13.3
Impact Factor
Theranostics 2017; 7(13):3354-3368. doi:10.7150/thno.19704 This issue Cite
Research Paper
Fluorocarbons Enhance Intracellular Delivery of Short STAT3-sensors and Enable Specific Imaging
1. Laboratory of Molecular Imaging Probes, Department of Radiology, University of Massachusetts Medical School, Worcester MA;
2. Department of Chemistry, Moscow State University, Moscow, Russian Federation;
3. A. Martinos' Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown MA 02129 USA.
* These authors contributed equally to this work.
Received 2017-2-17; Accepted 2017-7-12; Published 2017-8-3
Abstract
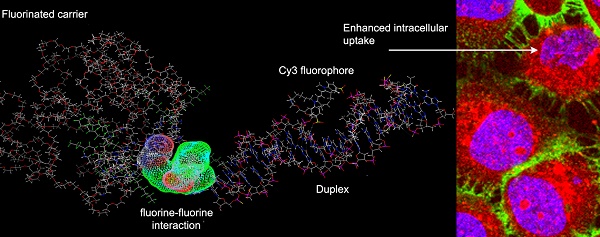
Short oligonucleotide sequences are now being widely investigated for their potential therapeutic properties. The modification of oligonucleotide termini with short fluorinated residues is capable of drastically altering their behavior in complex in vitro and in vivo systems, and thus may serve to greatly enhance their therapeutic potential. The main goals of our work were to explore: 1) how modification of STAT3 transcription factor-binding oligodeoxynucleotide (ODN) duplexes (ODND) with one or two short fluorocarbon (FC)-based residues would change their properties in vitro and in vivo, and if so, how this would affect their intracellular uptake by cancer cells, and 2) the ability of such modified ODND to form non-covalent complexes with FC-modified carrier macromolecule. The latter has an inherent advantage of producing a 19F-specific magnetic resonance (MR) imaging signature. Thus, we also tested the ability of such copolymers to generate 19F-MR signals.
Materials and Methods. Fluorinated nucleic acid residues were incorporated into ODN by using automated synthesis or via activated esters on ODN 5'-ends. To quantify ODND uptake by the cells and to track their stability, we covalently labeled ODN with fluorophores using internucleoside linker technology; the FC-modified carrier was synthesized by acylation of pegylated polylysine graft copolymer with perfluoroundecanoic acid (M5-gPLL-PFUDA).
Results. ODN with a single FC group exhibited a tendency to form duplexes with higher melting points and with increased stability against degradation when compared to control non-modified ODNs. ODND carrying fluorinated residues showed complex formation with M5-gPLL-PFUDA as predicted by molecular dynamics simulations. Moreover, FC groups modulated the specificity of ODND binding to the STAT3 target. Finally, FC modification resulted in greater cell uptake (2 to 4 fold higher) when compared to the uptake of non-modified ODND as determined by quantitative confocal fluorescence imaging of A431 and INS-1 cells.
Conclusion. ODND modification with FC residues enables fine-tuning of protein binding specificity to double-strand binding motifs and results in an increased internalization by A431 and INS-1 cells in culture. Our results show that modification of ODN termini with FC residues is both a feasible and powerful strategy for developing more efficient nucleic acid-based therapies with the added benefit of allowing for non-invasive MR imaging of ODND therapeutic targeting and response.
Keywords: fluorocarbon, oligonucleotide, magnetic resonance imaging, confocal microscopy, STAT3.
Introduction
Short oligonucleotides and their mimics (such as siRNA, oligonucleotide decoys, aptamers, locked and peptide- nucleic acids and others [1]) are the essential components of a plethora of emerging therapeutics designed for targeting and treating multiple diseases [2]. These therapies aim to manipulate the transcription and translation of a growing number of genes and proteins identified as causally linked to many diseases and some oligonucleotides may serve equally well as diagnostic agents via their capacity to be imaged [3-5]. Non-viral delivery of therapeutic and theranostic oligonucleotide-based constructs and their mimics generally requires protecting small oligonucleotides (ODN) from rapid removal and/or degradation in vivo which occurs due to their short circulation blood half-lives [6, 7]. In general, ODN are protected from the biological environment by either chemical modification of nucleotide units [8], or by encapsulation and/or by formation of complexes with synthetic carrier macromolecules with concomitant partial condensation into smaller structures. The latter approach most frequently relies on the attractive forces between the positively charged binding elements within the carrier and the negatively charged ODN resulting in polyionic interactions and may not be suitable for peptide nucleic acids and morpholinos. The latter could be induced by polyadenylation [9], or non-covalent addition of cell-penetrating peptides [10]. In most cases polyionic interactions can be fine-tuned to support the required formation of the protective complex and as a prerequisite for efficient interaction with the negatively charged cell surface. The limitation of this approach is that polyionic complexes show toxicity in vivo [11-13]. Within the last two decades a significant synthetic effort was made to provide an alternative to polyionic interactions. In particular, lipid and sterol- modification of ODN (reviewed in [14, 15]) as well as more complex multi-component systems of interaction-promoting mediators [16] showed promise in improving ODN delivery into cells [15, 17, 18]. A separate and highly attractive emerging approach is to utilize the interactions between fluorinated moieties introduced into the ODN and ODN-like molecules [19-21]. For decades, fluorine substituents served as common components in pharmaceuticals, particularly for applications in oncology in which about 50% of the marketed drugs were fluorinated, including multiple antineoplastic agents (e.g. fluorinated taxane and 5-fluorouracil) [22]. Fluorinated molecules tend to favor fluorine-dominated phases and engage in highly specific fluorine-fluorine interactions as a result of simultaneous hydrophobic and lipophobic electronegativity of fluorinated groups that have large molecular cross-sectional areas [23]. Such interactions facilitate interfacial assembly of nanoparticles and facile enrichment of engineered biomolecules carrying fluorinated tags [24]. As a consequence of fluorine-fluorine interactions, macromolecules carrying FC moieties exhibit a tendency to form supramolecular structures which were initially postulated to facilitate their penetration into the cells [19]. The internalization is also facilitated by a decrease of the diameters of nanoparticles as in the case of peptide nucleic acid carrying FC moieties [25]. Recent research demonstrated that fluorinated residues introduced into single-component or multi-component positively charged branched carriers resulted in improved transfection [26] and enabled gene editing [27]. Therefore, the majority of research so far has been focused on self-assembly of fluorinated single-strand ODN into nanoparticles or on fluorination of dendrimeric polycations. We endeavored to further enhance the therapeutic potential of these gene delivery constructs by incorporating two additional and as yet untested properties of fluorinated residues i.e., 1) the use of fluorine-fluorine interactions in maintaining the integrity of non-viral theranostic oligonucleotide-based constructs; 2) the potential of fluorinated molecules to act as highly specific MR imaging probes allowing detection and spatial localization of fluorinated compounds through the use of 19F selective magnetic resonance imaging (MRI), reviewed in [28, 29]. The utility of the latter has been suggested by demonstrations in vitro of 19F MR signal dependence on the presence of specific proteins [30] and on activity of a reporter enzyme [31]. Since cells internalize duplexes (ODND) much more efficiently than single-strand ODN [32], we hypothesized that fluorination would further benefit ODND delivery into cells. To test this hypothesis, we devised a hybrid, water-soluble macromolecular carrier to carry covalently linked FC residues and synthesized FC-modified ODND cargo molecules (Figure 1). The roles of ODND cargo in our research were assigned to STAT3-binding ODND constructs due to their potential for clinical development as cancer therapeutics. Constitutively activated STAT3 is being increasingly recognized as playing a pivotal role in progression of aggressive neoplasias such as difficult to treat head and neck variants and triple-negative breast cancer [33-35]. We report here on drug delivery and 19F-magnetic resonance imaging (MRI) properties of ODND modified with FC groups and their complexes with fluorinated macromolecular carriers.
A scheme showing major structural elements of fluorescent, Cy3 labeled oligonucleotide duplexes (ODND) used in this work. ODND contained either a single- fluorinated 5'-linked perfluorooctylpropyl group (1FO-ODND-Cy3) or two FO groups (2FO-ODND-Cy3). Fluorinated residues are shown in bright green, Cy3 fluorophore is shown in magenta. Yellow color of ODN chains shows the areas where phosphodiester bonds were replaced with phosphorothioates in hybrid (PS) ODNs. This design can be easily modified for use with different spectral frequencies because any other fluorophore (e.g. IRDye 800CW) can be used in the place of Cy3 for labeling ODND.
