13.3
Impact Factor
Theranostics 2013; 3(12):1004-1011. doi:10.7150/thno.4580 This issue Cite
Review
In vivo Monitoring of microRNA Biogenesis Using Reporter Gene Imaging
1. Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea;
2. Department of Nuclear Medicine, Seoul National University Boramae Hospital, Seoul, Republic of Korea;
3. Institute of Radiation Medicine, Medical Research Center, and
4. WCU, Department of Molecular Medicine and Biopharmaceutical Science, Graduate School of Convergence Science and Technology, Seoul National University, Seoul, Korea
Received 2012-8-23; Accepted 2013-1-4; Published 2013-12-8
Abstract
MicroRNAs are small noncoding RNAs regulating gene expression, through base paring with their target mRNAs, which have been actively investigated as key regulators in a wide range of biological processes. Conventional methods such as Northern blot are generally time-consuming, non-repeatable, and cannot be applied in vivo due to the requirement for cell fixation. Therefore, a noninvasive imaging system is required for the monitoring of microRNA biogenesis to understand the versatile functions of microRNAs in vivo as well as in vitro and to accelerate the clinical application of microRNA-based therapy. In this paper, we reviewed noninvasive molecular imaging systems for the monitoring of the biogenesis of microRNAs and post-transcriptional regulation of genes by microRNAs and the roles of microRNAs in various biological processes.
Keywords: MicroRNA biogenesis, MicroRNA imaging, Optical reporter gene, In vivo optical imaging
Introduction
MicroRNAs are defined as a class of single-stranded noncoding small RNAs, composed of approximately 22 nucleotides in length. microRNAs are negative gene regulators at the post-transcriptional level, participating in various biological processes, including developmental stages, cellular proliferation/differentiation, and cell death [1-4]. microRNAs exert their function on plant or animal genomes through the degradation of target mRNAs or translational inhibition of the target mRNAs by means of complete or partial binding to the 3' untranslated regions (UTRs) [5].
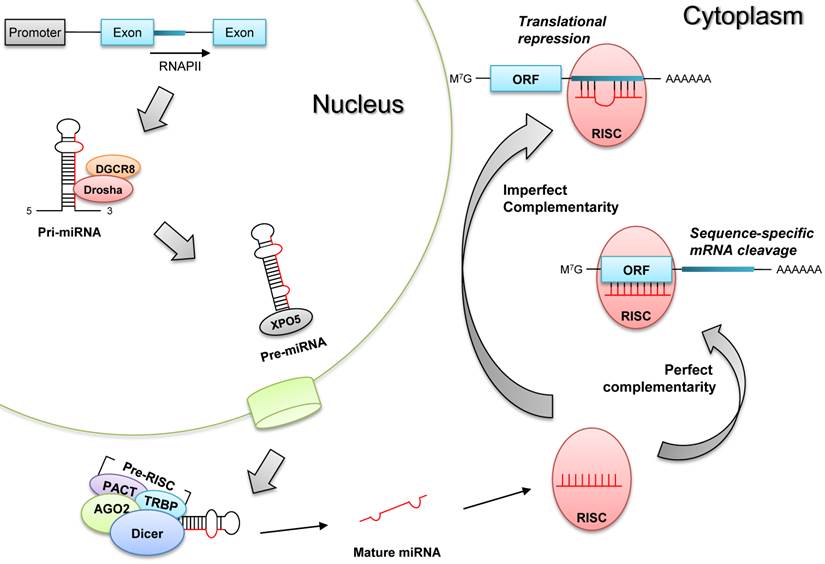
The biogenesis of microRNAs undergoes sequential processes to generate mature microRNAs (Fig.1). First, primary microRNAs (pri-miRNAs) are mostly transcribed from the intragenic or intergenic regions by RNA polymerase II [6], and these pri-miRNAs are excised by Drosha (double-stranded RNA-specific ribonuclease) in the nucleus to produce pre-miRNAs having hair-pin structures [7]. After being translocated to the cytoplasm, pre-miRNAs are further processed by Dicer (RNase III endonuclease) into mature single-stranded microRNAs [8]. Finally, these mature microRNAs are incorporated into RISC (RNA-induced silencing complex) to elicit target mRNA degradation or translational repression [9].
