13.3
Impact Factor
Theranostics 2012; 2(2):139-147. doi:10.7150/thno.3696 This issue Cite
Review
Enzymatic Self-Assembly of Nanostructures for Theranostics
CAS Key Laboratory of Soft Matter Chemistry, Department of Chemistry, University of Science and Technology of China, 96 Jinzhai Road, Hefei, Anhui 230026, China
Received 2011-10-23; Accepted 2011-11-10; Published 2012-2-2
Abstract
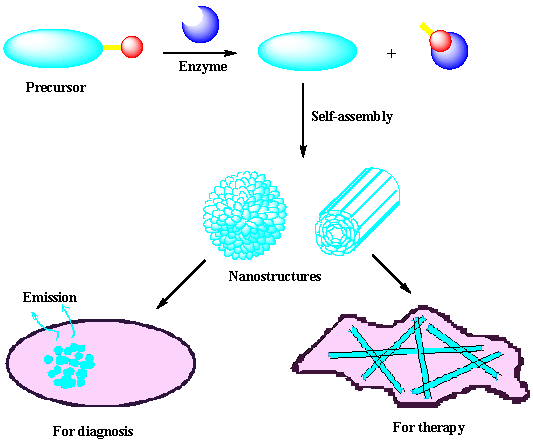
Self-assembly of small molecules or macromolecules through non-covalent or covalent bonds to build up supramolecular nanostructures is a prevalent and important process in nature. While most chemists use small molecules to assemble nanostructures with physical or chemical perturbations, nature adopts enzymes to catalyze the reaction to assemble biological, functional nanostructures with high efficiency and specificity. Although enzymatic self-assembly of nanostructures has been remained challenging for chemists, there are still a few examples of using important enzymes to initiate the self-assembly of nanostructures for diagnosis or therapy of certain diseases because down-regulation or overexpression of certain enzymes always associates with abnormalities of tissues/organs or diseases in living body. Herein, we introduce the concept of enzymatic self-assembly and illustrate the design and application of enzyme-catalyzed or -regulated formation of nanostructures for theranostics.
Keywords: Enzyme, Self-assembly, Nanostructures, Diagnosis, Therapy, Theranostics.
Introduction
Enzymes are a class of proteins that exhibit high efficiency and specificity when catalyzing a myriad of reactions in cellular environment [1]. Self-assembly is a prevalent process in nature which plays important roles in biology from maintaining the integrity of cells to performing cellular functions and inducing abnormalities that cause diseases [2-6]. Compared with physical or conventional chemical perturbations (e.g., pH, temperature, ionic strength, ligand-receptor interactions, etc.) for initiating the self-assembly process, enzymatic regulation shows promising applications because it integrates the self-assembly with biological events [7-8]. Generally, enzymatic self-assembly of small molecules into nanostructures includes two fundamental steps: the bond cleavage or formation of the precursors by the action of enzymes, and the self-assembly of the amphiphilic product generated from the first step to yield three-dimensional fibrillar/spherical nanostructures via supramolecular interactions (π-π interactions, hydrogen bonding, and charge interactions among the molecules, among others). As the expression and dispersion of the so many kinds of enzymes vary in cells or in different parts at different states of a body, enzymatic self-assembly allows people to control the delivery, function, and response of functional nanostructures according to a specific biological condition or environment, which provides accurate information about the health condition of a body for diagnosis as well as therapy.
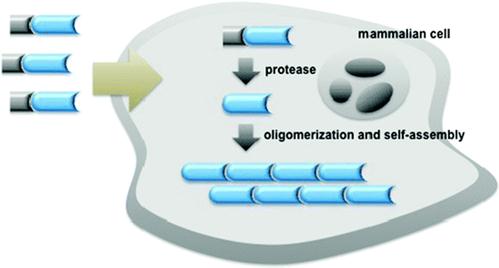
Figure 1 shows an enzymatic self-assembly process in cells. After entering the cell, the precursor with a substrate motif (normally a peptide sequence) for specific enzyme will be catalyzed by the enzyme (e.g., hydrolyzed by a protease) to yield the amphiphilic product which self-assembles into nanostructures inside cells. If the enzymatic product is not hydrophobic enough for self-assembly, Liang and co-workers designed another oligomerization reaction to generate the amphiphilic oligomers for self-assembly. This enzymatic polymerization and self-assembly can be used to trap an imaging agent (or drug) at the site of enzyme activity if optical properties (or therapeutic effect) of the new material are not strongly attenuated [9].
