13.3
Impact Factor
Theranostics 2017; 7(3):634-646. doi:10.7150/thno.16840 This issue Cite
Research Paper
MicroRNA Detection Using a Double Molecular Beacon Approach: Distinguishing Between miRNA and Pre-miRNA
1. Division of Cardiology, Emory University School of Medicine, Atlanta, GA, USA;
2. Department of Biomedical Engineering, Rice University, Houston, TX, USA;
3. Atlanta Veterans Administration Medical Center, Decatur, GA, USA.
Received 2016-7-13; Accepted 2016-11-14; Published 2017-1-15
Abstract
MicroRNAs (miRNAs) are small, noncoding RNAs that post-transcriptionally regulate gene expression and are recognized for their roles both as modulators of disease progression and as biomarkers of disease activity, including neurological diseases, cancer, and cardiovascular disease (CVD). Commonly, miRNA abundance is assessed using quantitative real-time PCR (qRT-PCR), however, qRT-PCR for miRNA can be labor intensive, time consuming, and may lack specificity for detection of mature versus precursor forms of miRNA. Here, we describe a novel double molecular beacon approach to miRNA assessment that can distinguish and quantify mature versus precursor forms of miRNA in a single assay, an essential feature for use of miRNAs as biomarkers for disease. Using this approach, we found that molecular beacons with DNA or combined locked nucleic acid (LNA)-DNA backbones can detect mature and precursor miRNAs (pre-miRNAs) of low (< 1 nM) abundance in vitro. The double molecular beacon assay was accurate in assessing miRNA abundance in a sample containing a mixed population of mature and precursor miRNAs. In contrast, qRT-PCR and the single molecular beacon assay overestimated miRNA abundance. Additionally, the double molecular beacon assay was less labor intensive than traditional qRT-PCR and had 10-25% increased specificity. Our data suggest that the double molecular beacon-based approach is more precise and specific than previous methods, and has the promise of being the standard for assessing miRNA levels in biological samples.
Keywords: molecular beacon, microRNA, microRNA detection, cardiovascular disease, PCR.
Introduction
MicroRNAs (miRNAs) are short (~19-23 nucleotides), conserved, non-coding RNAs that repress translation or induce degradation of specific messenger RNAs (mRNAs) [1-3]. MiRNAs have been identified across every plant and animal species and have been shown to regulate a multitude of cellular processes. MiRNA biogenesis begins in the nucleus where primary miRNA transcripts (pri-miRNAs) are generated from various genomic loci [4] by RNA polymerase II and III [5]. Subsequently, the Microprocessor complex, consisting of the RNase II enzyme, Drosha, and its cofactor DGCR8 (DiGeorge Syndrome Critical Region 8), cleaves the pri-miRNA. The resulting product is a pre-miRNA, which varies in length between 60-70 nucleotides. The pre-miRNA has a stem-loop hairpin structure that consists of a double stranded stem (approximately 33 nucleotides) and a single stranded loop [6]. Pre-miRNA is exported from the nucleus to the cytoplasm by Exportin-5-Ran-GTP where a second RNase, Dicer, cleaves the pre-miRNA terminal loop to produce a transient double stranded miRNA. The two strands separate into two complimentary mature miRNAs (5'-3' and 3'-5') [7], either of which may become complexed with the RNA-induced silencing complex (RISC). Importantly, through variations in miRNA processing, a single miRNA genomic locus can generate multiple mature miRNAs that differ by only a few nucleotides; these variants are known as isomiRs (miRNA isoforms). miRNAs guide the RISC to specific mRNA targets through base pairing of the miRNA seed sequence, defined as nucleotides 2-8 of the 5' end, with the complementary sequence in the target mRNA. This interaction can either inhibit target mRNA translation or induce its degradation [8]. Since miRNA-mediated gene suppression typically requires only seven to eight nucleotides, a single miRNA can regulate the expression of hundreds of mRNAs [9] and many different miRNAs can regulate a single mRNA [10].
The discovery of miRNAs and their function has enhanced our understanding of the complexity of gene regulation in health and disease. Physiologic concentrations of mature and precursor forms of miRNA vary depending on species, tissue, cell type and disease state with ranges from nanomolar to fetamolar concentrations [11]. To date, a multitude of different techniques have been used to detect and quantitate miRNA, including northern blotting, qRT-PCR, miRNA microarrays, next generation sequencing, and molecular beacons. Each quantitative method has advantages and disadvantages. Northern blotting is the oldest of these techniques but it has low throughput, it is time consuming and requires large quantities of RNA (8 μg or more) [4]. Most northern blotting techniques are less sensitive than newer miRNA detection methods; approximately 100,000 copies of RNA sequence are required for detection by blot hybridization compared to a single copy of RNA needed in other methods. In addition, northern blotting does not multiplex well. qRT-PCR is currently the standard for quantitating miRNA. It is simple and typically very specific and sensitive. However, qRT-PCR often lacks the ability to distinguish isomiRs unless the qRT-PCR protocol is altered, thereby increasing its cost and making it less time-efficient [12, 13]. Microarrays have emerged as a high throughput, comprehensive option for quantitating miRNA, but they are expensive and have limitations in sensitivity and specificity. Also, microarray data still need to be validated by other methods such as qRT-PCR or northern blot. Next generation deep sequencing (NGS) is a highly sensitive and specific method for miRNA quantification. Using current technology, NGS is able to generate high quality data from as little as 10 ng of miRNA. However, NGS is expensive and requires a significant amount of bioinformatics and biostatistical support. As with miRNA microarrays, NGS results should be validated by northern or qRT-PCR analysis. Therefore, there is a need for new miRNA detection and quantification assays that are dynamic, sensitive and specific, but also time- and cost-efficient. Molecular beacons (MBs) have shown to have these features.