Since microRNAs are considered crucial in a variety of cellular pathogenesis, especially in tumorigenesis, intensive efforts have been made to develop molecular diagnostics and therapeutics using microRNAs [10-12]. During the past few years, a few therapeutics using microRNAs have entered clinical trials. The understanding of microRNA biogenesis and its processing and their contribution to molecular pathogenesis will contribute to revolutionizing the treatment of human disease. Thus, efficient and reliable methods to detect microRNAs and evaluate the biogenesis and regulation of microRNAs are essential to investigate their possible roles in various biological or pathological processes, as well as to accelerate clinical application of microRNA-based therapy.
Conventionally, Northern blot, real-time PCR, microarray, etc., have been widely used to detect and evaluate the endogenous production of microRNAs. These current methods, however, are generally time-consuming, laborious, non-repeatable, and not clinically relevant because they require the fixation or lysis of cells. Therefore, a noninvasive imaging system for the monitoring of microRNA biogenesis and regulation is necessary to investigate the microRNA expression patterns and their versatile functions in human diseases. Recently, molecular imaging using an optical reporter gene system has been developed and utilized in assessing microRNA biogenesis, both in vitro and in vivo [13-17], because optical imaging system is advantageous to monitor microRNAs among various kinds of molecular imaging methods (Table 1). Here, we reviewed molecular imaging systems used in the investigations of microRNA biogenesis, in particular, reporter gene systems that monitored microRNA biogenesis and its post-transcriptional regulation in vivo as well as in vitro.
Biogenesis of microRNAs. Pri-miRNAs are transcribed by RNA polymerase (RNAPII), and they become pre-miRNAs after being processed by Drosha and its partner DGCR8 in the nucleus. Pre-miRNAs are translocated to the cytoplasm by exportin 5 (XPO5), and they are processed into mature single stranded miRNAs by Dicer-containing pre-RISC (RNA-induced silencing complex). These matures miRNAs are incorporated into RISC to produce post-transcriptional gene silencing, according to the complementarity to target mRNAs; target mRNA degradation (perfect complementarity) or translational repression (imperfect complementarity).
Comprehensive comparison of advantages and disadvantages of molecular imaging methods.
| Molecular imaging methods | Advantages | Disadvantages | |
|---|---|---|---|
| Optical imaging | Fluorescence-based imaging | High sensitivity (10-9-10-12 mol/L) Multiplex detection available Subcellular data available | Depth limit (< cm) Relatively difficult for in vivo tomography High background |
| Bioluminescence-based imaging | Highest sensitivity (10-15-10-17 mol/L) Low cost Low background Quick and simple method | Depth limit (1-2 cm) Limited translational study | |
| Positron emission tomography (PET) | High sensitivity (10-11-10-12 mol/L) Quantitative analysis No limit of depth Translation to human Tomographic data available | Relatively low resolution High cost Cyclotron needed | |
| Magnetic resonance imaging (MRI) | High resolution (25-100 µm) No limit of depth Possible for quantitative analysis Tomographic data available Translation to human | Poor sensitivity (10-3-10-5 mol/L) High cost | |
Imaging strategies
1. Optical reporter gene system for transcript production of microRNA
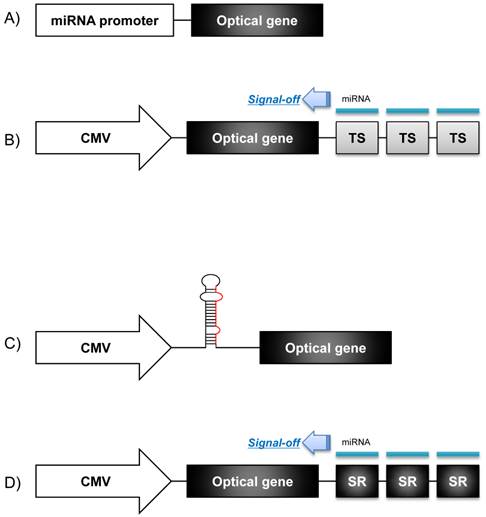
