKK1, a repressor of Wnt signaling, was repressed by endogenous cyclin D1, and expression of a cyclin D1 transgene in the mammary gland of transgenic mice induced expression of Wnt-related gene expression.
Emerging evidence has demonstrated interactions between cyclin D1 and miRNAs (miR-17/20, let-7 and miR-15/16) in control of human cancer [16, 34, 35]. The current studies demonstrated that cyclin D1 enhanced the expression of several onco-miRNAs. The miR-17-92 cluster of miRNAs on human chromosome 13 and its cluster paralogs (miR-106b-25, on chromosome 7) and the miR-106a-363 cluster on the X chromosome are known to act as oncogenes [36]. The miRNAs induced by cyclin D1 included the oncomiR miR-17-20 and all of its paralogs [36] (Figure S1). Components of a miRNA polycistron (miR-506, 513a, 518f, 519d, 520b and 524) [37], which are amplified in tumors [38] and contribute to tamoxifen resistance, were induced by cyclin D1. It is of interest that every miRNA of the oncomiR clusters was induced by cyclin D1. The cyclin D1 gene undergoes alternative splicing, resulting in two distinct isoforms encoding alternate carboxyl terminus C, with distinct properties in the DNA damage response and cell migration [39, 40].
Cyclin D1 activated Wnt signaling in the mammary gland in vivo. (A) Schematic representation of the TOP-LUC and Tcf site mutant reporter (FOP-Luc). Tcf activity was assessed in MCF-7 cells using the Top-Flash-Luc reporter, with activity shown as mean ± SEM. The assays indicate higher Wnt signaling activity in cyclin D1+ MCF-7 cells compared with cyclin D1- MCF-7 cells. (B) Schematic representation of the tetracycline-inducible mammary gland epithelial cell-targeted cyclin D1a transgenic mice. Mammary gland mRNA was analyzed after 1 week from three transgenic mice. The expression and p value of Wnt signaling pathway genes regulated by the transgene are shown. (C) TCGA analysis showed the upregulation of cyclin D1 and Wnt 5A and downregulation of DKK1 in 1085 breast cancer patients compared to 291 normal control samples. (D) TCGA analysis showed the positive correlation between cyclin D1 and Wnt 4/Wnt 5A expression, and negative correlation between cyclin D1 and DKK1 expression in breast cancer patients.
(A) ChIP of cyclin D1 followed by ChIP sequencing (ChIP-Seq) indicated the binding of cyclin D1 on the promoter region of indicated miRNAs that were identified within the cyclin D1-regulated miRNA signature. (B) Schematic representation of the mechanisms through which cyclin D1 induces tumorigenesis in breast cancer by repression of DKK1 and induction of Wnt signaling, which is at least partly mediated by miRNAs.
The current studies conducted hierarchical clustering of miRNA from 459 breast cancers and thereby identified 4 distinct types of breast cancer with distinct survival outcomes, G1 being favorable and G4 associated with poor outcome. A comparison between miRNA-based classification and mRNA-based classification of the same tumors evidenced only moderate overlap with some enrichment of the basal mRNA expression subtype amongst the G4 miRNA subtype. The cyclin D1-mediated miRNA signature was correlated with the G2 subtype and anti-correlated with the G4 subtype. Together these findings suggest independent drivers or independent evolution of the miRNA vs. mRNA expression within an individual patient's breast cancer.