Materials and Methods
Synthesis of ODNs with CF3(CF2)7(CH2)3- group on the 5'-end
All synthesized ODNs were 21-mer oligodeoxyribonucleotides (for simplicity index d in the formulas below was omitted). Two strategies for linking fluorine-containing groups to (or introducing them synthetically into) the ODNs were used:
1) the synthesis of FO-oligonucleotides (where FO is CF3(CF2)7(CH2)3O-) by using direct chemical synthesis on an Expedite 8909 DNA synthesizer and a phosphoramidite monomer C8F17CH2CH2CH2OP(OCH2CH2CN) N(iPr)2 (fluorous propanol CEP, Berry&Associates, Dexter MI) as the last monomer during the synthesis;
2) the synthesis of FCN-oligonucleotides (where FCN is CF3(CF2)7(CH2)3 NHCO(CH2)9O-) by using post synthetic modification of a fully protected ODN on controlled pore glass (CPG) by linking of 3-(perfluorooctyl)propylamine to the activated carboxy group on ODN.
Strategy 1 Method
For standard coupling we used Ultramild CE Phosphoramidites (Glen Research), dC*- or T*-amidite- (synthons with the TFA-protected amino-linkers) synthesized as described before [36]. Monomer phosphoramidite CF3(CF2)7(CH2)3O-P(OCH2CH2CN) N(iPr)2 (250 µmoles) were dissolved in 3 ml of acetonitrile and used at the last step of the synthesis [37]. For synthesis of phosphorothioates the oxidation and the sulfurization steps were performed as described in [38]. After the synthesis, CPG with attached oligonucleotides was treated with NH4OH at 40oC for 2 h. Oligonucleotides were re-precipitated in ethanol-sodium acetate twice before loading onto HPLC columns. The isolation of FO-oligonucleotide was accomplished on C18 reversed phase-HPLC (Discovery C18, 250x4.6 mm, 5 µm) using a gradient of buffer A (2% acetonitrile in 0.1 M TEAA) and buffer B (20% of buffer A, 80% acetonitrile) at 1 ml/min. A gradient was formed by ramping 100% buffer A to 80% B in 22 min followed by 2 min 80% B. The typical chromatograms of oligonucleotide mixture analysis are shown in Fig.S1A and S1B as an example of the analysis of CF3(CF2)7(CH2)3-O-AGAGATTTAC*GGGAAATGGCT, where * stands for the internucleoside amino linker -(OCH2CH2)2OCH2CH2NH2 attached to phosphorus. The following oligonucleotides with the FO group on the 5'-end of the oligonucleotide were synthesized using Method 1:
1. 5' FO-AGCCAT*TTCCCGTAAATCTCT 3' FO-PS-STAT-SE-T6
2. 5' FO-AGAGATT*TACGGGAAATGGCT 3' FO-PS-STAT-AN-T7
3. 5' FO-AGCC*ATTTCCCGTAAATCTCT 3' FO-PO-STAT-SE-C4
4. 5' FO-AGAGATTTAC*GGGAAATGGCT 3' FO-PO-STAT-AN-C10
5. 5' FO-AGAGATTTAC*GGGAAATGGCT 3' FO-PS-STAT-AN-C10
The above oligonucleotides were designed in a way that sense and antisense strands could form decoys for STAT3. The positions of bases linked via phosphorothioate bonds are indicated in bold, whereas phophodiester-linked bases are shown with plain text and the numbers after the base indicate the position of the internucleoside amino linker.
Strategy 2 Method
The synthesis of fluorinated ODN does not necessarily require the use of fluorinated synthons. Fluorinated residues were also introduced by using ODNs terminated with activated carboxyls as precursors resulting in FCN-oligonucleotides bearing heptadecafluoroundecylcarbamoylnonyl groups. First, ODNs with N-hydroxysuccinimide esters (5'-NHS) on the 5'-end oligonucleotides were synthesized by using standard monomers and 5'-carboxy-modifier C10 (Glen Research) for the last coupling step. Second, 3-(perfluorooctyl)propylamine was linked to the activated oligonucleotide directly after the synthesis. 0.12 ml of methanol solution of 3-(perfluorooctyl)propylamine (50 mg/ml) and 4 µl triethylamine were added to 15-25 mg CPG with fully protected oligonucleotide carrying 5'-NHS group, and after 3-5 hours at room temperature CPG was washed with methanol. Deprotection was performed by using concentrated NH4OH at 55°C overnight. Oligonucleotides were re-precipitated by ethanol-sodium acetate twice before HPLC purification (see Fig.S2).
The following oligonucleotides with FCN on 5'-end (FCN-ODN) were synthesized using Method 2:
1. 5' FCN-CGATGTGGGACTTTCCAGGCT 3' FCN-PS-IGL-AN
2. 5' FCN-AGCCTGGAAAGTCCCACATCG 3' FCN-PS-IGL-SE
3. 5' FCN-CGAGATTTACGGGAAATGGCT 3' FCN-PS-STAT-AN
4. 5' FCN-AGCCATTTCCCGTAAATCTCG 3' FCN-PS-STAT-SE
HPLC analysis of fluorocarbon-modified ODNs was performed by using either C18, or CN columns with characteristic differences in retention times noted for all tested 21-mer ODNs (Table S1).
ODN modification with cyanine fluorophores
To 5 nmol of FO-AGAGATTTAC*GGGAAATGGCT, FO-AGAGATTTAC*GGGAAATGGCT or FO-AGCC*ATTTCCCGTAAATCTCT dissolved in 35 µl of 0.1 M NaHCO3 were added 5-6 µl of 5 mM Cy3-NHS ester (GE Healthcare Life Sci.) or 7.5 mM IRDye 800CW NHS ester (Li-COR Biosciences). The reaction mixture was kept at room temperature overnight with agitation. After adding 30 µl H2O, ODN were loaded on Bio-Spin P6 columns (Bio-Rad) and purified according to the standard protocol described above. HPLC analysis of FO-AGAGATTTAC*Cy3GGGAAATGGCT is shown in Figure S3.
Molecular dynamics simulations
Molecular dynamics simulations in a molecular operation environment (MOE) was accomplished by using MOE2015.10 and MOE2016 software (Chemical Computing Group, Montreal CA) [39]. The models of ODND probes were placed in a simulated NaCl/water cell environment with n=8 margin and after calculating partial charges and potential energy minimization by applying assisted model building with energy refinement (Amber10:EHT) force field. This is an all-atom forcefield combining EHT and Amber10, parameterized for proteins and nucleic acids as well as for small molecules according to 2D-extended Hückel's theory conventions [40]. Simulations were performed by using the Nosé-Poincaré-Andersen (NPA) algorithm with a 0.001 ps time step.
ODN probes, spectroscopy and melting behavior
ODND probes were prepared by mixing approximately equimolar amounts of the corresponding complementary ODN dissolved in 25 mM Hepes, 1 mM MgCl2, 50 mM NaCl, pH 7.4. One of the complementary ODN used in these experiments contained the covalently linked fluorophore. The solutions were heated to 95°C for 5 min to dissociate intra-strand secondary structures and allowed to cool at room temperature to attain equilibrium. Fluorescence emission spectra of ODND or single-strand ODN (λem=500-650 nm, Cary Eclipse spectrofluorometer) were collected at a concentration of 2 µM by using λex=488 nm. The fluorescence intensity of ODNs and ODND was normalized by the concentration of Cy3 determined from the absorbance spectra (λmax=552 nm, ε552=150000 [M.cm]-1). Melting behavior was studied using a Hitachi U-2900 spectrophotometer equipped with a thermoelectric cell holder and a temperature controller. ODND were diluted in 10 mM sodium phosphate, 0.15 M NaCl, pH 7.4 for melting experiments.
Stability in cell culture medium and electrophoretic mobility shift assay (EMSA)
The conditioned DMEM cell culture medium was obtained after culturing A431 cells (human epidermioid carcinoma, ATCC CRL-1555) for 1-2 days after which it was filtered through a 0.22µm Millipore Express PLUS Membrane and stored at 4oC. ODND at 0.5 µM dilution was incubated in 50% conditioned medium either at RT or at 37oC. For EMSA, IRDye 800CW-labelled ODND (0.04 pmol, final concentration 3-4 nM) was incubated with 10-20 µg A431 nuclear extract (isolated as described in [41]) in 20 mM HEPES, 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 25% v/v glycerol, 0.5 mM phenylmethylsulfonyl fluoride, 5 μg/mL leupeptin, 100 μg/mL aprotinin, 1 μg/mL pepstatin for 30 min RT in a volume of 10-15 µl in the presence of protein binding buffer (10 mM Tris, 100 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 0.1 mg/mL tRNA, 10% v/v glycerol, 0.25 mM DTT, pH 7.5). Samples were loaded and run on 15% TBE Ready Gels (Bio-Rad Laboratories, Hercules CA) with 0.5×TBE buffer. The gels were imaged and digitized using an Odyssey Infrared Imaging system (Li-Cor Biosciences, Lincoln NE) using simultaneous solid diode excitation at 685 nm (IRDye-680 STAT3 probe, Li-Cor) and 785 nm (800CW-labeled ODND). TIFF images (16 bit) were analyzed, colorized and fused using ImageJ software [42].