A molecular imaging system using an optical reporter gene can utilize various imaging strategies according to the purposes of the imaging. An optical reporter gene vector under the control of a specific microRNA promoter can be used for monitoring the pri-miRNA expression during microRNA biogenesis. This imaging strategy is based on the fact that the transcription of microRNA is controlled by its own promoter [6]. To constitute a microRNA promoter-restricting reporter gene system, the 5' regulatory promoter region of a microRNA is located upstream of a cDNA fragment encoding fluorescent or bioluminescent proteins (Fig. 2A), which proportionally reflects the endogenous expression of a pri-miRNA [18]. Using this strategy, Ko et al. monitored the biogenesis of neuronal specific miR-9 in P19 cells during neuronal differentiation, both in vivo and in vitro [14]. They identified the 5' regulatory upstream sequence of the pri-miR-9 through a database analysis, and this upstream sequence was fused into the cassette of a promoterless firefly luciferase (Fluc) vector to examine the pri-miR-9 expression patterns. In addition, Lee et al. devised a microRNA promoter-restricting reporter gene imaging system using a Fluc gene vector that was located downstream to the miR-23a~27a~24-2 gene cluster promoters [13]. In this study, the bioluminescence signal intensity represented the concentration of the pri-miRNA at the transcription level since it was controlled by the same 5' regulatory promoter of the microRNA. Using this imaging strategy, they demonstrated that the expression of pri-miR-23a increased in P19 cells once neuronal differentiation was induced by retinoic acid.
2. Optical reporter gene system containing microRNA target sequence
In general, an optical reporter gene imaging system consists of a regulatory (constitutive or cell/ gene-specific) promoter and an optical gene vector encoding a fluorescent protein or bioluminescent protein. As for a constitutive regulatory promoter, retroviral vectors such as the cytomegalovirus (CMV) promoter or lentiviral vectors are chosen since they are strong promoters and constitutive in expression in a broad range of cell types. Reporter genes are constitutively expressed under these strong promoters if transfection was successful.
When we try to image the expression of mature microRNAs, microRNA target sequences are fused with the optical reporter gene vector in its 3' UTR (Fig. 2B). These microRNA target sequences are designed to reside between reporter exon and their poly(A) tail, and if microRNA are bound to target sequences, optical reporter gene is down-regulated just like the endogenous target genes of these microRNAs. And fortunately, over-expression of the exogenous reporter genes containing the perfectly matching target sequences of the microRNA was not reported to saturate endogenous microRNA processing apparatus [19]. Thus, the transfected reporter genes could be used as tracers of mature microRNA activity. In this system, the fluorescence or bioluminescence signals decreased in the presence of the endogenous microRNAs, which can be called as a “signal-off system.” Many studies have been performed in vitro to investigate post-transcriptional regulation using this imaging strategy.
Schematics of optical reporter gene systems for in vivo monitoring of microRNA biogenesis. A) The 5' regulatory region of a pri-miR is fused into a promoterless optical gene vector to examine the pri-miR expression level. B) A constitutive promoter such as a CMV promoter or lentiviral vector is chosen to control transcription of an optical gene vector, and 3 or 4 microRNA targeting sequences (TS) are connected between the optical gene vector and poly(A) tails to examine mature microRNA action. C) Oligonucleotides of a pri-miRNA/ pre-miRNA are inserted between the constitutive promoter and the start codon of the optical gene vector. D) The 3' UTR of a microRNA containing a seed region (SR) of a target microRNA is located downstream to the optical gene vector, under the control of the constitutive regulatory promoter. Signal extinction of the reporter gene means that microRNA acted on the SR of target mRNA of that microRNA.