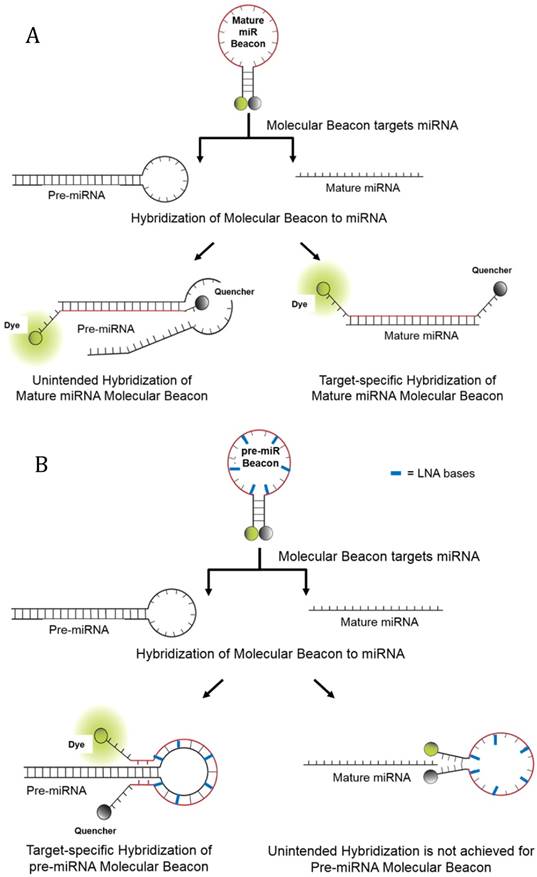
Molecular beacons are single stranded oligonucleotide (DNA or RNA) probes labeled with a reporter fluorophore at one end and a quencher at the other end [14]. In the absence of a complementary target nucleic acid sequence, they form a stem-loop hairpin structure that results in quenching of the fluorophore's fluorescence [14-16]. Hybridization of the molecular beacon's loop sequence with target nucleic acid opens the hairpin and physically separates the fluorophore from quencher, allowing a fluorescence signal to be emitted upon excitation, which can be assessed in a standard fluorescence plate reader (Figure 1). For detection of a mature miRNA, the molecular beacon hybridizes to matching nucleotides in the mature miRNA. The detection of the pre-miRNA occurs when the pre-miRNA molecular beacon hybridizes to the loop sequence of the pre-miRNA stem-loop complex. In theory, the molecular beacon elicits a signal only upon direct hybridization to the complementary RNA sequence. This enables a molecular beacon to function as a sensitive and specific probe with a better signal-to-background noise ratio [17-24]. In addition, RNA abundance is directly correlated to molecular beacon fluorescence.
The mature miRNA sequence is also present in its pri-miRNA and pre-miRNA forms. Therefore, methods that are not able to readily distinguish between mature and precursor forms of miRNA, such as qRT-PCR and miRNA microarray, may report falsely high values for miRNA abundance in samples that contain a mixture of different miRNA forms. This is a particular problem for low abundance miRNAs. Furthermore, the ability to distinguish between mature and precursor forms of miRNA is important for studies that examine regulation of miRNA processing. Our group recently demonstrated that molecular beacons can readily distinguish mature and precursor miRNA and reliably quantify their expression in the same sample. In contrast to qRT-PCR and microarrays, molecular beacons can also distinguish between different isomiRs of a consensus miRNA as well as between miRNA family members that have similar sequences [25]. Intrinsic limitations in molecular beacon affinity for target miRNA can be overcome by the incorporation of LNA backbone chemistry into molecular beacon design. Here, we describe a novel molecular beacon-based assay that overcomes limitations in molecular beacon specificity for mature and precursor forms of miRNA. We demonstrate that quantitation of multiple mature miRNAs and pre-miRNAs in a single sample can be accomplished through multiplex assays with molecular beacons that target different miRNA forms and contain fluorophores with non-overlapping spectra.
Schematics of Molecular Beacon Hybridization to target mature and precursor miRNA. In the absence of complementary target, molecular beacons form a stem-loop structure that brings the quencher in close proximity to the fluorophore, thereby quenching the fluorescence emission. Hybridization of the molecular beacons to its target mature or precursor miRNA opens the molecular beacon hairpin, thereby leading to physical separation of the fluorophore from the quencher and allowing fluorescence to be measured upon excitation. (A) Hybridization of molecular beacons to the mature miRNA target sequence and unintended hybridization of the mature molecular beacon to pre-miRNA. (B) Hybridization of pre-miRNA molecular beacons to the pre-miRNA loop sequence. Also depicted is the inability for mature miRNA to hybridize to the pre-miRNA molecular beacon, though 2-3 nucleotide bases from the pre-miRNA molecular beacon may hybridize to the mature miRNA. However, in this latter example there is not enough hybridization to separate the quencher from the fluorophore. The use of LNA bases allows the pre-miRNA molecular beacon to have a relatively higher affinity for its intended pre-miRNA target then to complementary mature miRNA sequences.
Materials and Methods
Design of miRNA Sequences used in Hybridization Assays
To determine the specificity of the molecular beacons, the sequences of wild-type mature miR-21 and miR-27b, and pre-miR-21 and pre-miR-27b were synthesized by Integrated DNA Technologies as synthetic miRNA targets. As shown in Table 1, each wild-type target also had a complementary LNA/DNA sequence designed that was used as a blocker in the hybridization assays. Specifically, for the mature miR-21 blocker, eight of its 22 nucleotides were LNA, while mature miR-27b blocker had six of its 20 nucleotides as LNA. Similarly, pre-miR-21 blocker and pre-miR-27b blocker had six LNA bases in each of their sequences. All synthetic nucleotide sequences were suspended in nuclease-free water (1 nmol/µL) and stored at -80ºC.
Molecular Beacon Design
DNA or DNA/LNA molecular beacons complementary to either the mature consensus nucleotide sequence of miR-21/miR-27b or the hairpin loop region of the pre-miR-21/pre-miR-27b were designed. Molecular beacons targeting mature miRNAs, previously described in Baker et al [25] contained at least 22 complementary bases, a 6FAM or Cy3 fluorophore on the 5' end, and a Black Hole Quencher 1 (BHQ1) or BHQ2 on the 3' end. Additional mature molecular beacons designed to target miR-27b that were created specifically for this study contained 28 DNA/LNA bases with Cy3 or HEX fluorophore on the 5' end, and a BHQ2 or Dabcyl (Dab) on the 3' end. Similarly, DNA/LNA molecular beacons complementary to the loop nucleotide sequences of pre-miR-21 and pre-miR-27b were designed with 6FAM, Cy3, Hex, or Cy5 fluorophores on the 5' end and BHQ1, BHQ2, Dab or an Iowa Black quencher (IAblk) on the 3' ends. For the LNA molecular beacon, a LNA base was placed every third nucleotide after the stem (Table 2). All molecular beacons were synthesized by Integrated DNA Technologies, suspended in nuclease-free water (1 µg/µL), and stored in an opaque tube and wrapped in foil at -80ºC.