M5-gPLL-PFUDA synthesis
The synthesis, properties and purification of M5-gPLL graft copolymer were described in detail in [43, 44]. Briefly, methoxy PEG succinimidyl carboxymethyl ester (m.w. 5000, JenKem Technology) was used to modify 30% of TNBS-reactive amino groups of poly-L-lysine (m.w. 21 KDa, DP 100, PDI 1.12, Alamanda Polymers) with subsequent purification of the product on UFP-100 ultrafiltration cartridges (GE Healthcare Life Sciences). Thirty mg (approximately 0.15 µmol) of M5-gPLL was dissolved, with heating in 1 ml of dry acetonitrile. Ten µl (60 µmol) of diisopropylethylamine (DIPEA) and 8 µl of 15.5 mM (124 nmol) Alexa Fluor 488 NHS ester (ThermoFisher Scientific) were added to M5-gPLL solution and incubated for 2 h under argon; equimolar amounts of perfluoroundecanoic acid (PFUDA) and HOBt (80 µmol each) were combined with 160 µmol EDC (Sigma-Aldrich) in 2 ml dry acetonitrile and mixed until complete solubilization. The solution was added to M5-gPLL, followed by 60 µmol DIPEA, sealed under argon and mixed overnight. The solvent was removed in vacuum and solids were solubilized in 10 ml water. After the addition of 20 ml of acetone and 40 ml of ethyl acetate, the lower phase was separated and the extraction procedure repeated once. Final purification was accomplished by dialysis against 30% isopropanol/water using 10 kDa cutoff membrane cartridges (ThermoFisher Scientific). The purity of M5-gPLL-PFUDA was verified by size-exclusion HPLC on Superose 6 Increase 10/300 GL column (GE Healthcare Life Sciences) eluted with 0.1 M ammonium acetate.
ODN duplex M5-gPLL-PFUDA binding experiments
Ten μL of 5 μM ODND solution was mixed with M5-gPLL-PFUDA solution at a ratio of 1-20 moles of M5-gPLL-PFUDA per ODN (ether modified with FO, or control) in a total of 35μL HBS, pH 7.5 and incubated at room temperature for 2h. Samples were loaded and subjected to electrophoresis on 2% agarose gel in 0.5XTBE buffer for 45min, 100v. The gels were imaged on an EpiChem III system (UVP BioImaging System, Upland, CA 91786) equipped with a Hamamatsu CCD camera and the obtained TIF images of gels were analyzed using ImageJ (NIH software) [42]. Fluorescence lifetime was measured as described in [45, 46] using 520-540 nm excitation and 580 nm long-pass filter; time resolved fluorescence was registered using a CCD camera coupled to a gated-image-intensifier (LaVision Gmbh, Germany) that provides a 300 ps time resolution.
ODN duplex cell uptake experiments
A431 cells were maintained in DMEM medium (Gibco)/ 10% fetal bovine serum (FBS) (Invitrogen). INS-1 cells [47] (generously provided by Dr. Anna Moore, Massachusetts General Hospital, Charlestown MA) were maintained in RPMI medium (Gibco) supplemented with 10% FBS. Cells were seeded on coverslips in 12-well plates. After the cells were 60-70% confluent, ODND were added to the cells at a final concentration of 0.5 μM and incubated overnight. The cells were washed three times with PBS and fixed by incubating for 15 min in 4% formaldehyde. For individual cell delineation, plasma membranes of fixed cells were stained using anti-EGFR (Cetuximab)- Alexa Fluor 488 conjugate, or, alternatively, by 5-min incubation in the presence of 5 µM Alexa Fluor 488 hydroxysuccinimide ester. The cells were mounted in Vectashield medium with DAPI (Vector Laboratories, Burlingame, CA) and sealed on slides. The slides were examined using a Leica TCS SP8 confocal microscope (Leica Microsystems, Buffalo Gove, IL) using a 63x/1.4NA lens and 95.6 µm pinhole (at 1AU, λem=580 nm) resulting in an approximate optical section thickness of dz = 1.4 µm. For Cy3 fluorescence excitation/registration we used a DPSS 561 nm laser line and hybrid photon counting HyD detector (active band 563-668 nm). Three to four fields of view were imaged in three channels and saved as 16-bit tiff files which were analyzed by using ImageJ software as follows: first, cell boundaries within the field of view were outlined individually with the outline following the green channel fluorescence signal representing anti-EGFR-Alexa Fluor 488 conjugate binding to cell membrane. The ROIs derived from the outlines of individual cells were then pasted onto the “red” channel images and the corresponding signals in the “red” (Cy3) channel were calculated. Fluorescence intensities were determined for 30 to 40 cells in three to four non-overlapping fields of view and the uptake of Cy3-labeld ODND was expressed as mean intensity/cell. The fluorescence intensity values were compared by using Mann-Whitney non-parametric test (Prism 6, GraphPad Software Inc.).
Magnetic resonance imaging
Phantoms were prepared by diluting 100 mM stock solution of PFUDA in acetonitrile at various concentrations. Before imaging M5-gPLL-PFUDA alone, the complex of 2FO-PO-Cy3 with M5-gPLL-PFUDA were solubilized either in acetonitrile or water, respectively. Imaging was performed either in NMR spectroscopy tubes or in 0.2 ml polypropylene PCR tubes. 19F NMR spectra were acquired using a Varian Mercury 200 MHz (4.7T) NMR Spectrometer and a 3T Achieva whole-body MRI scanner (Philips Medical Systems, Best, Netherlands). The 19F chemical shifts were referenced to sodium trifluoroacetate at 0 ppm. All 1H and 19F MRI experiments were performed at 3.0 T (128 MHz for 1H) by using a custom-made solenoid T/R coil and modular 19F Gateway Interface (Clinical MR Solutions, LLC Brookfield, WI). The coil was first tuned to the proton resonant frequency and a turbo spin echo (TSE) pulse sequence was used to acquire 1H images. The 1H imaging parameters were: 8 slices with slice thickness of 4 mm; field of view (FOV) = 30 mm × 30 mm with matrix size of 76 × 72; TR/TE = 3000/70 ms; flip angle (FA) = 90°; TSE-factor = 8; NSA = 8. Subsequently, the coil was tuned to 19F resonant frequency and 1H images were used to plan the 19F MR imaging plane. A 3D ultrashort echo time (UTE) pulse sequence was used to acquire 19F MR images to maximize signal from 19F, which has a very short T2, and thus rapidly decaying MR signal. Feasibility 19F MR images were acquired by using the following parameters: TR/TE = 300/0.12 ms; flip angle (FA) = 60°; NSA = 32; 5 slices with slice thickness of 4 mm; field of view (FOV) of 30 mm × 30 mm with matrix size of 32 × 32, the total acquisition time for 19F imaging was 51 minutes. The 19F MR images of Cy3-labeled 2FO-ODND complex with M5-gPLL-PFUDA were acquired by using UTE (TR/TE=600/0.12 ms; FA= 60°), 9 slices with slice thickness of 0.9 mm; voxel size=1x1x4 mm, field of view (FOV) of 30 mm × 30 mm (11.5 min acquisition time with 6xNSA) or with a smaller voxel size of 0.5x0.5x2 mm (47 min acquisition time with 6xNSA). Raw images were imported and reconstructed using ImageJ. Signal-to-noise ratios were calculated by normalizing the region-of-interest mean intensity values by standard deviation of signal noise.
Results
ODN synthesis, structure analysis and properties of ODNs and ODND
We synthesized several sense and anti-sense ODN encoding STAT3 transcription factor binding interferon-gamma activated sequence (GAS), TTNCNNNAA. To provide the building blocks of 5'-perfluorooctylpropyl(FO)- containing ODND probes, the sense ODN (FO-ODN) was synthesized using a perfluorooctyl-terminated phosphoramidite. Several sense ODN were also synthesized using an alternative technique that involved linking 3-(perfluorooctyl)propylamine to 5'-NHS-terminated ODN-CPG followed by the cleavage of ODN product (FCN-ODNs) from CPG. The ODNs carried a hydrophilic internucleoside amino-linker [36, 48] which enabled covalent linking of Cy3 dye to ODNs with approximately the same stoichiometric yields (0.8-1.1 mol/mol) regardless of their composition and, importantly, the presence of ODN 5'-end-modification (Table 1).
Further, we investigated whether the modification of ODNs with either one (1FO), or two FO (2FO) fluorinated residues resulted in ODND destabilization. This was accomplished by comparing melting temperatures (Tm) of FO-ODND and control ODND (Table 2 and 2S). As expected, introducing three consecutive phosphorothioate bonds at the 3' and 5'- ends of ODN resulted in the lowering of Tm of ODND from approximately 71°С for 100% phosphodiester-bond linked ODN to 64°С for phosphodiester-phosphorothioate hybrid ones (abbreviated as PS further on in the text). However, the modification of a duplex with a single FO “tail” as well as linking of a single Cy3 fluorophore to the internucleoside amino linker did not yield any significant destabilization of ODND structure (according to the measured changes of Tm) in the presence of 0.15M NaCl and the destabilizing effect in the absence of NaCl was also negligible (Table 2). Changing the position of the fluorophore by linking Cy3 to a complementary strand also did not result in major changes to Tm values (Table S2). In 2FO-ODND samples the analysis of Tm data showed a decrease of Tm only if PS bonds were additionally introduced during the ODN synthesis (Table 2).