1. Fluorescence-based microRNA imaging
The optical imaging system using a fluorescent protein has widely been used to investigate various biological processes since green fluorescent protein (GFP) was first isolated from the jellyfish, Aequorea victoria, in 1962. Recently, a few studies have adopted a fluorescence protein as an optical reporter gene to investigate microRNA biogenesis and regulation [19-21]. Brown et al. reported that the activity of miR-142 differed according to tissue types, using a fluorescence imaging system that consisted of a lentiviral vector encoding a GFP and target sequences in its 3'UTR [19]. In this study, the GFP expression from miR-142 was effectively suppressed only in hematopoietic lineages since mature miR-142 was expressed in these lineages.
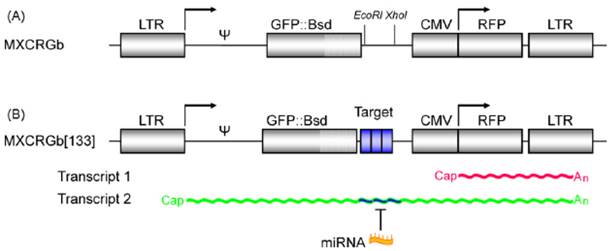
The fluorescent protein-based optical imaging can constitute a multi-color imaging system since there are diverse fluorescent proteins with unique emission wavelengths without much overlapping in emission spectrum, such as GFP, yellow fluorescent protein (YFP), red fluorescent protein (RFP), etc. Using these multiplexed fluorescent proteins, it was possible to perform real-time monitoring of the dynamics of the microRNA activity. Kato et al. developed a two-color fluorescence imaging system to investigate the dynamic changes of miR-133 activity in living cells during myogenic differentiation [21]. In this system, they designed a proviral vector, which is transcribed as two mRNAs encoding GFP and RFP from a single transcript. The 5' long term repeat (LTR) was linked to GFP, and then three copies of the binding sequences with complementarity to miR-133 were located downstream of the GFP gene. Downstream to the GFP genes, the CMV promoter-regulated RFP cassette was also inserted under the control of the same retroviral vector (Fig. 3). The expression of endogenous miR-133 reduced the GFP signal without affecting the RFP signal. In this study, RFP worked as an internal reference for normalization of transfection efficiency. And the authors could evaluate the activity of expressed miR-133 during myogenesis through real-time dual-color imaging.
A schematic illustration of a dual-color fluorescent imaging system. (A) Two different mRNAs encoding GFP and RFP are inserted in the proviral form of the lentiviral vector based on the Moloney murine leukemia virus (M-MLV). (B) The 5' LTR is linked to GFP connected to three copies of the binding sequences with complementarity to a miR, and the CMV promoter-regulated RFP cassette follows, which is used as internal reference for data normalization. (Adapted from reference [21] with permission)
Despite these successes in vitro, in vivo application of fluorescence imaging faces with several inherent limitations such as a high background auto-fluorescence, or limited tissue penetration of the light in vivo [22]. In particular, a signal-off system works poorly in vivo due to the ambiguity of signal loss in the presence of high background auto-fluorescence. To overcome the limitations of fluorescence-based imaging systems, new probe chemistries were adopted and new imaging strategies were developed [23]. Among these efforts, a signal-on system for microRNA imaging was developed using a fluorescent nanoparticle-based molecular beacon (MB) having surface-bound nucleic acids [24, 25]. The nanoparticle surface-bound MB had oligonucleotides that are complementary to a microRNA of interest, having a fluorophore (nanoparticle) at one end and a quencher at the other end [26]. The MB-based imaging system worked as follows; binding of endogenous microRNAs to the oligonucleotide makes the quencher dissociate from the nanoparticle and recover fluorescence (“signal-on”), while fluorescence had been absorbed by the quencher in the absence of endogenous microRNAs. This signal-on MB-based imaging system, however, is not reporter gene-based one and cannot be applied to proliferating cells in vivo as well as in vitro.