Sequences of mature, precursor miRNA targets and oligo blockers used in molecular beacon hybridization assays.
| Target Name | Sequence (5'-3') |
| Wild-type mature miR-21 | UAGCUUAUCAGACUGAUGUUGA |
| Wild-type precursor miR-21 | UGUCGGGUAGCUUAUCAGACUGAUGUUGACUGUUGAAUCUCAUGGCAACACCAGUCGAUGGGCUGUCUGACA |
| Wild-type mature miR-27b | UUCACAGUGGCUAAGUUCUGC |
| Wild-type precursor miR-27b | ACCUCUCUAACAAGGUGCAGAGCUUAGCUGAUUGGUGAACAGUGAUUGGUUUCCGCUUUGUUCACAGUGGCUAAGUUCUGCACCUGAAGAGAAGGUG |
| Oligo Blockers Name | Sequence (5'-3') |
| Mature miR-21 Blocker | TCAA+CA+TC+AG+TC+TG+AT+AA+GCTA |
| Precursor miR-21 Blocker | GCCATGA+GA+TT+CA+AC+AG+TCAACATCAGTC |
| Mature miR-27b Blocker | GCAGA+AC+TT+AG+CCA+CT+GTGAA |
| Precursor miR-27b Blocker | AGAACT+TAGCCACTG+TGAACAAAG+CGGAAACCA+ATCACTGTTC+ACCAATCAG+CTAAGC |
Bold = Mature miRNA Blocker and Mature miRNA Molecular Beacon binding sequence; Italics = Precursor miRNA Blocker binding sequence;
Underline= Precursor miRNA Molecular Beacon binding sequence; + = Locked Nucleic Acid (LNA) Base.
Characteristics of Molecular Beacons.
| Probe Names | Sequence (5'-3') | Dye | Quencher | Probe Length | Stem Length |
|---|---|---|---|---|---|
| miR21 Mature 1* | AGCGTCAACATCAGTCTGATAAGCTACGCT | Cy3 | BHQ2 | 30 | 5 |
| miR21 Precursor 1 | CGCCATGAGATTCAACAGTCGGCG | FAM | BHQ1 | 24 | 4 |
| miR21 Precursor 2 | GGTGTTGCCATGAGATTCAAGAGTCAACACC | FAM | Dab | 30 | 6 |
| miR21 Precursor 3* | CCTGTTGCC+AT+GA+GA+TT+CA+AC+AG+TCAACAGG | FAM | IABLK | 30 | 7 |
| miR27b Mature 1* | CGCAGAACTTAGCCACTGTGAATCTGCG | FAM | BHQ1 | 28 | 6 |
| miR27b Mature 2 | CGCAGAACTTAGCCACTGTGAATCTGCG | Cy3 | BHQ2 | 28 | 6 |
| miR27b Mature 3 | CGCAGAACTTAGCCACTGTGAATCTGCG | HEX | Dab | 28 | 6 |
| miR27b Precursor 1* | CGGCCAAAGC+GG+AA+AC+CA+AT+CACTGGCCG | Cy3 | BHQ2 | 30 | 6 |
| miR27b Precursor 2 | CGGCCAAAGC+GG+AA+AC+CA+AT+CACTGGCCG | HEX | Dab | 30 | 6 |
| miR27b Precursor 3 | CGGCCAAAGC+GG+AA+AC+CA+AT+CACTGGCCG | Cy5 | BHQ1 | 30 | 6 |
+ = Locked Nucleic Acid (LNA) Base
Stem Underlined
BHQ1 = Black Hole Quencher 1; BHQ2 = Black Hole Quencher 2; Cy3 = Cyanine 3; Cy5 = Cyanine 5; Dab = Dabcyl; FAM = (Tetrachloro)fluorescein;
HEX = Hexachlorofluorescein; IABlk= Iowa Black Quencher.
* = Beacon used for Validation
RNA Isolation from Endothelial Cells and Whole Blood
Human aortic endothelial cells (HAECs) were cultured in EBM-2 media in the presence of 5% CO2 at 37°C and were used between passages 4-8. RNA from the cells was extracted using the miRNeasy kit (Qiagen) according to manufacturer's instructions. The miRNA abundance was assessed by the single or double molecular beacon assay and qRT-PCR.
RNA from whole blood was extracted using the PAXgene Blood RNA Kit (Qiagen) according to manufacturer's instructions. The miRNA abundance was assessed by qRT-PCR and double molecular beacon methods.
Testing of Molecular Beacon Design
Molecular beacon hybridization to synthetic miRNA targets was evaluated in a final volume of 50 µL containing PBS (pH 7.0) and 200 nM molecular beacon. Reactions were performed in a Tecan Safire plate reader (384-well flat bottom tray) with continuous shaking at 37ºC for 60 minutes before the intensity of the fluorescence was determined.
To determine the impact of backbone chemistry on molecular beacon sensitivity, verification and quantification assays were performed with the synthetic targets over a concentration range of 0-5 nM. Background signal (molecular beacon without target) was subtracted from the fluorescence signal recorded for the molecular beacon-miRNA/pre-miRNA duplexes. The molecular beacons with the greatest sensitivity i.e. the molecular beacon that detected the lowest concentration of corresponding targets or those molecular beacons previously selected based on Baker et al [25] were used in all subsequent experiments (Table 2; highlighted molecular beacons).
Mature molecular beacon specificity was tested in hybridization assays that included synthetic oligos designed to block (oligo blockers, 200 nM; Table 1) binding of molecular beacons to their target mature or pre-miRNAs. Hybridization conditions (200 nM of molecular beacon; 0-100 nM of mixed target miRNA) and fluorescence detection methods were similar to those described above. The oligo blocker sequences shown in Table 1 are comprised of LNA bases and are complementary to mature and precursor forms of specific miRNAs. The use of DNA/LNA blockers that were complementary to mature and precursor forms of specific miRNAs allowed for competitive binding of the oligo blockers to its target sequence. Thus, target sequences that were not bound to blockers could subsequently hybridize to a molecular beacon.
qRT-PCR assays
For qRT-PCR assessment of miRNA abundance, RNA (synthetic - 10 and 50 nM each; cellular - 20-200 nM; blood -~3 µg/mL) was reverse transcribed using the TaqMan MicroRNA Reverse Transcription kit (Ambion) as per manufacturer's instructions. PCR was performed using miRNA-specific TaqMan MicroRNA Assays (Ambion) and the pre-programmed standard curve protocol by StepOne™ Real-time PCR System.