Herein the miRNA induced by cyclin D1 were shown to align with the seed sequences in the 3' UTR of essentially all known genes of the Wnt pathway. Consistent with cyclin D1 activating the Wnt/β-catenin signaling pathway, a surrogate measurement of this pathway's activity in reporter gene assays, TCF activity was maintained by endogenous cyclin D1 in breast cancer cells. Furthermore, acute induction of a cyclin D1 transgene induced Wnt activity in vivo as assessed by TCF4 IHC in the murine mammary gland. In the current studies, cyclin D1-regulated miRNAs were predicted to induce a broad array of Wnt-activating components (Wnt1, Wnt3a, Wnt5A, Wnt10b, Wnt16, Vangl2, SIAH1, PP2A) and restraining components governing Wnt repression (Prickle 2, DKK1,2,4). In addition to the description of Wnt signaling above, the canonical Wnt pathway involves degradation of β-catenin by a β-TrCP1-containing E3 ligase, and alternative β-catenin degradation mediated by SIAH-1 (seven in absentia homolog-1) is responsible for the effect of CSN5 on β-catenin signaling [41]. Cross-talk occurs with other pathways including the PI3K/AKT [42, 43]. Active β-catenin is induced by the PI3K pathway and is dependent on the phosphatase activity of the protein phosphatase PP2A [43]. ERK phosphorylation of GSK3β is induced by IGF and TGFβ. ERK phosphorylates GSK3β at the Thr 43 residue, which primes GSK3β for its subsequent phosphorylation at Ser9 by p90RSK, resulting in inactivation of GSK3β and upregulation of β-catenin [42]. Vangl2, a core component in planar cell polarity, forms a Wnt-induced receptor complex with Ror2 to sense Wnt dosages [44]. Wnt signaling is retrained by DKK, and the 3' UTR of DKK1 is repressed by cyclin D1. Tumor suppression by PTEN and p27KIP1 involves restraint of β-catenin activity [45]. Cyclin D1 is a target of β-catenin/ TCF signaling [24] and β-catenin is a marker of poor prognosis in breast cancer [26]. In recent studies, cyclin D1 was shown to induce clusters of mRNA that govern Wnt/β-catenin transcriptional pathways [46]. Endogenous cyclin D1 enhanced Wnt and ES cell gene expression and expanded a prostate stem cell population. In these prior studies using chromatin immunoprecipitation sequencing, cyclin D1-occupied genes were found to govern stem cell expansion and induce their transcription. The current studies extend our understanding of cyclin D1, known to be a downstream target of Wnt/β-catenin signaling, by showing cyclin D1-induced miRNA clusters that engage the Wnt pathway.
Methods and Materials
Transgenic mice. Cyclin D1-/- mice were maintained in our lab on a mixed C57B1/6x129/SvJ background. The tetracycline-inducible mammary epithelial cell-targeted cyclin D1a transgenic mice established in the FVB strain (MMTV-rtTA-cyclin D1a transgenic mice) were previously described [47]. Eight to ten-weeks-old female mice was used for in vivo experiments. All mouse experiments were done in accordance with the guidelines for the care and use of laboratory animals at Thomas Jefferson University.
Cell culture. MCF-7 and Hs578T cells were originally from ATCC and were maintained in our laboratory. Cells were cultured in DMEM containing penicillin and streptomycin (100 mg of each/L) and supplemented with 10% fetal bovine serum (FBS).
siRNAs and transfection. All siRNAs were from Qiagen. The target sequence for cyclin D1 was AACAAGCTCAAGTGGAACCTG. The target sequence for the negative control siRNA was AATTCTCCGAACGTGTCACGT. Lipofectamine RNAiMax (Invitrogen) was used to transfect siRNA into cells following the manufacturer's instructions.
miRNA expression profiling and data analysis. Human miRNAs in cyclin D1 siRNA-treated MCF-7 cells and MCF-7 control cells were profiled in triplicate using the Exiqon microRNA Ready-to-Use PCR Human panel I+II (Product no:203607) and miRCURY LNATM Universal cDNA Synthesis Kit (Product no:203300) following the Manufacturer's instructions. miRNA expression measurements were estimated using the delta-delta Ct method. The set of miRNAs regulated by cyclin D1 was identified using a differential expression fold change threshold of 2.0 for comparison with the miRNA expression database in breast tumors. For the target gene prediction and pathway analysis, a threshold for significance as p<0.05 was applied to the above miRNA set regulated by cyclin D1. Predicted gene targets for the miRNAs were identified using the miRdbv4.0 (http://mirdb.org/miRDB/). These predicted targets were compared with the KEGG Wnt signaling pathway genes obtained from the KEGG pathway database (http://www.genome.jp/kegg/kegg2.html).