2. Bioluminescence-based imaging
Optical imaging using a bioluminescence reporter gene allows one to visualize molecular changes that are caused by microRNA. Moreover, it is advantageous over fluorescence-based imaging in terms of high signal to background ratios since the bioluminescence detects light emitted from the cells or tissues by an enzymatic reaction between a luciferase and its substrate; no light is emitted in the absence of the enzymatic reaction and luciferase is absent in mammalian cells.
Fluc, Renilla luciferase (Rluc), and Gaussia luciferase (Gluc) are the most commonly used optical gene vectors encoding bioluminescent proteins. They use the same (coelenterazine for Rluc or Gluc) or different substrates (luciferin for Fluc) and multiplexing with Rluc and Fluc or Gluc and Fluc is possible for imaging. Gluc was reported to emit a strong light, which is approximately hundreds fold greater than Fluc [27], but as its emission wavelength is shorter, the photons attenuate more in in vivo imaging.
Like dual-color fluorescence imaging, it is also possible to constitute a dual-luciferase reporter imaging system for microRNA biogenesis. In the previously described study using a microRNA promoter-restricting reporter gene imaging system [13], Lee et al. proposed a dual-luciferase reporter imaging system to simultaneously monitor endogenous microRNA biogenesis and its post-transcriptional regulation. For simultaneous monitoring, they designed two different reporter gene constructs: one with a Fluc gene for the monitoring of endogenous pri-miRNA production, and the other with a Gluc gene for the visualization of post-transcriptional regulation by microRNAs. As a result, the luciferase activity of Fluc increased when the pri-miRNA transcript was expressed, while the bioluminescence activity of Gluc diminished in the presence of the mature microRNAs. This dual reporter system for the simultaneous monitoring of microRNA biogenesis and its post-transcriptional regulation is noteworthy for elucidating the precise role of microRNA considering that the microRNAs are regulated by other peptides/ RNAs in their processing step from their primary transcripts into their active mature forms. Interestingly, in this result, the expression levels of the primary and mature miR-23 were different in 293 cells, while the expression level of the pri-miR-23 was positively correlated with that of the mature miR-23 in P19 cells and HeLa cells. An unknown regulatory mechanism that is responsible for the maturation of microRNA during Drosha-or Dicer-related process would cause this kind of discrepancy in the 293 cells. This dual reporter strategy using different luciferase reporter genes is suitable not only for the simultaneous detection of pri-miR expression and mature microRNA activity in individual cells, but also for the investigation of abnormalities in microRNA processing in cells.
Luciferase-reporting strategy was also used to assess simply the transfection efficiency of exogenous microRNA into cells. Oligonucleotides of a microRNA were designed having a hair-pin structure and fused with a Fluc gene vector under the control of constitutive promoter such as the CMV promoter [13] (Fig. 2C). This imaging system could easily track the production of transfected exogenous microRNA by observing the expression of reporter proteins in vivo as well as in vitro.
The endogenous targets (mRNAs) of a microRNA could be monitored by an optical reporter gene imaging system. A reporter gene imaging system for microRNA targets are constructed, instead of multiple copies of microRNA perfect target sequences, as the 3'UTR of probable target mRNA containing a seed region is fused downstream to a reporter gene vector under the control of a constitutive promoter (Fig. 2D). This optical reporter gene system has been utilized to find the probable target (mRNA) of a specific microRNA and to examine the functional relationship between specific microRNAs and their mRNA targets. For example, a reporter gene imaging system consisting of a Gluc vector fused with the putative miR-221 seed region in its 3'UTR of the homeobox B5 (HOXB5) was used to monitor the activity of miR-221 on the target mRNA of HOXB5 during the development of papillary thyroid cancer (PTC) [15].