Double Molecular Beacon Assay
For the double molecular beacon assays, a pair of molecular beacons (one mature and one pre-miRNA molecular beacon), containing fluorophores with non-overlapping spectra, were incubated with a sample containing a mixture of complementary target mature and pre-miRNA. Hybridization conditions (200 nM of molecular beacon; 0-100 nM of mixed target miRNA) and fluorescence detection methods were similar to those described above.
When quantitating pre-miRNA and mature miRNA abundance using the double molecular beacon assay in the whole blood samples, 500 nM of molecular beacon was use for hybridization while fluorescence detection methods were similar to those described above.
Data analysis and statistics
For both single and double molecular beacon assays, the background fluorescence signal from molecular beacon alone was subtracted from the fluorescence signal from the duplexes formed by the molecular beacon and (pre)miRNA. Paired t-tests were used to analyze differences between mature and pre-miRNA concentrations that were determined by different detection methods (molecular beacon vs. qRT-PCR).
Results
Design and Testing of Molecular Beacons Targeting Mature and Precursor miRNA
Molecular beacons may be composed of RNA, DNA or LNA backbones. It is known that LNA/RNA and LNA/DNA complexes have a higher affinity than RNA/RNA complexes, DNA/DNA complexes and DNA/RNA complexes, respectively [26, 27]. Previously, our group showed that molecular beacons with LNA bases have the highest affinity for miRNA targets [25]. Figures 2A and 2B show the relative fluorescence intensity of molecular beacons with different backbone chemistries hybridized to mature and precursor forms of miR-21 and -27b. For this study, all mature miRNA molecular beacons were designed with a DNA backbone, while the pre-miRNA molecular beacons are a combination of both DNA and LNA/DNA backbones (Table 2). The use of LNA/DNA backbone for the pre-miRNA molecular beacons was chosen to help increase the affinity of the molecular beacon to the target. Using the highest affinity backbone for pre-miRNA molecular beacon allows it to bind to its targeting pre-miRNA despite the predominantly double stranded structure. A DNA backbone was used for the mature miRNA beacon because, in a sample containing both mature and precursor miRNA, the lower affinity of the mature molecular beacon is more desirable in mixed miRNA populations. For example, a molecular beacon targeting mature miRNA may bind to sequence in the stem of the precursor miRNA, producing an erroneously high fluorescence in a mixed population of mature and precursor miRNAs. In this scenario, a DNA molecular beacon would be less likely to bind to the double stranded stem sequence of the pre-miRNA, resulting in less unintended hybridization and florescence.
Determining Molecular Beacon Sensitivity for each Target. Differences in hybridization of molecular beacons to target miRNA with varying backbones consisting of DNA and LNA/DNA bases. Molecular beacons (200 nM) were incubated with each synthetic target separately (pre-miR-21 or miR-21 [A] and pre-miR-27b or miR-27b [B] [0-5 nM]) in PBS at 37°C for 60 min prior to recording the fluorescence intensity; three pre-miRNA LNA/DNA molecular beacons were designed and tested while previously verified mature miRNA DNA molecular beacons were tested for sensitivity only. For each experiment, background fluorescence was subtracted from signal produced by hybridization of the molecular beacon to its target miRNA.
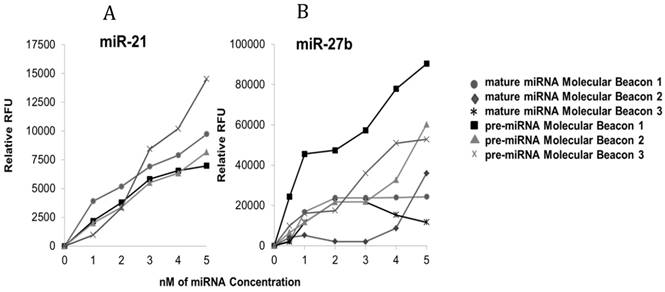
The molecular beacons containing different DNA and LNA/DNA backbones were tested in hybridization assays with their respective synthetic target miRNAs at low concentrations (0-5 nM). These data show that molecular beacons with DNA and DNA/LNA backbones can detect target miRNA in concentrations as low as 0.5 nM. Only one molecular beacon targeting mature miR-21, previously characterized by our group, was tested and used [25]. However, three mature miRNA molecular beacons were tested for miR-27b. The mature miRNA molecular beacons for miR-21 and -27b that were chosen for subsequent analysis contained DNA bases only. Specifically, mature molecular beacon miR-21 was 30 bases long and its stem consisted of 5 bases (10 bases total), while the mature molecular beacon chosen for miR-27b was 28 bases long with a stem of 6 bases (12 bases total). Additionally, three pre-miRNA DNA and DNA/LNA molecular beacons were assessed for their binding to their respective targets (pre-miR-21 and -27b). The pre-miRNA molecular beacons chosen for subsequent analysis of pre-miR-21 and -27b both contained LNA bases. For pre-miR-21, precursor molecular beacon 3, which had a total of eight LNA bases in its backbone, demonstrated the best signal to noise ratio. In contrast, precursor miR-21 molecular beacon 1 and 2, contained no LNA bases and did not achieve a signal to noise ratio that distinctly distinguished the hybridized beacon from non-hybridized beacon. For pre-miR-27b, precursor molecular beacon 1 was determined to be the best candidate. This molecular beacon contained six LNA bases and had a better signal to noise ratio, compared to the other molecular beacons designed for pre-miR-27b.