Nowadays, nanomaterials are widely used for theranostics [11-15]. Why nanostructures? An obvious source of inspiration for the design of nanoscale materials from the bottom up is provided by Nature. Life has produced a range of self-assembling nanoscale systems based on lipids, nucleic acids and amino acids. In these systems, disordered mixtures of molecular building blocks spontaneously arrange themselves to form highly organized structures with well defined properties. Numerous practical examples of self-assembly have long being existed in Nature, the 5-6-nm-thick cell membrane to maintain the integrity of a cell, the 2-nm-wide DNA double stranded chains to store genetic information, and the 25-nm-wide microtubules to govern cellular mitosis, to name just a few [4]. Their small molecular precursors share some common features, such as amphiphilicity and supramolecular interactions, which provide us with directions on precursor design.
The specificity of enzyme determines that one certain enzyme can only be functional towards one substrate or a class of compounds which share a similar structure. A vast array of natural enzymes are known, most catalyze a defined reaction but the degree of specificity to their substrate varies from highly specific to non-specific. Enzymatic self-assembly, therefore, calls for rational design of a specific structured substrate to be recognized by the certain enzyme, either the same as its natural substrate or natural substrates derivatives. After enzyme catalyzing the synthesis of a molecule, or removing a blocking group from a molecule, the as-formed intermediates should have the tendency to aggregate and form more stable nanostructures. Peptides have been exploited the most as building blocks for self-assembling nanomaterials. Table 1 gives several enzymes and their corresponding substrates that have been explored so far for experimental and clinical applications.
Schematic illustration of enzyme-controlled polymerization and/or self-assembly of nanostructures in cells [9-10].
Examples of enzymes, corresponding substrates, and potential applications of enzymatic self-assembly of nanostructures.
| enzyme(s) | substrate | applications |
|---|---|---|
| alkaline phosphatase | Fmoc-Y-P(O)(OH)2 | biomineralization [16] |
| acid phosphatase | Fmoc-Y-P(O)(OH)2 | inhibitor screening [17] |
| phosphatase | Nap-βPhg-βPhg-Y-P(O)(OH)2 | drug delivery [18] |
| kinase/phosphatase switch | Nap-FFGEY-P(O)(OH)2 | drug delivery [19] |
| β-lactamase | Nap-FF-NH(CH2)2S-ACLH | assay bacteria [20] |
| thermolysin | Fmoc-F and -FF | tissue engineering [21-22] |
| esterase | Nap-FF-NH(CH2)2OCO(CH2)2CO2H | control the fate of cells [23] |
| human tyrosine phosphatase | Nap-FFY-P(O)(OH)2 | bacterial inhibition [7] |
| caspase-3 | Ac-DEVDC-CBT | cell apoptosis [9] |
| furin | Ac-RVRRC-CBT | breast cancer [24] |
Regarding enzymatically controlled self assembling system based on peptides, two general categories are available, natural and unnatural. The former one utilizes the basic conformational units of naturally existence: proteins, β-sheets and turns, α-helices and coiled coils. The examination of protein sequences has made it possible for researchers to explore basic rules that promote the formation of one of the basic conformational units, which helps to create a sequence of the peptides. The latter category usually covalently links amino acids to other molecules, either an alkyl chain to form a peptide amphiphile, or to an aromatic group to create π-π interactions between the aromatic groups [25]. Design of other systems based on lipids, which primarily act as barriers, or polynucleic acid for information storage and retrieval mechanisms has also been explored [26-27].
Enzymatic self-assembly of nanostructures for diagnosis
Disease often comes along with abnormal expression of some enzymes, and the abnormality happens before the symptoms emerge. Since enzymatic self-assembly could assist people to examine the existence, content, and distribution of enzymes, which have close relationship with pathological changes, it has great potential to be applied to clinical diagnosis. Some recent examples of examining the irregular expression of enzymes are listed to illustrate the mechanisms for diagnosis.