ODND molecular dynamics simulations
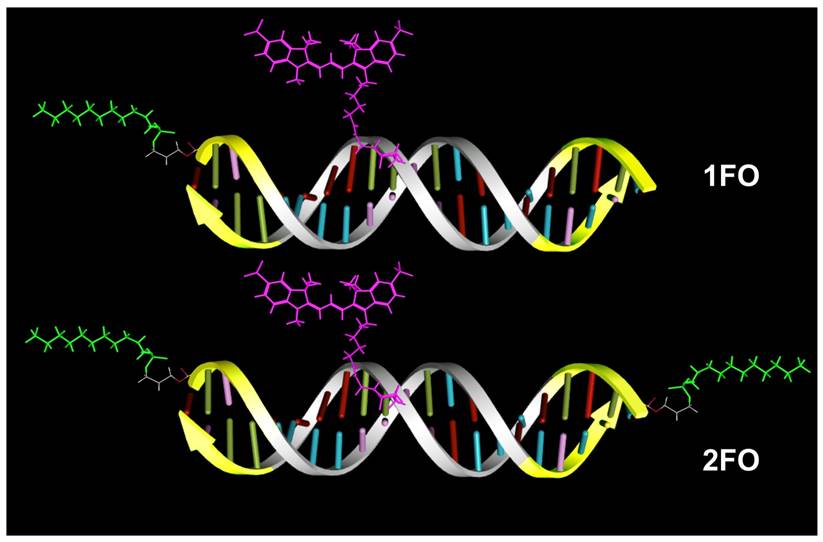
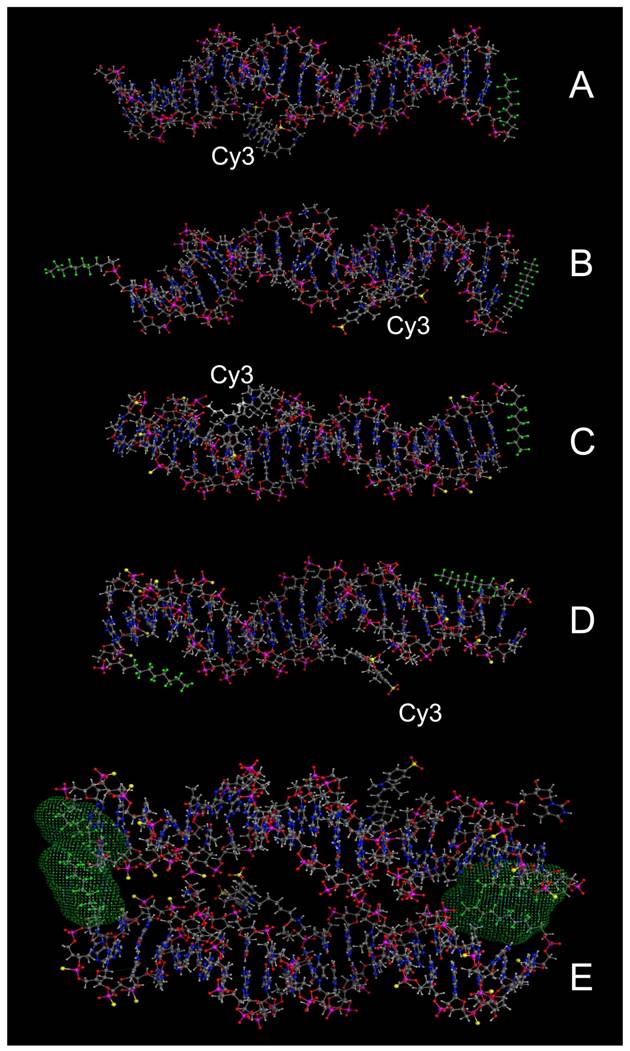
We selected four different Cy3-modified, red fluorescent ODND for further testing (Table 2). The structures of these ODND (Fig. 2) were built in silico and molecular dynamics in a water-filled cell was simulated in molecular operation environment (MOE) using an elliptical water/NaCl filled cell with n=8 solvent margin. Simulation results pointed to minimal changes in the structure of phosphodiester duplexes after the modification of ODND with one or two fluorinated residues, Fig. 2 (B-D). In the case of PO/PS ODND, we noted the formation of duplex structures with partially unwound terminus or termini at the equilibrium, which were favored by simulation algorithm only if two FO groups were present. In addition, simulations performed in a combined water/NaCl solute cell containing two FO-carrying ODND pointed to a high probability of inter-duplex fluorine-fluorine interactions resulting in the formation of dimers (Fig. 2, E).
Properties of 21-mer synthetic oligonucleotides with and without covalently linked fluorinated moieties.

* FO = CF3(CF2)7(CH2)3O-
** ODN samples were analyzed by LC/MS using C18 column separation and Synapt G2si instrument. Column: Waters OST C18, 2.5 µm 2.1x 50mm; Flow rate: 0.2ml/min
Sample: 10pmol Gradient: mobile phase A: 20mM ammonium acetate; mobile phase B: acetonitrile
*** The ratios were determined by using the following extinction coefficients: Cy3 (in [mol .cm]-1: ε260=4930; ε552=150000; ODN extinction was determined by using the following formula: ε260 = ((nA × 15.4) + (nC × 7.4) + (nG × 11.5) + (nT × 8.7)) × 900. The position of an aminodiethoxyethyl linker is shown by | symbol.
The sequences of control and ODN duplexes carrying FO groups with Cy3 fluorophore linked to sense or antisense ODNs used in this study.

a) ODNs were labeled with Cy3 at internucleoside linker position (shown in red). The position of an amino linker on a complementary ODN in a duplex is shown by |;
b) shown as fluorescence intensity (λex=488 nm, λem=552 nm, in AU) normalized by Cy3 concentration (µM).
Molecular modeling of PO ODND (A,B) and PS ODND: (A) 1FO-PO-Cy3; (B) 2FO-PO-Cy3; (C) 1FO-PS-Cy3; (D) 2FO-PS-Cy3 (see Table 2). A hypothetical dimer of 2FO-PS-Cy3 ODND is shown in E. Fluorine atoms are rendered in green and sulfur atoms are rendered in yellow. Van-der-Waals surfaces (4.5 Å) of interacting fluorinated residues were calculated and rendered in green, Cy3 dye is shown covalently linked to the amino groups of internucleoside phosphate triethylene glycol amino linkers. Amino linkers of complementary ODNs in these duplexes were left non-modified in all simulations. Oligonucleotides carry GAS STAT3 recognition sequence (TTCCCGTAA). The structures were obtained by using molecular dynamics simulations in MOE by applying assisted model building with energy refinement (Amber10:EHT) force field using a simulated elliptical water/NaCl solvent cell (n=8 margin).
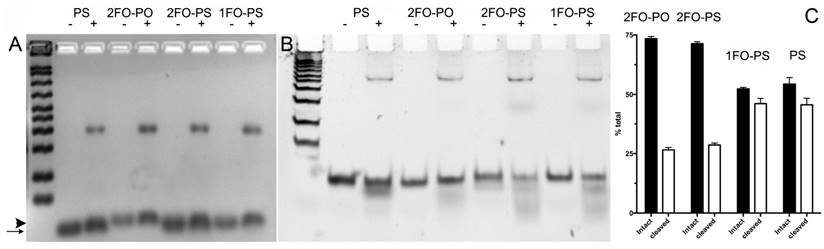
Electrophoretic analysis of Cy3-labeled ODN duplex probe stability in conditioned cell culture medium. A- 3% agarose gel electrophoresis of FO-ODND or control ODND samples (2 µM ODND) incubated in the presence (+) or absence (-) of A431 cell conditioned medium for 16 h at room temperature; B- 15% polyacrylamide gel electrophoresis (0.5xTBE) of the same samples incubated for 16 h at 37oC. The first lane of the gels shows a pre-stained 50-200 bp ODN ladder. The images were acquired using SybrGold filters and are inverted for better viewing. The arrow shows the position of ODND and arrowhead shows the position of 2FO-ODND migration, respectively; C- quantification of intact and degraded fractions of ODND as determined by using ROI measurements of integrated band intensity.