Beacon Specificity: Using Oligo Blockers
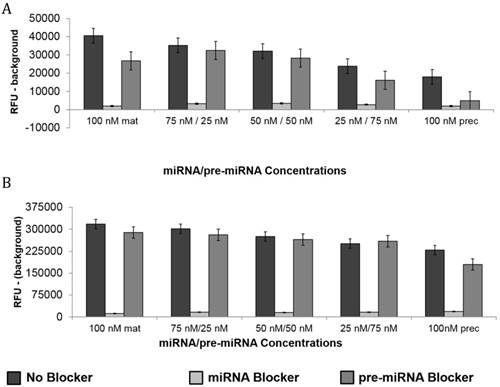
To determine the specificity of molecular beacons designed to target mature and precursor miR-21 or -27b, molecular beacon hybridization assays were performed with the addition of competitive oligo blockers. Figure 3A and 3B demonstrates that mature miRNA molecular beacons targeting mature miR-21 and -27b had low fluorescence signal when co-incubated with oligos that were anti-sense to the mature miRNA sequence (mature miRNA blocker). However, both of these mature miRNA molecular beacons hybridized well to their synthetic targets when no blockers were present or when the pre-miRNA oligo blockers were present. It should be noted that, in a mixed population of mature and precursor targets, the mature miRNA molecular beacon has the potential to produce a falsely high signal due to its ability to hybridize to the mature miRNA sequence within pre-miRNA targets. Therefore, the addition of a pre-miRNA blocker with the mature molecular beacon in an assay containing a mixture of miRNA targets is a more accurate representation of the mature molecular beacon binding.
Additionally, these data also reveal the differences in the affinity of each molecular beacons for their targets. Specifically, in samples with mixed miRNA populations that contained mostly pre-miRNA, the mature miR-27b molecular beacon produced fluorescence signal, despite the presence of a pre-miRNA blocker. Although the fluorescence signal is mitigated by the presence of the pre-miRNA molecular blocker, additional strategies, such as utilizing a standard concentration curve, may be needed to ensure accurate quantification of the mature miRNA only by the mature miR-27b molecular beacon.
In the supplementary data (Figure S1 A and B), pre-miR-21 and -27b molecular beacons, respectively, showed very high fluorescence signal (indicating hybridization of molecular beacon to target) when incubated with varying concentrations (0-200nM) of synthetic target miRNA. As expected, the fluorescence signal of the molecular beacons targeting each pre-miRNA was not significantly decreased when oligos complementary to the mature miRNA sequences (mature miRNA blockers) were included in the hybridization assay. In contrast, oligos anti-sense to the pre-miRNA sequences (pre-miRNA blockers) dramatically inhibited hybridization and resultant fluorescence from molecular beacons targeting pre-miRNA.
Double Molecular Beacon Assay
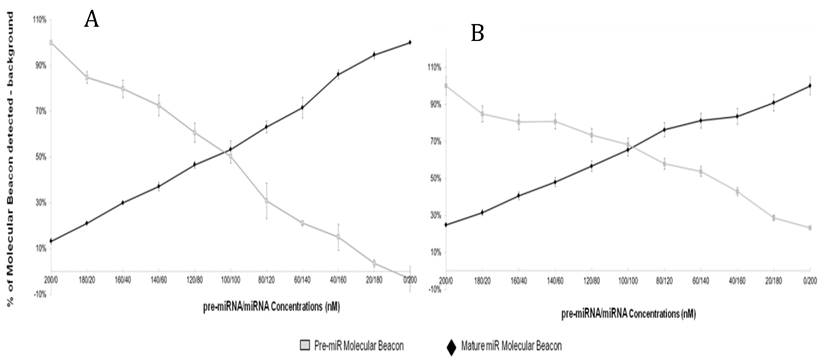
The results of the previous experiment suggest that the specificity of molecular beacons targeting mature miRNA is enhanced in the presence of precursor miRNA blockers. We hypothesized that incubation of both mature and precursor molecular beacons with a mixed population of miRNA would enable simultaneous measurement of both mature and precursor miRNA targets while enhancing specificity of the assay. Figure 4A and B shows the results of the “Double Molecular Beacon Assay”, where concentrations of synthetic mature and precursor miRNA targets were varied from 0-200nM.
Molecular Beacon Hybridization to Targets in Presence of Oligo Blockers. The molecular beacons were designed to hybridize mature miRNA targets. The presence of oligo blockers (complementary or competitive) demonstrated the specificity of each molecular beacon. Mature miRNA molecular beacons targeting miR-21(A) and -27b (B) were incubated with mixtures of synthetic mature and pre-miRNA at varying concentrations (0-200nM). The design of each mature miRNA molecular beacon allowed for the unintended hybridization to precursor miRNA. Therefore, the presence of a mature miRNA blocker inhibited signal from the molecular beacon while the presence of pre-miRNA blocker showed a modest change in fluorescence intensity. Each bar represents mean ± SE of 3-6 separate experiments.
Double Molecular Beacon Assay. Hybridization of molecular beacons to mature or precursor miRNA. The mature miRNA molecular beacon (200 nM) and pre-miRNA molecular beacon (200 nM) (A: miR-21; B: miR-27b) were incubated with mixtures of mature and pre-miRNA targets at the indicated concentrations (60 min at 37°C). The relative fluorescence signal from each molecular beacon alone in PBS was used as a background measurement and subtracted from the sample signal. Each data point represents mean ± SEM of three separate experiments.
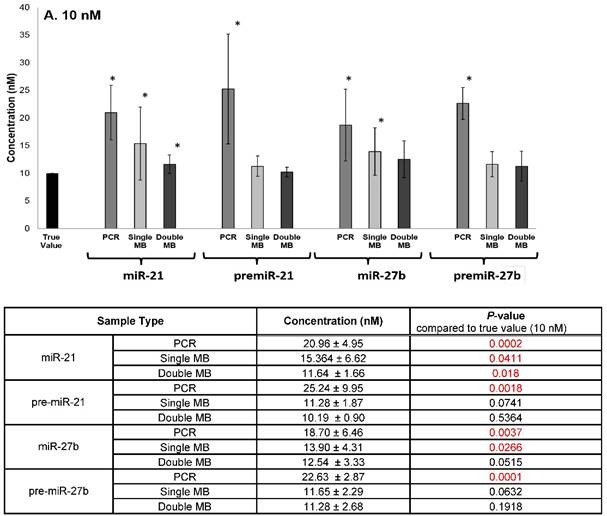
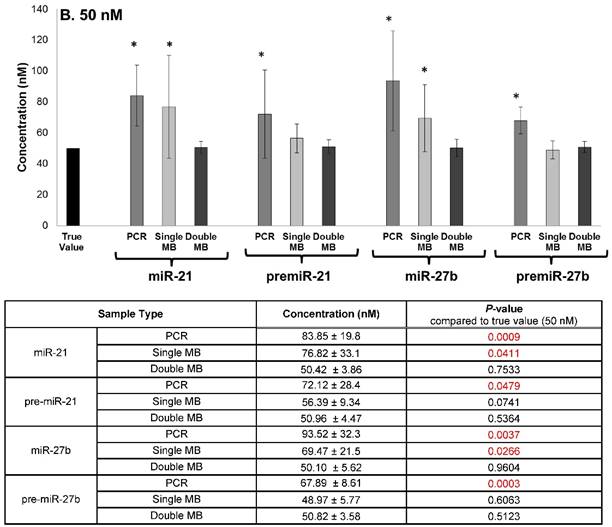
The specificity of the double molecular beacon assay compared to other methods (single molecular beacon and qRT-PCR) was assessed. Equimolar mixtures of synthetic mature and precursor miRNA (10 nM and 50 nM) were quantitated using qRT-PCR, the single molecular beacon assay, and the double molecular beacon assay. As shown in Figure 5A (10nM) and B (50 nM), the double molecular beacon assay was 87.5% accurate in assessing mature and precursor miRNA values. On the other hand, the single molecular beacon overestimated the mature miRNA levels for miR-21 and -27b while the qRT-PCR consistently overestimated the mature and precursor miRNA levels for miR-21 and -27b. These data validate the ability of the double molecular beacon assay to accurately quantify mature and precursor miRNA in a mixed population at different concentrations. Furthermore, the data indicate the double beacon assay has greater specificity compared to qRT-PCR and single molecular beacon methods.