Applications of microRNA imaging
1. Investigation of microRNA biogenesis during organogenesis
Optical reporter gene imaging system was used primarily to investigate the biogenesis of microRNAs during a variety of biological processes including neurogenesis, carcinogenesis, or myogenesis [13, 16, 17, 25]. The first step of microRNA biogenesis was mainly studied using tissue-specific microRNA promoter and this promoter-driven reporter gene. However, pri-miRNAs, once produced, proceeds sequentially to generate mature microRNAs to regulate target mRNAs in the cytoplasm. Each step of the microRNA biogenesis and processing is to be understood in detail and can be monitored by microRNA imaging. For instance, simultaneous examination of the expression of pri-miRNAs and mature microRNA activity could disclose concordance or discordance of expression and action of microRNAs in differentiating cells and their meaning is yet to be interpreted in both biological and mechanistic aspects [13, 18]. The functional activity of the Dicer enzyme, which is dicing pre-miR in the cytoplasm, could also be evaluated by a MB-based fluorescence imaging system [18, 28].
2. Elucidation of endogenous targets of microRNAs
MicroRNA contains an anti-sense “seed region” 7-8 nucleotide long among its 22 nucleotides that generally binds the sense seed region in the 3' UTR of their target mRNAs. Imperfect binding of microRNA seed region to mRNA's seed region is necessary to elicit mRNA degradation or translational inhibition. As each microRNA has a seed region specific to multiple mRNA sequences [29], microRNAs can repress multiple target mRNAs and signaling pathways, which has been supported by observations that various target mRNAs were down-regulated by a specific microRNA. Accordingly, the identification of bona fide target mRNAs is essential to understand the precise role of microRNAs in biological processes. An optical reporter gene system for finding microRNA targets can identify the target mRNAs and elucidate the functional relationship between the microRNAs and their target mRNAs. As described above in 'Imaging strategies' section, the molecular imaging strategy to elucidate the endogenous miRNA target is based on the use of seed sequence of target genes situated in their 3' UTR for miRNA. Reporter gene construct containing multiple copies of seed sequence of miRNA target located in its downstream region can be used to image the miRNA targets directly. Consequently, HOXB5 was found as an endogenous target of an abundant miR-221 in thyroid cancer cells by Kim and colleagues [15]. They could elucidate the target, HOXB5, using a reporter gene imaging system to monitor the activity of miR-221 during the development of PTC. The same group also applied this 3'-UTR seed region-based reporter gene imaging successfully to monitor miR-124a-mediated repression of the target gene chromosome 14 open reading frame 24 (c14orf24) during neurogenesis [17].
3. Characterizing the roles of microRNAs in cancer
While microRNAs play a key role in the fine-tuning of mRNA expression, and regulating various biological processes under normal physiological conditions and development, the role of microRNAs has recently been spotlighted in the pathogenesis of human cancers. Fifty percent of the microRNAs are known to be located at fragile sites that are often mutated in cancers [30, 31]. It is hypothesized that microRNAs repressing tumor suppressor genes are over-expressed or microRNAs repressing oncogenes are downregulated during the development of cancers [12].
The discovery of the close relationship between microRNAs and tumorigenesis shall enable better understanding of tumor biology and/ or provide a new target for cancer therapeutics. Optical reporter gene imaging systems were used to identify microRNA candidates related with certain types of cancers for the development of novel cancer therapeutics. An in vivo luciferase imaging system was used to monitor miR-221 biogenesis which was considered to play a key role in the development of PTC [16]. In this study, the optical reporter gene imaging system enabled in vitro quantitative assessment and in vivo qualitative visualization of miR-221 biogenesis in cancer.
The potential of microRNA imaging as a biomarker increases since novel microRNA-based therapeutic strategies have emerged in the field of oncology. Ponomarev et al. developed a cytometry-based imaging protocol allowing simultaneous visualization and quantification of microRNAs and their putative targets in neuroblastoma cells [32]. Though this method is suitable for analyzing the expression and cellular localization of microRNAs and target proteins, this method can only be applied for small cell subsets. On the other hand, Würdinger et al. demonstrated that the therapeutic effect of anti-miR-296 inhibiting tumor angiogenesis could be monitored by fluorescence imaging in tumor-xenografted mice [33]. The tumor size was reduced and the vasculature changed, however, miR-296 expression level was not directly visualized in living animals.