Biological stability of ODND and STAT3 binding affinity
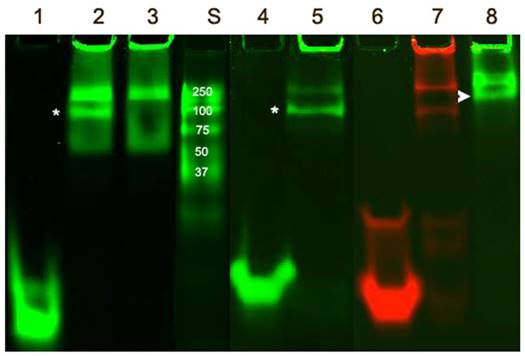
The remarkable differences in simulated ODND structures at equilibrium in solution at ambient temperatures suggested potential differential stability to degradation in biological milieu. To investigate this we first analyzed the stability of the four chosen ODND structures against degradation using the medium conditioned by A431 human squamous cell carcinoma culture. Agarose electrophoresis showed that all four duplexes were partially associated with the medium components to a similar extent regardless of the presence or absence of FO groups. In addition, agarose electrophoresis also showed a somewhat slower migration of FO-modified ODND suggesting a potential interaction with the gel matrix (Fig. 3A). More detailed analysis of the same samples incubated at 37oC overnight in the presence of conditioned medium instead of room temperature showed that all ODND exhibited at least some degree of degradation (Fig. 3B). All ODND tested showed association with cell culture media proteins that ranged between 7.5% and 12%. In the samples containing 2FO-PO duplex (Table 2) the degradation did not exceed 20% (Fig. 3B), whereas duplexes 2FO-PS and 1FO-PS, were more affected by the degradation. While only 33-35% of FO-containing PS ODND were degraded, in the absence of FO groups, degradation of ODND exceeded 50% (Fig. 3). As the next step we studied the effect of fluorination on binding of GAS-sequence encoding ODND to STAT3 in the nuclear lysates of A431 cells overexpressing this transcription factor. By conjugating a near-infrared NIR Dye 800CW to an internucleoside amino linker instead of Cy3 we achieved high sensitivity of ODND detection enabling the tracking of ODND binding with STAT3 by using electrophoretic shift mobility assay (EMSA, Fig. 4). The assays demonstrated that all PS ODN devoid of FO groups (green-pseudocolor fluorescence) and a control commercially available near-infrared ODND probe (red pseudo color fluorescence) showed similar patterns of electrophoretic mobility shift in the presence of nuclear lysate.
The addition of an excess of the non-labeled ODND resulted in the elimination of the band corresponding to STAT3-ODND complex (marked with an asterisk, Fig. 4, lane 3) which allowed determination of the specificity of ODND-STA3 binding. The addition of anti-STAT3 antibody to the mixture resulted in detectable supershift Fig. 4, lane 8, arrowhead). The introduction of a single FO group resulted in an increase of ODND binding specificity as the intensity of a non-specific band decreased (Fig. 4, lane 5), while 1FO-ODND was still able to recognize STAT3. However, unlike ODND and single FO-group modified ODND, two FO groups in the composition of 2FO-ODND probes resulted in a loss of binding either to STAT3, or to non-specific components in the lysate.
Cell culture experiments
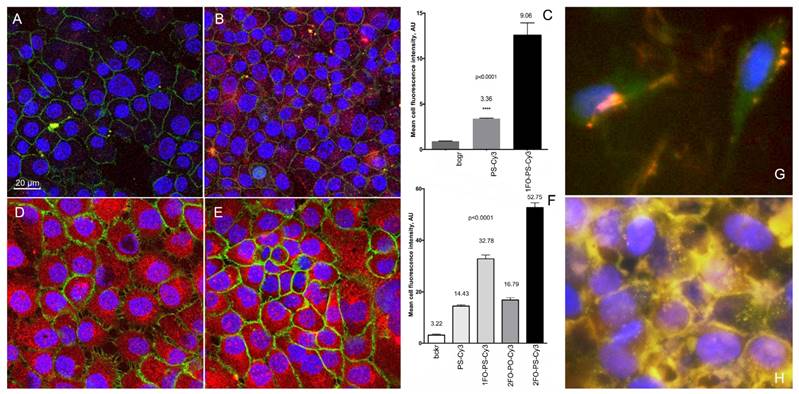
We further compared the intracellular fluorescence intensity of individual cells to determine whether Cy3-labeled ODND exhibit any measurable differences in cell uptake depending on the presence or absence of FO residues in their composition. We examined monolayer cell cultures after incubation at 37oC overnight in the presence of various 0.5 µM Cy3-labeled ODND probes. The comparison of Cy3-labeled PS, 1FO-PS, 2FO-PS and 2FO-PO ODND probes showed that the FO-modified ODND were internalized into cells of two tumor cell lines (A431 and INS-1) at significantly higher levels than control ODND that were lacking FO groups (Fig. 5). The efficacy of ODND probe internalization (expressed as mean fluorescence intensity of a single cell optical section) in INS-1 cells showed higher uptake of fluorinated ODND probes. The mean cell fluorescence in INS-1 cells was approximately 3-times higher in the case of 2FO-PS than PS ODND probe, Fig. 5A-C. The levels of uptake of all ODND were higher in A431 cells then in INS-1. For example, in PS ODND the addition of each FO residue to ODND resulted in statistically significant doubling of intracellular fluorescence reflecting the enhanced efficacy of uptake, with 2FO-PS modified duplex probes showing at least a 5-times higher uptake than the PS control based on mean cell fluorescence values (Fig. 5D-F).
Electrophoretic mobility shift assay performed using FO-containing and control ODN duplex probes. IRDye 800 CW-labeled ODND, i.e. PS, 1FO-PS, 2FO-PO as well as a control duplex (4 pmol IRDye-680 STAT3 probe, Li-Cor) were incubated in the presence (lanes 2,3,5,7,8) or the absence (lanes 1,4,6) of 24µg A431 nuclear extract and resolved in 15% polyacrylamide TBE gels. A 100-fold molar excess of non-labeled ODND competitive inhibitor was added to the sample of lane 3. Three µg of rabbit polyclonal anti-STAT3 antibody was added to the sample of lane 8. Lanes 1-3 and 8 - PS ODND; lanes 4-5 - 1FO-PS; lanes 6-7 - IRDye-680 STAT3 probe (positive control). Asterisks mark STAT3 band (position determined by using competitive inhibition); arrowhead points to a supershifted band. S- IRDye 800CW-labeled protein molecular weight markers.
The uptake of Cy3-labeled ODND (0.5 µM, 18h) with or without 5'-linked FO groups in cell culture of human cells. (A, B) confocal multichannel fluorescence microscopy of insulinoma INS-1 cells after an overnight incubation with PS-Cy3 (A) or 1FO-PS-Cy3 (B) ODND. (D, E) confocal microscopy of human squamous cell carcinoma A431 cells after incubating with 1FO-PS-Cy3 (D) or 2FO-PS-Cy3 (E) Blue - DAPI. Green - anti-EGFR-AF488 (A431 cells) or AF488-NHS (INS-1). Red - Cy3 (ODND probes). (C, F) quantification of the uptake using mean intensity measurements of individual cells (C) INS-1; (F) A431; acquired in the red channel as described in Materials and Methods. Column statistics and P values were determined by using non-parametric Mann-Whitney test, n=30-60 cells; (G, H) fluorescence microscopy of 2FO-PS-Cy3 (red) complex with M5-gPLL-PFUDA (AF488-labeled, green) after a 2 h (G) or 24h (H) incubation with A431 cells.
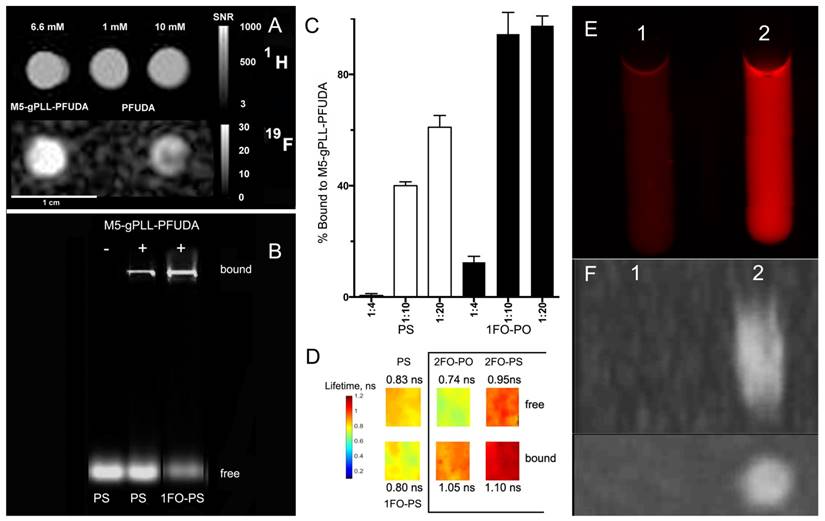
19F/1H MR imaging and FO-ODN binding to M5-gPLL-PFUDA carrier. A- 1H (TSE pulse sequence) and corresponding 19F MR images (UTE pulse sequence) of M5-gPLL-PFUDA (at 6.6 mM PFUDA linked to M5-gPLL) and two PFUDA standards (1 and 10 mM). Note the higher MR signal in M5-gPLL-PFUDA at lower PFUDA concentration than in the 10 mM PFUDA standard acquired as described in Materials and Methods. Calibration scales showing SNR values are on the left, 1 cm bar is shown at the bottom; B- agarose electrophoresis results showing the comparative binding of near-infrared IR Dye 800CW fluorescent-labeled PS- and 1FO-PS ODND to M5-gPLL-PFUDA; C- quantification of Cy3-labeled ODN binding to M5-gPLL-PFUDA using near-infrared agarose gel imaging and analysis. The ratios are shown as mol ODN/mol M5-gPLL-PFUDA. D- fluorescence lifetime maps of Cy3 fluorophore linked to ODND measured in 0.1 µM samples of PS, 1FO-PS ODND as well as free and M5-gPLL-PFUDA bound 2FO-PO and 2FO-PS ODND (showed in a box). The average FL (measured in ns) is shown next to the maps and the pseudocolor lifetime scale is shown on the left; E- the comparison of red fluorescence of 4.5 µM 2FO-PO-Cy3 (sample 1) and 4.5 µM 2FO-PO-Cy3/M5-gPLL-PFUDA, 2 mM PFUDA (sample 2); F - Maximum intensity pixel projection reconstruction 19F MR image (n=6 slices) of samples 1 and 2 acquired by using UTE pulse sequence in two projections: sagittal (above) and transverse (below).