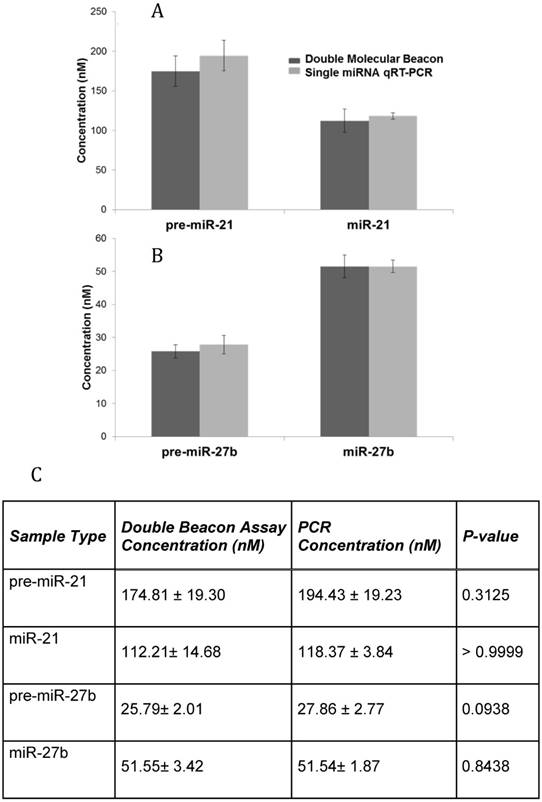
Additionally, the sensitivity and specificity of the double molecular beacon method to quantify mature miRNA and pre-miRNA from biological samples was tested. Our previous studies have demonstrated the use of the single molecular beacon method to quantitate cellular pre-miRNA and mature miRNA [25]. Figure 6 displays the quantitation of cellular pre-miRNA and mature miRNA using the novel double molecular beacon compared to qRT-PCR. RNA was isolated from human aortic endothelial cells (HAECs) and standard concentration curves were used to determine exact concentrations for both double molecular beacon and qRT-PCR. Mature and precursor miRNA concentrations obtained using qRT-PCR and the double molecular beacon assay are listed in Figure 6C. These findings are aligned with our previous studies of pre-miRNA/miRNA ratio [28] and are also supported by Lee et al. which indicates the ratio of mature to precursor miRNA varies by miRNA and between different tissues and cells[29]. In addition, this experiment supports the validity of using the double molecular beacon-based method to simultaneously assess precursor and mature miRNA expression in cells and tissues.
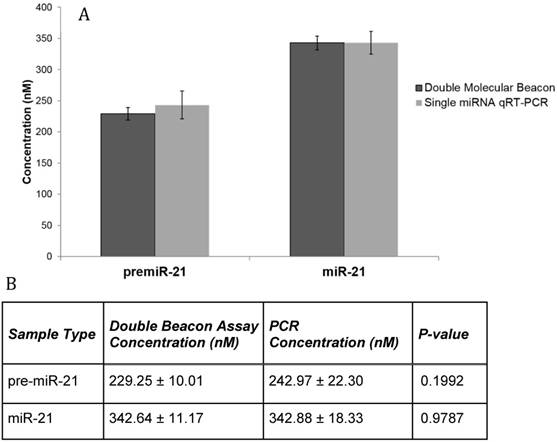
Lastly, the clinical relevance of the double molecular beacon method for simultaneously quantitation of pre-miRNA and mature miRNA in a complex biological matrix was tested. We incubated the mature miR-21 molecular beacon and the pre-miR-21 molecular beacon with total RNA that had been isolated from whole blood. Figure 7A shows the fluorescent signals from hybridization assays using the miR-21 double molecular beacon method compared to single qRT-PCR assays. Mature and precursor miRNA concentrations obtained using qRT-PCR and the double molecular beacon assay are listed in Figure 7B. This assay needs to be repeated with molecular beacons targeting other miRNAs, but these results support the use of this method in clinical samples as it is more time-efficient and as specific as the gold-standard qRT-PCR.
Comparison of Double Molecular Beacon Assay to qRT PCR and Single Molecular Beacon Assay. Comparison of double molecular beacon assay with qRT-PCR and single molecular beacon assay in simultaneously measurement of mature and precursor miRNAs. Assays were performed on solutions containing mixed, equimolar concentrations (A. 10 nM and B. 50 nM) of mature and precursor forms of miR-21 and miR-27b. For single and double MB assays, background fluorescence was subtracted from hybridized beacon signal. For qRT-PCR standard TaqMan protocol was followed. Standard concentration curves were used to determine concentration in all methods. Each data point represents mean ± SD of six separate experiments. Tables display the numerical concentrations and the P-values (paired t-tests P > 0.05) (significance indicated in red).