Gene reporter assays. The WT and mutated DKK1-3'UTR luciferase reporter vectors were created in our lab. The TOP-LUC / FOP LUC reporters were previously described [25]. For cellular transfection with reporter DNA, actively growing cells were seeded on 12-well plates at a density of 1×105 cells/well. The next day, cells were co-transfected using lipofectamine 2000 (Invitrogen) with 0.5 μg of Luc vector and 0.1 μg of CMV/ß-galactosidase plasmid. Twenty-four hours after transfection, luciferase activities were measured by AutoLumat and normalized with ß-galactosidase.
Compilation of miRNA expression data across platforms. Four datasets of microRNA expression profiles of breast tumors linked to corresponding clinical data were collected from the ArrayExpress database (ArrayExpress Identifiers: E-GEOD-22220, E-GEOD-7842, E-GEOD-19783, E-GEOD-15885) [20, 48-51]. In addition to expression data, sample annotation information and array design information were obtained in the form of ArrayExpress .sdrf and .adf files, respectively. Pre-processed data was used if available. If pre-processed data was not available, raw data was normalized by: calculating an average value for foreground and background intensity of replicate spots mapping to the same miRNA, log2 transformation of foreground and background signals, background correction by subtracting the background signal from each foreground signal, thresholding to set missing expression values to an arbitrary low value (0.1), and quantile normalization. In order to identify common features across array platforms, miRNA probes were mapped to the mirbase mature miRNA accession identifier [52]. For array platforms annotated with sequence information, an exact match of the target sequence to the mirbase sequence was used to ensure accurate annotation. Array platforms without sequence information were incorporated only when the specific release of the mirbase database used for platform design was reported. For these platforms, miRNA name was used for annotation based on the corresponding mirbase release. Cross-platform normalization was used to estimate and minimize the effects from dataset-specific noise. In this process, arrays are filtered to obtain expression data associated with the phenotype of interest. Following array filtering, datasets are transformed from probe set space to miRNA space by calculating the median expression value of all probes mapping to a given miRNA. Next, miRNA are filtered to retain only the miRNA that are represented on all array platforms. Array-filtered, transformed and miRNA-filtered data is normalized across platforms using the XPN algorithm [53] implemented with the CONOR package [54] in the R Statistical Environment. Data summarization plots, including boxplots, two-dimensional PCA projections, and hierarchical clustering were used for quality control of cross-platform normalization to ensure good mixing of data across platforms.
Evaluation of clinical relevance of cyclin D1-regulated miRNA. The clinical implications of cyclin D1-mediated miRNA expression were investigated by examining the association between cyclin D1-regulated miRNA and clinical covariates. Cyclin D1-regulated miRNA were identified as described above. A signature of cyclin D1 activity in clinical tumor samples was obtained based on the expression profile of the cyclin D1-regulated miRNA. Expression data was median centered such that tumor expression profiles reflected the expression of a given miRNA relative to the average expression of the miRNA across all tumors. The value of the cyclin D1-mediated miRNA expression signature for each tumor was determined by calculating the average expression of cyclin D1-regulated miRNA in each tumor. Tumors were then sorted according to expression of the cyclin D1-mediated miRNA expression signature and association with clinical covariates was tested using non-parametric statistical hypothesis testing. Association with relapse-free survival was assessed by first classifying tumors on the basis of miRNA expression, calculating the association of cluster membership with relapse-free survival and calculating the association between the cyclin D1-mediated miRNA expression signature and the miRNA clusters. Clusters were defined using hierarchical clustering using Pearson correlation and average linkage. The first two major bifurcations of the sample dendrogram (excluding two small clusters of outliers) were used to identify miRNA clusters. Cox proportional hazards analysis was used to associate miRNA clusters with relapse-free survival. Association of the cyclin D1-mediated miRNA expression with miRNA clusters was determined as described above.