ODND binding and delivery by M5-gPLL-PFUDA
We used covalent linking of a fluorinated “adaptor molecule” to M5-gPLL, a conjugate of polylysine backbone and grafted methoxyPEG chains [43] to synthesize a carrier for the delivery of fluorinated ODND into cells. By using acetonitrile as a solvent for both M5-gPLL and perfluoroundecanoic acid (PFUDA) we achieved efficient coupling of PFUDA to primary amino groups of M5-gPLL via a two-step carbodiimide activation and coupling procedure. The resultant purified conjugate of M5-gPLL and PFUDA was fully soluble in acetonitrile. While PFUDA alone is insoluble in water, the purified M5-gPLL-PFUDA was water-soluble if the initial pegylation degree was 25-30% of total available primary amino groups of PLL. Fluorine nuclear magnetic resonance (NMR) spectra of M5-gPLL-PFUDA solutions and PFUDA standards were then performed at high filed to determine PFUDA concentrations. Integration of aliphatic fluorine resonances (from -35 to -45 ppm, with exception of the line of perfluoromethyl fluorines set at zero ppm shift, see Fig. S5A) allowed determination of the concentration of PFUDA in M5-gPLL-PFUDA (Fig. S5E). An expected broadening of peaks was observed in both NMR spectra due to the increased viscosity of polymer carrier (Fig. S5 C, D). On T1 weighted images the 19F MR signal intensity obtained by using ultra-short echo pulse sequence was higher at lower concentrations of fluorine than 19F MR intensity signal of PFUDA (Fig. 6A) suggesting the decreased mobility of perfluorinated acyl chains in the vicinity of PLL backbone due to the high local viscosity compared to PFUDA solutions in acetonitrile. As a result, we observed a 5.7-fold reduction in T1 relaxation time (2000 ms vs. 350 ms) and this change translated into a higher MR signal intensity. The comparison of normalized signal to noise ratios (SNR) showed a 1.6-times higher SNR in the case of M5-gPLL-PFUDA than 10 mM PFUDA standard (20.5 vs. 13.0). We then made an observation that both duplexes, whether they were modified with 3-perfluorooctylpropyl residues or not, were forming complexes with M5-gPLL-PFUDA carrier polymer (Fig. 6B). However, only ODNs bearing FO residues such as 1FO-PS were bound at 100% to M5-gPLL-PFUDA at the molar ratio of 1:20 (ODND/carrier) while control PS was binding to the carrier at lower levels (Fig. 6C). These results were obtained by integrating fluorescence signals of the fraction of free ODND probes covalently labeled either by using Cy3 or 800CW fluorophores. This was necessary because both cyanine fluorophores used for coupling to ODNs showed a considerable increase of fluorescence intensity, i.e. ODN-linked Cy3 showed a 20% increase and NIRDye 800CW-linked showed a 50% signal intensity increase after binding to M5-gPLL-PFUDA compared to the free 1FO-PS ODND. These results suggest a change in fluorophore microenvironment resulting in higher fluorescence yields. Our measurements of Cy3 fluorescence lifetime (FL) showed that after the binding to M5-gPLL-PFUDA, the lifetime of Cy3 increased from 0.74±0.04 to 1.05±0.02 ns in the case of 2FO-PO and from 0.95±0.04 to 1.10±0.06 ns in the case of 2FO-PS (Fig. 6D). The relative increase of 2FO-PO-linked Cy3 fluorescence intensity was also present when two samples containing equal concentrations of 2FO-PO-Cy3 were subjected to fluorescence imaging either in the absence of (sample 1, Fig. 6E) or in the presence of 50 mg/ml M5-gPLL-PFUDA (sample 2, Fig 6E) at an approximate molar ratio of 1:1000 (ODND/carrier). Sample 2 had 3.8-times higher fluorescence intensity of Cy3 than sample 1. 19F MRI of samples 1 and 2 demonstrated that the bound complex of 2FO-PO-Cy3 with M5-gPLL-PFUDA carrier was detectable with imaging at a concentration of PFUDA in the range of 1.5-3 mM (Fig. 6 F). Cell uptake experiments (Fig. 5G, H) performed by combining 2FO-PO-Cy3 with M5-gPLL-PFUDA carrier showed that the complex between 2FO-PO-Cy3 had very high stability and that both ODND and M5-gPLL-PFUDA were localized inside the same intracellular compartments after a prolonged incubation in cell culture.
Discussion
Numerous reports suggest that covalent modification of oligonucleotides with sterols [49, 50] or conjugation with lipids (reviewed in [15, 51]) can serve as a means of increasing affinity to plasma membranes as an alternative to more common ODN charge neutralization with polycations. There is an important distinction between lipids and/or sterols [52], and fluorocarbon residues as ODN modifiers as the latter requires only simple, relatively short fluorinated carbon chains due to the strength of the hydrophobic/lipophobic effect of fluorine [53, 54]. Therefore, we explored a combined approach to fluorescent ODND probe delivery and protection by using fluorocarbon (FC) chemistry. We expected that FC tails would associate non-covalently with each other to create transient defects in plasma membrane resulting in the internalization of individual ODN or their supramolecular associates and furthermore that hydrophobic interaction of FC chains with plasma membrane would result in an increase intracellular accumulation of ODND. In addition, we assumed that fluorine-fluorine interactions could serve as a driving force capable of promoting the association of fluorinated ODN with fluorinated polymers that would then act as carriers of ODND (showed schematically in Fig. S4). We reasoned that by modifying the residual amino groups of graft copolymer M5-gPLL with FC residues, the resulting copolymer would be capable of participating in a spontaneous complex formation with FO-ODND due to non-covalent interactions between FO-ODND and M5-gPLL carrying PFUDA “tails”. The advantage of fluorinated carriers and their complexes with FO-ODNs is in that these complexes or M5-gPLL-PFUDA alone are potentially detectable in the tissue by using 19F-NMR spectroscopy or MRI. 19F MR imaging is lower than sensitivity of proton MRI assisted with traditional paramagnetic contrast agents and it is unlikely that the sensitivity of detection of fluorinated polymer-ODND complexes at realistic doses required for ODND delivery in vivo will be on par with 19F NMR and MRI of previously studied perfluorocarbon emulsions with much higher content of fluorine [55-57]. However, the sensitivity can be improved by using building blocks containing fluorine atoms with equivalent chemical shifts, e.g. perfluoro-tert-butyl groups which are useful for a range of diverse applications, from cell tracking in vivo [58] to drug delivery applications of fluorinated polymers [59]. Parallel efforts directed at improving sensitivity and acquisition times, i.e. the implementation of clinically acceptable higher field strengths in conjunction with pulse sequence optimization [60, 61] and the use of three-dimensional compressed sensing [62], are transforming 19F MRI into a clinically acceptable modality [63].
The choice of ODN sequences used in our work was based on transcription factor-binding ODND decoys [64]. Such decoys were previously devised for blocking or attenuating STAT3- dependent transcription of genes associated with cancer progression [65, 66] which are implicated in poor cancer prognosis [33]. Therefore, to compare cellular uptake of ODND we chose cells overexpressing STAT3, which is involved in regulation of their survival [67, 68].
The experiments performed with ODND modified with perfluorocarbons and Cy3 fluorophores (Table 2) showed results that were consistent with minimal loss of thermal and thermodynamic stability reported by others [69]. Moreover, FO residues provided protection from ODND degradation in conditioned cell culture medium (Fig. 3) while preserving the ability to bind STAT3 with higher specificity than FO-free ODND (Fig. 4). While two FO residues interfered with STAT3 binding, one could use reactive functional groups on the ODN ends for generating reversible end caps [70] that dissociate after the uptake by live cells thereby abrogating such interference.