Comparison of qRT-PCR and Double Molecular Beacon Assay in assessing mature and precursor miRNA levels in total cellular RNA. Comparison of qRT-PCR and double molecular beacon assay in assessing miRNA concentrations in total RNA isolated from HAECs. MiRNA levels (A. miR-21 and B. miR-27b) in total RNA extracted from HAECs was determined. For each qRT-PCR reaction, the standard TaqMan protocol was used using 1 µg of total RNA. For the double molecular beacon hybridization assays, 200 nM of each molecular beacon and 1 µg of total RNA were used. Standard concentration curves were used to determine concentration in both methods. Additionally, for the double molecular beacon assay background fluorescence was subtracted from beacon signal. Each data point represents Mean ± SD of six separate experiments. C. Table displaying the numerical concentrations and the p-values (paired t-tests). (P > 0.05).
Discussion
In this study, we demonstrated that the double molecular beacon assay could distinguish between mature miRNA and pre-miRNA species as well as effectively and efficiently assess miRNA levels in a sample containing a mixture of mature and precursor forms of miRNA. This assay has the potential to be a powerful tool for quantifying miRNA expression in clinical specimens. Specifically, we showed that the double molecular beacon assay was effective for quantifying different miRNAs forms within a concentration range of 0.5-200 nM.
Backbone chemistry is an important component of molecular beacon design and in the double molecular beacon assay, the mature and pre-miRNA molecular beacons had different backbone chemistries. Since targeted pre-miRNA can hybridize to both pre-miRNA and mature molecular beacons (Figure 1), we developed pre-miRNA molecular beacons that contained LNA bases, thereby enhancing their binding to pre-miRNA target relative to the mature molecular beacon, which contained a DNA backbone without LNA bases. Additionally, the LNA bases in the pre-miRNA molecular beacon should reduce steric hindrance by stabilizing the hairpin pre-miRNA target; and can also be useful in future applications as LNA designed beacons have the ability to distinguish between single nucleotide differences within the mature miRNA and pre-miRNA sequences.
Comparison of qRT-PCR and Double Molecular Beacon Assay in assessing mature and precursor miRNA levels in whole blood RNA. Comparison of qRT-PCR and double molecular beacon assay in assessing miR-21 concentrations in total RNA isolated from whole blood (obtained from healthy volunteer). MiRNA levels (A. miR-21) in total RNA extracted from whole blood was determined. For each qRT-PCR reaction, the standard TaqMan protocol was used with 3 µg of total RNA. For the double molecular beacon hybridization assays, 500 nM of each molecular beacon and 3 µg of total RNA were used. Standard concentration curves were used to determine concentration in both methods. Additionally, for the double molecular beacon assay background fluorescence was subtracted from beacon signal. Each data point represents Mean ± SD of six separate experiments. B. Table displaying the numerical concentrations and the p-values (paired t-tests) (P > 0.05).
In addition, in samples containing a mixture of mature and precursor miRNAs, we did observe that the affinity of the mature molecular beacon with a DNA backbone was strong enough to bind when the complementary mature miRNA was present, however, the mature miRNA molecular beacon did not “lock” or create as tight of a bond with the much longer pre-miRNA targets. This is evidenced by decreased mature miRNA molecular beacon signal when incubated with pre-miRNA blocker or pre-miRNA beacon in samples containing predominantly pre-miRNA target (Figure 3 and Figure 4). Furthermore, the pre-miRNA blockers/pre-miRNA molecular beacons contained LNA/DNA bases that allowed them to competitively bind to the pre-miRNA in the sample and facilitate the binding of the mature miRNA molecular beacon only to mature miRNA target (instead of to both the mature and precursor miRNA target). This finding led to development of the innovated double molecular beacon assay for measurement of mature miRNA and subsequently pre-miRNA in a sample containing a mixed population of miRNA targets.
The important features of the double molecular beacon assay are: i) the usage of mature and pre-miRNA molecular beacons with different backbone chemistries and target binding affinities; and ii) the complementary blocking of the pre-miRNA target by the precursor miRNA molecular beacon and the intentional binding of the mature miRNA molecular beacon to the mature miRNA targets. These features were essential for attenuating falsely high background signal related to the binding of the mature miRNA molecular beacon to the target sequence in the stem of the pre-miRNA target during the double beacon assay (Figure 1A).
Again, in the double molecular beacon assay, a pre-miRNA molecular beacon is used to block the unintended hybridization of pre-miRNA target to mature molecular beacon (Figure 4), thereby allowing for accurate measurement of different miRNA forms in a mixed population. In contrast, we found that qRT-PCR and the single molecular beacon assay both overestimated miRNA levels in samples containing mixed miRNA populations. For qRT-PCR, this is likely due to nonspecific binding of the RT primers to the mature and pre-miRNA targets, allowing for nonspecific reverse transcription and amplification of RT products by PCR primers [30, 31]. For a single molecular beacon, the overestimation was observed when measuring mature miRNA in a mixed miRNA population and was caused by the unintended binding of the pre-miRNA to the mature miRNA molecular beacon with DNA backbone. As pointed out earlier, we found that the presence of pre-miRNA blockers enhanced the specificity of the mature molecular beacons for their targets during a single molecular beacon assay (Figure 3). Thus, unintentional binding in the single molecular beacon assay and the nonspecific binding and broad amplification of miRNAs seen when performing qRT-PCR resulted in a quantifiable overestimation of both pre-miRNA and mature miRNA.
The double molecular beacon assay does have certain shortcomings. When performing the double molecular beacon assay on synthetic miRNA targets (Figure 5), we found that the double beacon assay also tended to overestimate miRNA levels, although this was significantly less than qRT-PCR and the single molecular beacon assay. Overestimation of miRNA levels by the double molecular beacon assay was also less pronounced in assays using cellular RNA or RNA from whole blood (Figure 6 and Figure 7). Overestimation by the double molecular beacon assay is likely caused by unintentional hybridization of pre-miRNA to mature molecular beacon at high concentrations and was exacerbated when quantitating miRNA in samples containing synthetic targets due to the purity of the targets in these samples. In addition, it should be noted that some molecular beacons have higher background signal, as observed with mature and pre-miRNA 27b molecular beacons (Figure 4B). To correct for this high background signal, double molecular beacon data should be reported as relative fluorescence or by using a standard curve and reporting miRNA concentration levels. Lastly, in this study, we have validated the use of the double molecular beacon assay for both miR-21 and -27b, but additional optimization experiments will be required for developing double molecular beacon assays for other miRNAs.