1. Caspase 3-controlled self-assembly of nanoparticles for detecting cell apoptosis
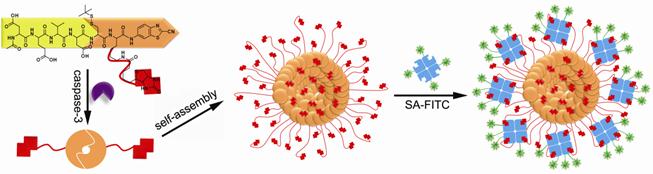
Many toxic agents, including most compounds used to treat cancer, induce apoptosis. Hence, factors that influence apoptotic cell death may contribute to the outcome of cancer therapy. Caspases are the executioners of cell apoptosis, a major form of cell death. Activation of caspases during apoptosis results in the cleavage of critical cellular substrates, including poly(ADP-ribose) polymerase and lamins, so precipitating the dramatic stereotypic morphological changes of cells [28]. Caspase-3, an important protease of caspase family, is tightly associated with cell apoptosis. Detection of caspase-3 activity provides people with an essay of assessing the chemotherapeutic effect of anti-cancer drug during the treatment [29-30]. A peptide sequence, DEVD, is a specific substrate for caspase-3 cleavage [31]. Inspired by this and employing a biocompatible condensation platform, Liang and co-workers designed a molecular probe for caspase-3 controlled self-assembling fluorescent nanoparticles to examine caspase-3 activity at low concentration [32]. As showed in figure 2, the probe Acetyl-Asp-Glu-Val-Asp-Cys(StBu)-Lys(Biotin)-CBT contains a DEVD substrate for caspase-3 cleavage, disulfided cysteine motif for supplying 1,2-aminothiol group for condensation, Lys(Biotin) for capturing FITC-labelled streptavidin (SA-FITC). After the reduction of its disulfide bond by tris(2-carboxyethyl)phosphine (TCEP) and removal of DEVD sequence by caspase-3 cleavage, the precursor yields an intermediate which condenses to form amphiphilic dimmers and subsequently self-assembles into biotinylated nanoparticles. The biotin arms standing on the surface of the nanoparticles can capture the FITC-labelled streptavidin (SA-FITC) via the specific interaction between biotin and streptavidin and turn on the fluorescence thereafter. Unlike other enzymatic self-assembly systems which need hundreds units of enzyme and millimolar substrates to launch the self-assembly, this protocol only requires several units of caspase-3 and micromolar substrates to self-assemble nanostructures (i.e., nanoparticles). In terms of caspase-3 detection on the other hand, this protocol provides people with a new method of sensing the caspase-3 activity at lower concentrations and may find its potential application for evaluating the in vivo chemotherapeutic effect of an anti-cancer agents [33].
A caspase-3 controlled condensation was applied to self-assemble biotinylated nanoparticles for capturing FITC-labelled streptavidin and subsequently turning on the fluorescence signal or on the other hand, detecting caspase-3 associated cell apoptosis.
2. Furin-controlled self-assembly of nanoparticles for early detection of cancer
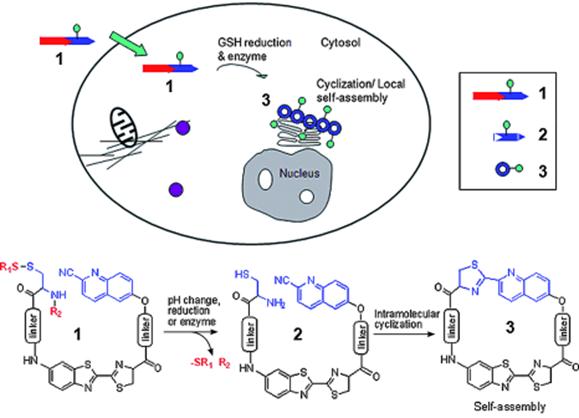
The trans-Golgi protease furin is a protein convertase that plays crucial roles in homeostasis, and in diseases ranging from anthrax and Ebola fever to Alzheimer's disease and cancer [31,34-36]. Furin is upregulated in several cancers, including non-small-cell lung carcinomas, squamous-cell carcinomas of the head and neck, and glioblastomas [37]. Moreover, the increase of furin in tumors correlates with an increase of membrane type 1 - matrix metalloproteinase (MT1- MMP), one of furin's substrates [38-39]. MT1-MMP activates extracellular pro-MMP2 to induce rapid tumor growth and metastasis [40]. Thus, the overexpression of furin offers people with a useful hint of early development of certain cancers. One big advantage for chemists to study furin is that furin preferentially cleaves Arg-X-Lys/Arg-Arg↓X motifs, where Arg is arginine, Lys is lysine, X can be any amino acid residue and ↓ indicates the cleavage site [41]. Inspired by this enzymatic reaction, Rao and co-workers designed a furin based macrocyclization reaction to self-assemble nanoparticles intracellularly for imaging furin activity in cells [24]. As demonstrated in figure 3, after entering a breast cancer cell MDA-MB-468 which overexpresses furin, the disulfide bond of a fluorescent probe 1 will be reduced by intracellular glutathione (GSH) and its Arg-Val-Arg-Arg motif will be cleaved by furin to yield intermediate 2. Then the 1, 2-aminothiol group on the cysteine motif and the cyano group on the 2-cyano-6-hydroxyquinoline motif (CHQ) of 2 will condense to initiate the intramolecular macrocyclization and yield amphiphilic product 3 which self-assembles into fluorescent nanoparticles at the location of furin. Using a fluorescence microscope to record this intracellular macrocyclization, Rao and co-workers successfully imaged furin activity in live cancer cells.