The influence of fluorinated groups on cell uptake was investigated by using semi-quantitative confocal microcopy-assisted measurements of fluorescence intensity of cell optical sections [71]. There are several potential advantages of this strategy: 1) high sensitivity and specificity; 2) the uptake in individual attached cells could be compared by outlining cell membranes that define cell-specific region of interest; 3) lack of a contribution of non-specific fluorescence (e.g. as a result of binding to extracellular substrate). In our experiments fluorinated groups were clearly capable of assisting in transporting ODND into the cells through plasma membrane. The exact mechanism of such transport still has to be studied in more detail. Though silencing of ApoB mRNA by FC-end modified siRNA in HepG2 cells has been recently reported [21], FC-end modified silencing construct was delivered into the cells via transfection and not by a simple co-incubation.
It is also important to consider the use of water-soluble synthetic macromolecules as carriers of FO-ODND (see Fig. S4). Zero-energy Van der Waals interaction surfaces in the surroundings of FC residues suggested a high probability of interactions between the FO group of the ODND and M5-gPLL-PFUDA in simulated water solutions. These simulations (Fig. S4) were corroborated by the analysis of complex formation with M5-gPLL-PFUDA (Figs. 5 and 6). In addition, a shift to longer fluorescence lifetimes of Cy3 was consistently observed if FO-ODND were incubated in the presence of M5-gPLL-PFUDA suggesting that Cy3 fluorophore was transferred to an environment with higher local viscosity within the FC-modified backbone of the copolymer.
In summary, our experiments with fluorescent ODND probes carrying either one or two fluorinated “tails” show higher ODND stability against degradation and significantly higher uptake in cells over time in vitro when compared to control ODN duplexes. Our results point to a potential use of ODN probes with one or more fluorinated residues for improving intracellular delivery in vitro and in vivo by using non-ionic and non-covalent interactions between fluorinated groups and fluorinated macromolecular carriers. Such carriers with or without bound FO-ODN duplexes have the advantage of carrying 19F tags potentially detectable by MR in living systems with zero background, i.e. the resultant images are not contaminated with the proton signal from conventional MRI. The experiments designed to optimize the macromolecular carriers for fluorinated ODND cargo are underway.
Abbreviations
ODN: oligonucleotide;
ODND: oligonucleotide duplex;
FC: fluorocarbon;
FO- 5': perfluorooctylpropyl;
CPG: controlled pore glass;
PO: phosphodiester;
PS: phosphorothioate/phosphodiester hybrid ODN;
M5-gPLL: a graft copolymer of poly-L-lysine and methoxypolyethylene glycol5000;
PFUDA: perfluoroundecanoic acid; M5-gPLL-PFUDA - M5-gPLL: modified by acylation with PFUDA;
GAS: interferon-gamma activated sequence;
SE: spin-echo.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported in part by grants from the NIH: R01DK095728 (AB), R01EB000858 (AB) and R21EB017980 (SZ). We appreciate the access to custom oligonucleotide synthesis facility generously provided by Dr. David Tabatadze (ZATA Pharmaceuticals, Inc.). We are grateful to Dr. Mary Mazzanti for scientific and stylistic editing of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129-38
2. Juliano R, Alam MR, Dixit V, Kang H. Mechanisms and strategies for effective delivery of antisense and siRNA oligonucleotides. Nucleic Acids Res. 2008;36:4158-71
3. Bogdanov AA. Merging molecular imaging and RNA interference: Early experience in live animals. J Cell Biochem. 2008;104:1113-23
4. Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34:322-33
5. Hong H, Zhang Y, Cai W. In vivo imaging of RNA interference. J Nucl Med. 2010;51:169-72
6. Juliano RL, Carver K. Cellular uptake and intracellular trafficking of oligonucleotides. Adv Drug Deliv Rev. 2015;87:35-45
7. Juliano RL. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44:6518-48
8. Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur J Biochem. 2003;270:1628-44
9. Jain HV, Verthelyi D, Beaucage SL. Amphipathic trans-acting phosphorothioate DNA elements mediate the delivery of uncharged nucleic acid sequences in mammalian cells. RSC Advances. 2015;5:65245-54
10. Munoz-Alarcon A, Eriksson J, Langel U. Novel Efficient Cell-Penetrating, Peptide-Mediated Strategy for Enhancing Telomerase Inhibitor Oligonucleotides. Nucleic Acid Ther. 2015;25:306-10
11. Arbab AS, Bashaw LA, Miller BR, Jordan EK, Bulte JW, Frank JA. Intracytoplasmic tagging of cells with ferumoxides and transfection agent for cellular magnetic resonance imaging after cell transplantation: methods and techniques. Transplantation. 2003;76:1123-30
12. Banerjee P, Reichardt W, Weissleder R, Bogdanov A Jr. Novel hyperbranched dendron for gene transfer in vitro and in vivo. Bioconjug Chem. 2004;15:960-8
13. Toncheva V, Wolfert MA, Dash PR, Oupicky D, Ulbrich K, Seymour LW. et al. Novel vectors for gene delivery formed by self-assembly of DNA with poly(L-lysine) grafted with hydrophilic polymers. Biochim Biophys Acta. 1998;1380:354-68
14. Sarisozen C, Salzano G, Torchilin VP. Recent advances in siRNA delivery. Biomol Concepts. 2015;6:321-41
15. Sarisozen C, Salzano G, Torchilin VP. Lipid-based siRNA Delivery Systems: Challenges, Promises and Solutions Along the Long Journey. Curr Pharm Biotechnol. 2016;17:728-40
16. Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR. et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci U S A. 2010;107:1864-9
17. Dahlman JE, Barnes C, Khan OF, Thiriot A, Jhunjunwala S, Shaw TE. et al. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat Nanotechnol. 2014;9:648-55
18. Novobrantseva TI, Borodovsky A, Wong J, Klebanov B, Zafari M, Yucius K. et al. Systemic RNAi-mediated Gene Silencing in Nonhuman Primate and Rodent Myeloid Cells. Mol Ther Nucleic Acids. 2012;1:e4
19. Godeau G, Arnion H, Brun C, Staedel C, Barthelemy P. Fluorocarbon oligonucleotide conjugates for nucleic acids delivery. Med Chem Comm. 2010;1:76-8
20. Dolain C, Patwa A, Godeau G, Barthélémy P. Nucleic Acid Based Fluorinated Derivatives: New Tools for Biomedical Applications. Appl Sci. 2012;2:245-59
21. de Rochambeau D, Barłóg M, Edwardson TGW, Fakhoury JJ, Stein RS, Bazzib HS. et al. "DNA-Teflon” sequence-controlled polymers. Polym Chem. 2016;7:4998-5003
22. Menaa F, Menaa B, Sharts O. Importance of Fluorine and Fluorocarbons in Medicinal Chemistry and Oncology. J Mol Pharm Org Process Res. 2013;1:1000104
23. Dalvi VH, Rossky PJ. Molecular origins of fluorocarbon hydrophobicity. Proc Natl Acad Sci USA. 2010;107:13603-7
24. Brittain SM, Ficarro SB, Brock A, Peters EC. Enrichment and analysis of peptide subsets using fluorous affinity tags and mass spectrometry. Nat Biotechnol. 2005;23:463-8
25. Ellipilli S, Murthy R, Ganesh KN. Perfluoroalkylchain conjugation as a new tactic for enhancing cell permeability of peptide nucleic acids (PNAs) via reducing the nanoparticle size. Chem Commun. 2016;52:521-4
26. Wang M, Liu H, Li L, Cheng Y. A fluorinated dendrimer achieves excellent gene transfection efficacy at extremely low nitrogen to phosphorus ratios. Nat Commun. 2014;5:3053
27. Li L, Song L, Liu X, Yang X, Li X, He T. et al. Artificial Virus Delivers CRISPR-Cas9 System for Genome Editing of Cells in Mice. ACS Nano. 2017;11:95-111
28. Ruiz-Cabello J, Barnett BP, Bottomley PA, Bulte JW. Fluorine (19F) MRS and MRI in biomedicine. NMR Biomed. 2011;24:114-29
29. Cametti M, Crousse B, Metrangolo P, Milani R, Resnati G. The fluorous effect in biomolecular applications. Chem Soc Rev. 2012;41:31-42