TCGA database analysis. The GEPIA web server [55] was used to analyze the gene expression levels of cyclin D1, DKK1 and Wnt A in 1,085 breast cancer patients vs. 291 normal samples from TCGA and GTEx databases. The Pearson Correlation Coefficient was used to determine the strength of the relationship between cyclin D1 and Wnt signaling factors. Non-log scale for calculation and log-scale axis for visualization were applied.
Abbreviations
CCND1: cyclin D1; DKK: Dickkopf; ERα: estrogen receptor alpha; HDAC: histone deacetylase; 3' UTR: 3' untranslated region; RISC: RNA induced silencing complex; LPR: LDL receptor-related protein; RT-PCR: reverse transcription polymerase chain reaction; KEGG: kyoto encyclopedia of genes and genomes; TCF: transcription factor; LUC: luciferase; TOP: optimal TCF-binding site; FOP: far-from-optimal TCF-binding site; MEC: mammary epithelial cell; MEFs: mouse embryo fibroblasts; IHC: immunohistochemistry; IGF: insulin-like growth factor; TGFβ: transforming growth factor β; MMTV: mouse mammary tumor virus; ATCC: American type culture collection; DMEM: dulbecco's modified eagle's medium; Ct: cycle threshold; CMV: cytomegalovirus; TCGA: the cancer genome atlas; GTEx: genotype-tissue expression; SEM: standard error of the mean.
Acknowledgements
This work was supported by grants from the NIH (R01CA070896, R01CA086072) (R.G.P.), the Breast Cancer Research foundation (R.G.P.), the Dr. Ralph and Marian C. Falk Medical Research Trust (R.G.P.), the Pennsylvania Department of Health (R.G.P.) and the National Key Research and Development Program of China Stem Cell and Translational Research (2016YFA0101202); grants 81370175, 81572593 and 81372299 from Natural Science Foundation of China.
Supplementary Material
Supplementary figures and tables.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7:331-42
2. Lee RJ, Albanese C, Fu M, D'Amico M, Lin B, Watanabe G. et al. Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol Cell Biol. 2000;20:672-83
3. Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017-21
4. Arnold A, Papanikolaou A. Cyclin D1 in breast cancer pathogenesis. J Clin Oncol. 2005;23:4215-24
5. Barnes DM, Gillett CE. Cyclin D1 in breast cancer. Breast Cancer Res Treat. 1998;52:1-15
6. Peurala E, Koivunen P, Haapasaari KM, Bloigu R, Jukkola-Vuorinen A. The prognostic significance and value of cyclin D1, CDK4 and p16 in human breast cancer. Breast Cancer Res. 2013;15:R5
7. Pestell RG. New roles of cyclin D1. Am J Pathol. 2013;183:3-9
8. Sakamaki T, Casimiro MC, Ju X, Quong AA, Katiyar S, Liu M. et al. Cyclin D1 determines mitochondrial function in vivo. Mol Cell Biol. 2006;26:5449-69
9. Wang C, Li Z, Lu Y, Du R, Katiyar S, Yang J. et al. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc Natl Acad Sci U S A. 2006;103:11567-72
10. Hulit J, Wang C, Li Z, Albanese C, Rao M, Di Vizio D. et al. Cyclin D1 genetic heterozygosity regulates colonic epithelial cell differentiation and tumor number in ApcMin mice. Mol Cell Biol. 2004;24:7598-611
11. Fu M, Rao M, Bouras T, Wang C, Wu K, Zhang X. et al. Cyclin D1 inhibits peroxisome proliferator-activated receptor gamma-mediated adipogenesis through histone deacetylase recruitment. J Biol Chem. 2005;280:16934-41
12. Casimiro S, Luis I, Fernandes A, Pires R, Pinto A, Gouveia AG. et al. Analysis of a bone metastasis gene expression signature in patients with bone metastasis from solid tumors. Clin Exp Metastasis. 2012;29:155-64
13. Bienvenu F, Jirawatnotai S, Elias JE, Meyer CA, Mizeracka K, Marson A. et al. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature. 2010;463:374-8