Recently, Kim et al. proposed miR-221 MB-conjugated nanoparticles combined with a nucleolin aptamer as a cancer targeting theragnostics tool [34]. In this study, the aptamer was used for targeted delivery into cancer cells, and miR-221 MB was used for the detection and inhibition of miR-221. The therapeutic effects of this aptamer/ miR-221 MB-conjugated nanoparticles were demonstrated in various cancer cell lines. When MB signal was on, one can presume that endogenous miR-221 is produced and inhibitory sequence of miR-221MB is freed to bind and hopefully inhibit miR-221 within the cancer cells.
4. Roles of microRNAs in cellular differentiation
Since the regulatory functions of microRNAs were clarified in various stages of development and cellular differentiation, hundreds of microRNAs have been identified in association with neuronal differentiation and myogenesis [35-38]. In myogenic differentiation, miR-1, miR-133, and miR-206 are known to play an important role through post-transcriptional regulation. Kato et al. demonstrated that miR-133 enhanced the proliferation of myoblast, and the expression of miR-133 was induced prior to the formation of myotubes using a two-color fluorescence imaging system [21]. On the other hand, numerous microRNAs have been investigated in association with neuronal differentiation. Among them, miR-9, miR-9* (passenger microRNA; the opposite strand of microRNA), or miR-124a are known to play a major role in the regulation of neuronal development. In particular, both miR-9 and mir-9* are highly co-expressed and neuron-specific during brain development [38]. Ko et al. developed an optical reporter gene imaging system to elucidate miR-9/ miR-9* biogenesis during the neuronal differentiation of P19 cells, and they demonstrated that miR-9 was more highly expressed than miR-9* both in vivo and in vitro [14].
Conclusions
A growing body of evidence has supported the importance of microRNAs as a key regulator in a wide range of biological processes as well as various human diseases. The reporter gene based imaging system is fundamentally a noninvasive and repeatable method, and capable of quantitative visualization of a microRNA or its target mRNAs and their longitudinal changes without sacrificing animals. Therefore, a noninvasive imaging system could be a reliable and efficient tool to investigate microRNA biogenesis and post-transcriptional regulation, and to understand their versatile functions in human diseases.
Acknowledgements
This work was supported by the Nano-Biotechnology Project (Regenomics, 20100002086), Brain Research Center of the 21st Century Frontier Research Program (2009K001257), WCU project of the MEST and National Research Foundation (NRF) (R31-2008-000-10103-0), the future-based technology development program of the NRF funded by the MEST (20100028755). This study was supported by a grant (kiom-2010-2) from the Inter-Institutional Collaboration Research Program provided by the Korea Research Council of Fundamental Science &Technology (KRCF), Korea.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Reinhart BJ, Slack FJ, Basson M. et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901-6
2. Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25-36
3. Dostie J, Mourelatos Z, Yang M, Sharma A, Dreyfuss G. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA. 2003;9:180-6
4. Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790-5
5. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
6. Lee Y, Kim M, Han J. et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051-60
7. Lee Y, Ahn C, Han J. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415-9
8. Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;40:293-6
9. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-33
10. Negrini M, Ferracin M, Sabbioni S, Croce CM. MicroRNAs in human cancer: from research to therapy. J Cell Sci. 2007;120:1833-40
11. Sandhu S, Garzon R. Potential applications of microRNAs in cancer diagnosis, prognosis, and treatment. Semin Oncol. 2011;2011(38):781-7