Proposed enzyme-controlled (e.g., furin) macrocyclization reaction in cells. A cell-permeable probe 1 enters cells and is converted by reduction and/or enzymatic processing into an intermediate 2, which quickly undergoes intramolecular cyclization to generate the macrocyclic product 3. The self-assembly of 3 leads to the formation of nanoparticles accumulated locally at or near the enzyme (e.g., furin) location in cells [24].
3. Enzyme-switch-regulated self-assembly of nanofibers for detecting the activities of kinase and phosphatase
Many diseases (e.g., cancer, diabetes, Alzheimer's disease, and multiple sclerosis) are associated with the abnormal activities of phosphatases and/or kinases [42-45]. Conventional protocols for detecting the activities of phosphatases or kinases are immunoassays. For immunoassays, to detect the activities of phosphatase and kinase as one pair at the same time is time consuming and instrumental dependent. Therefore, developing a simple and easy method for quick measurement of the activities of phosphatase and kinase in vitro and even in vivo has remained challenging for chemists. By carefully designing and numerous test, Xu and co-workers developed a self-assembly/disassembly system of supramolecular hydrogel (composed of nanofibers) based on phosphatase/kinase switch and used this system to detect the activities of phosphatase and kinase in vitro and in vivo [19]. As shown in figure 4, they designed a pentapeptidic hydrogelator, Nap-FFGEY (1), which self-assembles into nanofibers and forms hydrogels at 0.6 wt %.
Schematic illustration of a phosphatase/kinase-switch-regulated self-assembly/disassembly of supramolecular hydrogel composed of nanofibers. Upper: Phosphorylation of the hydrogelator 1 under the action of ATP and kinase yields 2 and induces a gel-solution phase transition. Dephosphorylation of 2 under the action of phosphatase generates 1 and launches the self-assembling of 1 into nanofibers and the formation of hydrogels. Lower: Chemical structures of 1 and 2.
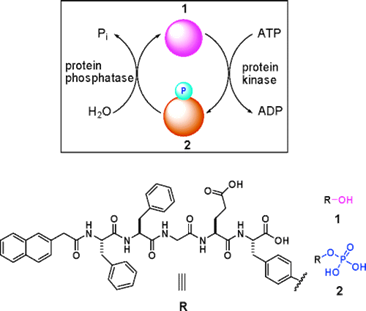
When a kinase is added to the hydrogel in the presence of adenosine triphosphates (ATP), the corresponding phosphate of 1 (i.e., 2) is generated, which disrupts the self-assembly to induce a gel-sol phase transition. Once the resulting solution is treated with a phosphatase, dephosphorylation of 2 yields the hydrogelator 1, thus restoring the self-assembly of 1 to form the hydrogel of nanofibers. By subcutaneously injecting the solution of 2 into mice, Xu and co-workers also successfully demonstrated this enzymatic conversion (i.e., from 2 to 1 under that action of phosphatase) and self-assembly of the enzymatic product to form nanofiber-based hydrogel in vivo.
Enzymatic self-assembly of nanostructures for therapy
The ultimate goal of scientific research is to serve us human-beings. Therefore, practical discoveries that can be applied to clinical treatment are much more valued. Up to date, a lot of precursors (or pro-drugs) based on enzymatic self-assembly have been developed to treat various kinds of diseases, especially cancers. The mechanisms of these disease treatments are either using enzymatic self-assembled nanostructures to disrupt the intracellular environment of the cytosol, or locally enhancing the concentration of a drug after enzymatic self-assembly, or self-assembling nanostructures as enzyme inhibitor. Nevertheless, they share a common feature that the self-assembly of nanostructures are under the catalysis of the over-expressed enzymes which correlate with the occurrence of a certain disease. Here are some examples listed below.