14. Casimiro MC, Di Sante G, Di Rocco A, Loro E, Pupo C, Pestell TG. et al. Cyclin D1 Promotes Androgen-Dependent DNA Damage Repair in Prostate Cancer Cells. Cancer Res. 2017;77:3391-3405
15. Li Z, Chen K, Jiao X, Wang C, Willmarth NE, Casimiro MC. et al. Cyclin D1 integrates estrogen-mediated DNA damage repair signaling. Cancer Res. 2014;74:3959-70
16. Yu Z, Wang C, Wang M, Li Z, Casimiro MC, Liu M. et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol. 2008;182:509-17
17. Yu Z, Wang L, Wang C, Ju X, Wang M, Chen K. et al. Cyclin D1 induction of Dicer governs microRNA processing and expression in breast cancer. Nat Commun. 2013;4:2812
18. Yu Z, Li Y, Fan H, Liu Z, Pestell RG. miRNAs regulate stem cell self-renewal and differentiation. Front Genet. 2012;3:191
19. Di Leva G, Cheung DG, Croce CM. miRNA clusters as therapeutic targets for hormone-resistant breast cancer. Expert Rev Endocrinol Metab. 2015;10:607-617
20. Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ. et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214
21. Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, Thiruchelvam P. et al. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci U S A. 2009;106:15732-7
22. Shibahara Y, Miki Y, Onodera Y, Hata S, Chan MS, Yiu CC. et al. Aromatase inhibitor treatment of breast cancer cells increases the expression of let-7f, a microRNA targeting CYP19A1. J Pathol. 2012;227:357-66
23. Ward A, Shukla K, Balwierz A, Soons Z, König R, Sahin O. et al. MicroRNA-519a is a novel oncomir conferring tamoxifen resistance by targeting a network of tumour-suppressor genes in ER+ breast cancer. J Pathol. 2014;233:368-79
24. Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R. et al. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A. 1999;96:5522-7
25. Albanese C, Wu K, D'Amico M, Jarrett C, Joyce D, Hughes J. et al. IKKalpha regulates mitogenic signaling through transcriptional induction of cyclin D1 via Tcf. Mol Biol Cell. 2003;14:585-99
26. Lin SY, Xia W, Wang JC, Kwong KY, Spohn B, Wen Y. et al. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci U S A. 2000;97:4262-6
27. Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP. et al. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225-30
28. Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y. et al. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407:530-5
29. Lewis SL, Khoo PL, De Young RA, Steiner K, Wilcock C, Mukhopadhyay M. et al. Dkk1 and Wnt3 interact to control head morphogenesis in the mouse. Development. 2008;135:1791-801
30. Ou L, Fang L, Tang H, Qiao H, Zhang X, Wang Z. Dickkopf Wnt signaling pathway inhibitor 1 regulates the differentiation of mouse embryonic stem cells in vitro and in vivo. Mol Med Rep. 2016;13:720-30
31. Li M, Lee KF, Lu Y, Clarke I, Shih D, Eberhart C. et al. Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell. 2009;16:533-46
32. Casimiro MC, Crosariol M, Loro E, Ertel A, Yu Z, Dampier W. et al. ChIP sequencing of cyclin D1 reveals a transcriptional role in chromosomal instability in mice. J Clin Invest. 2012;122:833-43
33. Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:18097-102
34. Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549-57
35. Deshpande A, Pastore A, Deshpande AJ, Zimmermann Y, Hutter G, Weinkauf M. et al. 3'UTR mediated regulation of the cyclin D1 proto-oncogene. Cell Cycle. 2009;8:3592-600
36. Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217-22
37. Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O. et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766-70
38. Li M, Wang J, Ng SS, Chan CY, He ML, Yu F. et al. Adenosine diphosphate-ribosylation factor 6 is required for epidermal growth factor-induced glioblastoma cell proliferation. Cancer. 2009;115:4959-72