The use of the double molecular beacon assay to quantitate miRNA is preferable to other methods because it can accurately assess differences in miRNA levels within mixed populations. The hybridization of molecular beacons to target miRNA and the measurement of fluorescence can be performed relatively quickly, in approximately 20-30 min, which is one-third the time of traditional qRT-PCR. Other conventional methods for miRNA detection, such as Northern Blotting, microarrays, and NGS, are able to quantitate miRNA expression in the fetamolar range, but they all have lower specificity with higher cost. These conventional methods also have not been thoroughly assessed for their ability to successfully multiplex for both mature and precursor forms of a miRNA while maintaining their sensitivity, specificity, time effectiveness, and cost [32].
In addition to the novelty of this method, it is also versatile. We demonstrated that this method can successful quantitate pre-miRNA and mature miRNA from synthetic targets, RNA isolated from cells (Figure 6) and even RNA isolated from whole blood (Figure 7). This versatility indicates that the method can not only be used as a laboratory research tool but also as a potential clinical tool.
Overall, the double molecular beacon assay is highly specific when detecting both mature and precursor forms of miRNA in a biological sample. Potential other applications of a double molecular beacon assay are quantitation of miRNA variants, such as isomiRs, which vary from the archetypal miRNA by 1-3 nucleotides. The advantage of quantitating isomiRs using molecular beacons lies in their ability to distinguish targets with single nucleotide differences and their ability to quantitate isomiRs that are expressed at very low concentrations. Therefore, this innovative double molecular beacon-based miRNA detection offers a hybridization assay format that is sensitive, specific, fast, easy to multiplex and capable of distinguishing mature and pre-miRNAs for both clinical and research use.
Supplementary Material
Figure S1.
Abbreviations
BHQ1: Black Hole quencher 1; CVD: cardiovascular disease; Dab: Dabcyl; DGCR8: DiGeorge Syndrome Critical Region 8: HAECs: Human aortic endothelial cells; IAblk: Iowa Black quencher; isomiR: microRNA variant or isoform; LNA: locked nucleic acid; miRNA: microRNA; mRNA: messenger RNA; MB: molecular beacon; NGS: next generation deep sequencing; pre-miRNA: precursor microRNA; pri-miRNA: primary microRNA transcript; qRT-PCR: quantitative real-time polymerase chain reaction; RISC: RNA induced silencing complex.
Acknowledgements
This work was supported by the National Heart Lung and Blood Institute of the National Institutes of Health as a Program of Excellence in Nanotechnology (HHSN268201000043C to GB) and a VA Merit Award (I01 BX000704 to CDS).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Dennis C. The brave new world of RNA. Nature. 2002;418:122-4
2. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835-40
3. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
4. van Rooij E. The art of microRNA research. Circulation research. 2011;108:219-34
5. Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nature structural & molecular biology. 2006;13:1097-101
6. Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Critical reviews in biochemistry and molecular biology. 2013;48:51-68
7. Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95-8
8. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-33
9. Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J. et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769-73
10. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome research. 2009;19:92-105
11. Wark AW, Lee HJ, Corn RM. Multiplexed detection methods for profiling microRNA expression in biological samples. Angewandte Chemie (International ed in English). 2008;47:644-52
12. Garibyan L, Avashia N. Polymerase chain reaction. The Journal of investigative dermatology. 2013;133:e6
13. Valasek MA, Repa JJ. The power of real-time PCR. Advances in physiology education. 2005;29:151-9
14. Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303-8
15. Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat Biotechnol. 1998;16:49-53
16. Vet JA, Majithia AR, Marras SA, Tyagi S, Dube S, Poiesz BJ, Kramer FR. Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc Natl Acad Sci U S A. 1999;96:6394-9
17. Bao G, Rhee WJ, Tsourkas A. Fluorescent probes for live-cell RNA detection. Annu Rev Biomed Eng. 2009;11:25-47
18. Guo J, Ju J, Turro NJ. Fluorescent hybridization probes for nucleic acid detection. Anal Bioanal Chem. 2012;402:3115-25
19. Nitin N, Bao G. NLS peptide conjugated molecular beacons for visualizing nuclear RNA in living cells. Bioconjug Chem. 2008;19:2205-11
20. Nitin N, Santangelo PJ, Kim G, Nie S, Bao G. Peptide-linked molecular beacons for efficient delivery and rapid mRNA detection in living cells. Nucleic acids research. 2004;32:e58
21. Santangelo P, Nitin N, Bao G. Nanostructured probes for RNA detection in living cells. Annals of biomedical engineering. 2006;34:39-50
22. Santangelo PJ, Nix B, Tsourkas A, Bao G. Dual FRET molecular beacons for mRNA detection in living cells. Nucleic acids research. 2004;32:e57
23. Tsourkas A, Behlke MA, Bao G. Structure-function relationships of shared-stem and conventional molecular beacons. Nucleic acids research. 2002;30:4208-15
24. Tsourkas A, Behlke MA, Xu Y, Bao G. Spectroscopic features of dual fluorescence/luminescence resonance energy-transfer molecular beacons. Anal Chem. 2003;75:3697-703
25. Baker MB, Bao G, Searles CD. In vitro quantification of specific microRNA using molecular beacons. Nucleic acids research. 2012;40:e13
26. Gerasimova YV, Hayson A, Ballantyne J, Kolpashchikov DM. A single molecular beacon probe is sufficient for the analysis of multiple nucleic acid sequences. Chembiochem: a European journal of chemical biology. 2010;11:1762-8
27. Wang L, Yang CJ, Medley CD, Benner SA, Tan W. Locked nucleic acid molecular beacons. Journal of the American Chemical Society. 2005;127:15664-5
28. Alexy T, Rooney K, Weber M, Gray WD, Searles CD. TNF-alpha alters the release and transfer of microparticle-encapsulated miRNAs from endothelial cells. Physiol Genomics. 2014;46:833-40
29. Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA. 2008;14:35-42
30. Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. Journal of molecular endocrinology. 2002;29:23-39
31. Freeman WM, Walker SJ, Vrana KE. Quantitative RT-PCR: pitfalls and potential. BioTechniques. 1999;26:112-22 24-5
32. Catuogno S, Esposito CL, Quintavalle C, Cerchia L, Condorelli G, De Franciscis V. Recent Advance in Biosensors for microRNAs Detection in Cancer. Cancers. 2011;3:1877-98
Author contact
![]() Corresponding author: amandamariejamesedu
Corresponding author: amandamariejamesedu
 Global reach, higher impact
Global reach, higher impact