12. Wilmott JS, Zhang XD, Hersey P, Scolyer RA. The emerging important role of microRNAs in the pathogenesis, diagnosis and treatment of human cancers. Pathology. 2011;43:657-71
13. Lee JY, Kim S, Hwang do W. et al. Development of a dual-luciferase reporter system for in vivo visualization of MicroRNA biogenesis and posttranscriptional regulation. J Nucl Med. 2008;49:285-94
14. Ko MH, Kim S, Hwang do W, Ko HY, Kim YH, Lee DS. Bioimaging of the unbalanced expression of microRNA9 and microRNA9* during the neuronal differentiation of P19 cells. FEBS J. 2008;275:2605-16
15. Kim HJ, Kim YH, Lee DS, Chung JK, Kim S. In vivo imaging of functional targeting of miR-221 in papillary thyroid carcinoma. J Nucl Med. 2008;49:1686-93
16. Kim HJ, Chung JK, Hwang do W, Lee DS, Kim S. In vivo imaging of miR-221 biogenesis in papillary thyroid carcinoma. Mol Imaging Biol. 2009;11:71-8
17. Ko HY, Lee DS, Kim S. Noninvasive imaging of microRNA124a-mediated repression of the chromosome 14 ORF 24 gene during neurogenesis. FEBS J. 2009;276:4854-65
18. Kim S, Hwang do W, Lee DS. A study of microRNAs in silico and in vivo: bioimaging of microRNA biogenesis and regulation. FEBS J. 2009;276:2165-74
19. Brown BD, Venneri MA, Zingale A, Sergi Sergi L, Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med. 2006;12:585-91
20. Mansfield JH, Harfe BD, Nissen R. et al. MicroRNA-responsive 'sensor' transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079-83
21. Kato Y, Miyaki S, Yokoyama S. et al. Real-time functional imaging for monitoring miR-133 during myogenic differentiation. Int J Biochem Cell Biol. 2009;41:2225-31
22. Troy T, Jekic-McMullen D, Sambucetti L, Rice B. Quantitative comparison of the sensitivity of detection of fluorescent and bioluminescent reporters in animal models. Mol Imaging. 2004;3:9-23
23. Rao J, Dragulescu-Andrasi A, Yao H. Fluorescence imaging in vivo: recent advances. Curr Opin Biotechnol. 2007;18:17-25
24. Hwang do W, Song IC, Lee DS, Kim S. Smart magnetic fluorescent nanoparticle imaging probes to monitor microRNAs. Small. 2010;6:81-8
25. Kang WJ, Cho YL, Chae JR, Lee JD, Choi KJ, Kim S. Molecular beacon-based bioimaging of multiple microRNAs during myogenesis. Biomaterials. 2011;32:1915-22
26. Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303-8
27. Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11:435-43
28. Davies BP, Arenz C. A fluorescence probe for assaying micro RNA maturation. Bioorg Med Chem. 2008;16:49-55
29. Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2006;3:e85
30. Calin GA, Sevignani C, Dumitru CD. et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999-3004
31. Volinia S, Calin GA, Liu CG. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257-61
32. Ponomarev ED, Veremeyko T, Barteneva NS. Visualization and quantitation of the expression of microRNAs and their target genes in neuroblastoma single cells using imaging cytometry. BMC Res Notes. 2011;28:517-21
33. Würdinger T, Tannous BA, Saydam O. et al. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;4:382-93
34. Kim JK, Choi KJ, Lee M, Jo MH, Kim S. Molecular imaging of a cancer-targeting theragnostics probe using a nucleolin aptamer- and microRNA-221 molecular beacon-conjugated nanoparticle. Biomaterials. 2012;33:207-17
35. Chen JF, Mandel EM, Thomson JM. et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2004;38:228-33
36. Shi Y, Zhao X, Hsieh J. et al. MicroRNA regulation of neural stem cells and neurogenesis. J Neurosci. 2010;30:14931-6
37. Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677-87
38. Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274-81
Author contact
![]() Corresponding author: Dong Soo Lee, MD, PhD. Department of Nuclear Medicine, Seoul National University Hospital, 28 Yongon-Dong, Jongno-Gu, Seoul 110-744, Korea. Tel: +82-2-20722501; Fax: +82-2-20727690; E-mail: dslac.kr
Corresponding author: Dong Soo Lee, MD, PhD. Department of Nuclear Medicine, Seoul National University Hospital, 28 Yongon-Dong, Jongno-Gu, Seoul 110-744, Korea. Tel: +82-2-20722501; Fax: +82-2-20727690; E-mail: dslac.kr
 Global reach, higher impact
Global reach, higher impact