39. Li Z, Jiao X, Wang C, Shirley LA, Elsaleh H, Dahl O. et al. Alternative cyclin d1 splice forms differentially regulate the DNA damage response. Cancer Res. 2010;70:8802-11
40. Li Z, Wang C, Jiao X, Katiyar S, Casimiro MC, Prendergast GC. et al. Alternate cyclin D1 mRNA splicing modulates p27KIP1 binding and cell migration. J Biol Chem. 2008;283:7007-15
41. Jumpertz S, Hennes T, Asare Y, Vervoorts J, Bernhagen J, Schütz AK. The beta-catenin E3 ubiquitin ligase SIAH-1 is regulated by CSN5/JAB1 in CRC cells. Cell Signal. 2014;26:2051-9
42. Ding Q, Xia W, Liu JC, Yang JY, Lee DF, Xia J. et al. Erk associates with and primes GSK-3beta for its inactivation resulting in upregulation of beta-catenin. Mol Cell. 2005;19:159-70
43. Persad A, Venkateswaran G, Hao L, Garcia ME, Yoon J, Sidhu J. et al. Active beta-catenin is regulated by the PTEN/PI3 kinase pathway: a role for protein phosphatase PP2A. Genes Cancer. 2016;7:368-382
44. Gao B, Song H, Bishop K, Elliot G, Garrett L, English MA. et al. Wnt signaling gradients establish planar cell polarity by inducing Vangl2 phosphorylation through Ror2. Dev Cell. 2011;20:163-76
45. Hulit J, Lee RJ, Li Z, Wang C, Katiyar S, Yang J. et al. p27Kip1 repression of ErbB2-induced mammary tumor growth in transgenic mice involves Skp2 and Wnt/beta-catenin signaling. Cancer Res. 2006;66:8529-41
46. Ju X, Casimiro MC, Gormley M, Meng H, Jiao X, Katiyar S. et al. Identification of a cyclin D1 network in prostate cancer that antagonizes epithelial-mesenchymal restraint. Cancer Res. 2014;74:508-19
47. Casimiro MC, Di Sante G, Crosariol M, Loro E, Dampier W, Ertel A. et al. Kinase-independent role of cyclin D1 in chromosomal instability and mammary tumorigenesis. Oncotarget. 2015Apr20;6(11):8525-38
48. Buffa FM, Camps C, Winchester L, Snell CE, Gee HE, Sheldon H. et al. microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 2011;71:5635-45
49. Enerly E, Steinfeld I, Kleivi K, Leivonen SK, Aure MR, Russnes HG. et al. miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One. 2011;6:e16915
50. Lowery AJ, Miller N, Devaney A, McNeill RE, Davoren PA, Lemetre C. et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009;11:R27
51. Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, Dylag M. et al. ArrayExpress update-an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011;39:D1002-4
52. Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154-8
53. Shabalin AA, Tjelmeland H, Fan C, Perou CM, Nobel AB. Merging two gene-expression studies via cross-platform normalization. Bioinformatics. 2008;24:1154-60
54. Rudy J, Valafar F. Empirical comparison of cross-platform normalization methods for gene expression data. BMC Bioinformatics. 2011;12:467
55. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017 Apr 12. doi: 10.1093/nar/gkx247. [Epub ahead of print]
Author contact
![]() Corresponding author: Richard G. Pestell, Pennsylvania Cancer and Regenerative Medicine Research Center, Baruch S. Blumberg Institute, Pennsylvania Biotechnology Center, 100 East Lancaster Avenue, Suite, 222, Wynnewood, PA., 19096. Email: richard.pestellcom or Zuoren Yu, Ph.D, East Hospital, Tongji University School of Medicine, 150 Jimo Road, Shanghai 200120, China, Email: zuoren.yuedu.cn
Corresponding author: Richard G. Pestell, Pennsylvania Cancer and Regenerative Medicine Research Center, Baruch S. Blumberg Institute, Pennsylvania Biotechnology Center, 100 East Lancaster Avenue, Suite, 222, Wynnewood, PA., 19096. Email: richard.pestellcom or Zuoren Yu, Ph.D, East Hospital, Tongji University School of Medicine, 150 Jimo Road, Shanghai 200120, China, Email: zuoren.yuedu.cn
 Global reach, higher impact
Global reach, higher impact