1. Esterase based self-assembly of nanofibers for regulating cell death
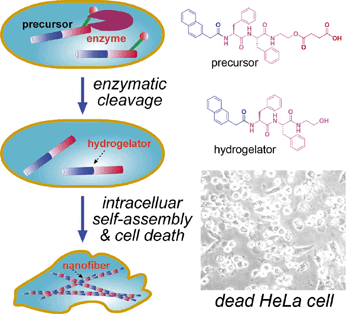
It was reported that some highly invasive melanoma cell lines (e.g., MUM-2B) secret high levels of active urokinase and esterase [46]. As mentioned above, intracellular artificial nanostructures can affect cellular behavior, and might induce cell death in the end. As shown in figure 5, Xu and co-workers designed a proper precursor which doesn't self-assemble extracellularly. After entering HeLa cells by perfusion, the precursor is converted into a hydrogelator by an endogenous enzyme (esterase in here) and self-assembles into nanofibers. The formation of nanofibers induces hydrogelation, which exerts stresses on the cell, and causes cell death [23]. To proof that the death of HeLa cells is caused by formation of intracellular nanofibers and hydrogelation, Xu and co-workers observed the intracellular nanofibers formed with the hydrogelator under a transmission electron microscope (TEM). Moreover, they incubated normal fibroblast cells NIH3T3 with the precursor at the same concentration and found that the precursor doesn't show any toxicity to the cells. Treating the two cell lines with an esterase indicator, Xu and co-workers found that the expression of esterase in HeLa cells is much higher than that in NIH3T3 cells, which proofs the death of HeLa cells is caused by intracellular esterase-controlled self-assembly of nanofibers and hydrogelation thereafter.
2. Phosphatase based self-assembly of taxol nanofibers for cancer therapy
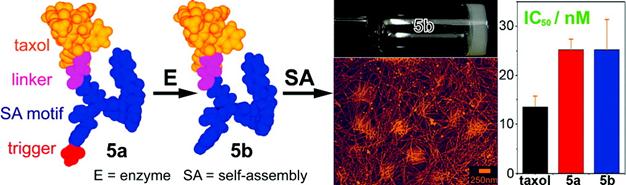
Taxol, an effective anti-cancer drug widely being used in clinic, is a well-established antineoplastic agent that binds specifically to the β-tubulin subunit of microtubules (MT) to arrest mitosis and result in programmed cell death (i.e., apoptosis) and has shown remarkable activity in the treatment of breast, lung, ovarian, bladder and head and neck cancers [47]. Because of its hydrophobicity, effective delivery of taxol to target tissues/organs without compromising its therapeutic effect has been remained challenging for pharmaceutics. Xu and co-workers developed an enzymatic self-assembly system to formulate this highly hydrophobic drug into an aqueous form (i.e., hydrogel) without comprising its activity for cancer therapy [48]. As shown in figure 6, they covalently connect taxol with a motif that is prone to self-assemble to generate the hydrophilic precursor (5a). Upon phosphatase cleavage, 5a changed into the hydrogelator (5b) which self-assembled into nanofibers without compromising the cytotoxicity of the taxol on HeLa cells. This approach promises a general method of enzymatic self-assembly to create nanofibers of therapeutic molecules that have a dual role, as both the delivery vehicle and the drug for chemotherapy.
Left: Schematic intracellular formation of nanofibers that leads to hydrogelation and cell death. Right upper: The chemical structures of the precursor and the hydrogelator. Right lower: Optical image of dead HeLa cells induced by intracellular self-assembly of nanofibers and hydrogelation.
Schematic illustration of the design of the precursor (taxol derivative, 5a), enzymatic cleavage of 5a by phosphatase to yield the hydrogelator (5b) which self-assembles into supramolecular hydrogel based on taxol nanofibers. Compared with that of taxol itself, both the precursor 5a and the hydrogelator 5b did not show compromised cytotoxicity on HeLa cells.