30. Takaoka Y, Kiminami K, Mizusawa K, Matsuo K, Narazaki M, Matsuda T. et al. Systematic study of protein detection mechanism of self-assembling 19F NMR/MRI nanoprobes toward rational design and improved sensitivity. J Am Chem Soc. 2011;133:11725-31
31. Kodibagkar VD, Yu J, Liu L, Hetherington HP, Mason RP. Imaging beta-galactosidase activity using 19F chemical shift imaging of LacZ gene-reporter molecule 2-fluoro-4-nitrophenol-beta-D-galactopyranoside. Magn Reson Imaging. 2006;24:959-62
32. Astriab-Fisher A, Fisher MH, Juliano R, Herdewijn P. Increased uptake of antisense oligonucleotides by delivery as double stranded complexes. Biochem Pharmacol. 2004;68:403-7
33. Johnston PA, Grandis JR. STAT3 signaling: anticancer strategies and challenges. Mol Interv. 2011;11:18-26
34. Gong W, Xiao Y, Wei Z, Yuan Y, Qiu M, Sun C. et al. Toward the use of precision medicine for the treatment of head and neck squamous cell carcinoma. Oncotarget. 2017;8:2141-52
35. McDaniel JM, Varley KE, Gertz J, Savic DS, Roberts BS, Bailey SK. et al. Genomic regulation of invasion by STAT3 in triple negative breast cancer. Oncotarget. 2017;8:8226-38
36. Tabatadze D, Zamecnik P, Yanachkov I, Wright G, Pierson K, Zhang S. et al. A novel thymidine phosphoramidite synthon for incorporation of internucleoside phosphate linkers during automated oligodeoxynucleotide synthesis. Nucleosides Nucleotides Nucleic Acids. 2008;27:157-72
37. Miriyala B. Fluorous Methods for the Synthesis of Peptides and Oligonucleotides. In: Horváth I, editor. Fluorous chemsitry, Top Curr Chem. Heidelberg Berlin: Springer Verlag. 2012;308:105-34
38. Iyer RP, Egan W, Regan JB, Beaucage SL. 3H-1,2-Benzodithiole-3-one 1,1-dioxide as an improved sulfurizing reagent in the solid-phase synthesis of oligodeoxyribonucleoside phosphorothioates. J Amer Chem Soc. 1990;112:1253-4
39. Chemical Computing Group Inc. SSW, Suite #910, Montreal, QC, Canada, H3A 2R7. Molecular Operating Environment (MOE). 2016 ed. 2017
40. Case D, Betz R, Botello-Smith W, Cerutti D, Cheatham T, Darden T. et al. AMBER 2016. University of California, San Francisco CA. 2016
41. Zhang S, Metelev V, Tabatadze D, Zamecnik P, Bogdanov A Jr. Near-infrared fluorescent oligodeoxyribonucleotide reporters for sensing NF-kappaB DNA interactions in vitro. Oligonucleotides. 2008;18:235-43
42. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671-5
43. Bogdanov AA, Mazzanti M, Castillo G, Bolotin E. Protected Graft Copolymer (PGC) in Imaging and Therapy: A Platform for the Delivery of Covalently and Non-Covalently Bound Drugs. Theranostics. 2012;2:553-76
44. Bogdanov AA Jr, Gupta S, Koshkina N, Corr SJ, Zhang S, Curley SA. et al. Gold nanoparticles stabilized with MPEG-grafted poly(l-lysine): in vitro and in vivo evaluation of a potential theranostic agent. Bioconjug Chem. 2015;26:39-50
45. Bogdanov AA Jr, Metelev V, Zhang S, Kumar AT. Sensing of transcription factor binding via cyanine dye pair fluorescence lifetime changes. Mol Biosyst. 2012;8:2166-73
46. Metelev V, Zhang S, Tabatadze D, Kumar AT, Bogdanov A. The three-dimensional context of a double helix determines the fluorescence of the internucleoside-tethered pair of fluorophores. Mol Biosyst. 2013;9:2447-53
47. Asfari M, Janjic D, Meda P, Li G, Halban PA, Wollheim CB. Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinology. 1992;130:167-78
48. Metelev V, Zhang S, Tabatadze D, Bogdanov A. Hairpin-like fluorescent probe for imaging of NF-kappaB transcription factor activity. Bioconjug Chem. 2011;22:759-65
49. Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK. et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. 2007;25:1149-57
50. Ly S, Navaroli DM, Didiot MC, Cardia J, Pandarinathan L, Alterman JF. et al. Visualization of self-delivering hydrophobically modified siRNA cellular internalization. Nucleic Acids Res. 2017;45:15-25
51. Roberts TC, Ezzat K, El Andaloussi S, Weinberg MS. Synthetic SiRNA Delivery: Progress and Prospects. Methods Mol Biol. 2016;1364:291-310
52. De Paula D, Bentley MV, Mahato RI. Hydrophobization and bioconjugation for enhanced siRNA delivery and targeting. RNA. 2007;13:431-56
53. Pohl NL. Fluorous tags catching on microarrays. Angew Chem Int Ed Engl. 2008;47:3868-70
54. Hong M, Zhou X, Lu Z, Zhu J. Nanoparticle-based, fluorous-tag-driven DNA detection. Angew Chem Int Ed Engl. 2009;48:9503-6
55. Mason RP, Antich PP, Babcock EE, Gerberich JL, Nunnally RL. Perfluorocarbon imaging in vivo: a 19F MRI study in tumor-bearing mice. Magn Reson Imaging. 1989;7:475-85
56. Sotak CH, Hees PS, Huang HN, Hung MH, Krespan CG, Raynolds S. A new perfluorocarbon for use in fluorine-19 magnetic resonance imaging and spectroscopy. Magn Reson Med. 1993;29:188-95
57. Hitchens TK, Ye Q, Eytan DF, Janjic JM, Ahrens ET, Ho C. 19F MRI detection of acute allograft rejection with in vivo perfluorocarbon labeling of immune cells. Magn Reson Med. 2011;65:1144-53
58. Srivastava AK, Kadayakkara DK, Bar-Shir A, Gilad AA, McMahon MT, Bulte JW. Advances in using MRI probes and sensors for in vivo cell tracking as applied to regenerative medicine. Dis Model Mech. 2015;8:323-36
59. Decato S, Bemis T, Madsen E, Mecozzi S. Synthesis and characterization of perfluoro-tert-butyl semifluorinated amphiphilic polymers and their potential application in hydrophobic drug delivery. Polym Chem. 2014;5:6461-71
60. Giraudeau C, Geffroy F, Meriaux S, Boumezbeur F, Robert P, Port M. et al. 19F molecular MR imaging for detection of brain tumor angiogenesis: in vivo validation using targeted PFOB nanoparticles. Angiogenesis. 2013;16:171-9
61. Colotti R, Bastiaansen JA, Wilson A, Flogel U, Gonzales C, Schwitter J. et al. Characterization of perfluorocarbon relaxation times and their influence on the optimization of fluorine-19 MRI at 3 Tesla. Magn Reson Med. 2016;77:2263-2271
62. Zhong J, Mills PH, Hitchens TK, Ahrens ET. Accelerated fluorine-19 MRI cell tracking using compressed sensing. Magn Reson Med. 2013;69:1683-90
63. Ahrens ET, Helfer BM, O'Hanlon CF, Schirda C. Clinical cell therapy imaging using a perfluorocarbon tracer and fluorine-19 MRI. Magn Reson Med. 2014;72:1696-701
64. Cho-Chung YS, Park YG, Lee YN. Oligonucleotides as transcription factor decoys. Curr Opin Mol Ther. 1999;1:386-92
65. Holschermann H, Stadlbauer TH, Wagner AH, Fingerhuth H, Muth H, Rong S. et al. STAT-1 and AP-1 decoy oligonucleotide therapy delays acute rejection and prolongs cardiac allograft survival. Cardiovasc Res. 2006;71:527-36
66. De Stefano D, De Rosa G, Carnuccio R. NFkappaB decoy oligonucleotides. Curr Opin Mol Ther. 2010;12:203-13
67. Mziaut H, Kersting S, Knoch KP, Fan WH, Trajkovski M, Erdmann K. et al. ICA512 signaling enhances pancreatic beta-cell proliferation by regulating cyclins D through STATs. Proc Natl Acad Sci USA. 2008;105:674-9
68. Oh YS, Lee YJ, Park EY, Jun HS. Interleukin-6 treatment induces beta-cell apoptosis via STAT-3-mediated nitric oxide production. Diabetes Metab Res Rev. 2011;27:813-9
69. Moreira BG, You Y, Behlke MA, Owczarzy R. Effects of fluorescent dyes, quenchers, and dangling ends on DNA duplex stability. Biochem Biophys Res Commun. 2005;327:473-84
70. Skrzypczynski Z, Wayland S. New reagents for the introduction of reactive functional groups into chemically synthesized DNA probes. Bioconjug Chem. 2003;14:642-52
71. Murray JM. Practical aspects of quantitative confocal microscopy. Methods Cell Biol. 2013;114:427-40
Author contact
![]() Corresponding author: S6-434, Department of Radiology, University of Massachusetts Medical School, 55 Lake Ave North, Worcester MA 01655 Tel. 508-856-5571 FAX 508-856-1860 Alexei.Bogdanovedu
Corresponding author: S6-434, Department of Radiology, University of Massachusetts Medical School, 55 Lake Ave North, Worcester MA 01655 Tel. 508-856-5571 FAX 508-856-1860 Alexei.Bogdanovedu
 Global reach, higher impact
Global reach, higher impact