3. Thermolysin catalyzed self-assembly of nanofibers for immobilizing laminin to treat ECM diseases
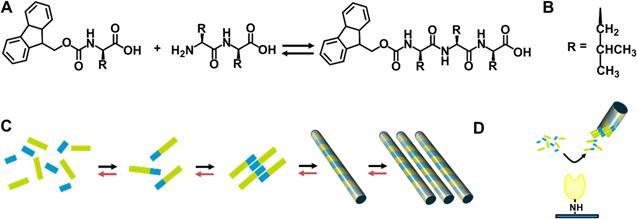
Genetic diseases of the extracellular matrix (ECM) have brought a considerable burden to our society. Most ECM diseases are identified as the result of a loss of function or a dominant negative effect of a single ECM protein mutated. A large number of ECM diseases including merosin deficient congenital muscular dystrophy (MDC1A) characterized by the symptom of loss of adhesion between cells are caused by mutations within the members of laminin gene family [49]. In order to treat ECM diseases, approaches including directly intramuscular delivery of free laminin to restore correct ECM function have been proposed [50-51]. However, this traditional approach is hampered by rapid transport of the injected free laminin away from the desired site of action, and incorporation into non-target tissues such as the microvasculature of the brain [52]. Williams and co-workers reported a new method of controlled delivery of laminin in an immobilized form [53]. As shown in figure 7, this system uses a protease enzyme (i.e., thermolysin) to act via reverse hydrolysis to produce fluorenylmethoxycarbonyl (fmoc) N-terminally derivatized tripeptide for self-assembling nanofibers. Entangled nanofibers at a large scale produce a clear hydrogel. This 3-dimensional network of nanofiber bundles enables the three dimensional distribution of laminins throughout the structure. They proposed that laminin binds with the residues of leucine on the nanofiber bundles via the formation of barrel-like structure rather than through simple electrostatic interactions. In order to study the stability and suitability of this type of material for in vivo application, they also utilized microinjection to deliver this material into a dystrophic zebrafish model which lacks laminin as a result of genetic mutation. Results indicated that the biomaterial remains structurally stable, and is confined spatially to the site of injection.
Thermolysin catalysed peptide Self-Assembly. A) Fmoc-amino acid reacts with a dipeptide in the presence of the enzyme, to yield the hydrogelator fmoc-tripeptide. B) The side chain, R, is Leucine. C) Schematic for the formation of a fmoc-tripeptide from disordered solution phase fmoc amino acids (blue) and dipeptide (green) to yield an fmoc-tripeptide building block. These interact though hydrophobic and hydrogen bonding interactions, yielding a nanoscale tubular structure. These align longitudinally, yielding a matrix which induces a gel/sol transition. The forward reaction (black arrows) and backward (red) take place to establish equilibrium. D) In the case of immobilized enzyme, localization of catalytic activity physically separating the assembled fmoc-tripeptide, preventing further reactions.
Conclusions
Enzymes, self-assembly, and nanostructures are probably the most common features of a cell and clearly associated with its behavior. Using enzymes to self-assemble nanostructures for diagnosis and therapy, we are just inspired by Nature and employ the knowledge of chemical biology to mimic the behavior of a cell and try our best to understand the mechanism of its function. Although a lot of methods have been successfully developed and utilized and some of them are explained here, there are still vast areas to be exploited considering the number of proteins in a single cell is up to one million. To get a better understanding of cellular behavior or a better theranostic outcome, people need multiple-enzyme or multiple-step involved self-assembly systems to build up more subtle, efficient probes/drugs for diagnosis/therapy, as Liang's and Xu's work listed in this paper. The features of these proposed probes/drugs are neither confined to the shapes as fiber or particle, nor sizes as nano-scale or micro-scale. They will have similar shapes and sizes as targeted proteins and simulate their behavior, participate or involve in the cell cycle during the development of a disease, and eventually treat the disease.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Walsh C. Enabling the chemistry of life. Nature. 2001;409:226-31
2. Whitesides GM, Mathias JP, Seto CT. Molecular self-assembly and nanochemistry - A chemical strategy for the synthesis of nanostructures. Science. 1991;254:1312-19
3. Timpl R, Brown JC. Supramolecular assembly of basement membranes. Bioessays. 1996;18:123-32
4. Schnur JM. Lipid tubules - A paradigm for molecularly engineered structures. Science. 1993;262:1669-76
5. Qi S, Groves JT, Chakraborty AK. Synaptic pattern formation during cellular recognition. Proc Natl Acad Sci U S A. 2001;98:6548-53
6. Scherzinger E, Sittler A, Schweiger K, Heiser V, Lurz R, Hasenbank R, Bates GP, Lehrach H, Wanker EE. Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: Implications for Huntington's disease pathology. Proc Natl Acad Sci U S A. 1999;96:4604-09
7. Yang Z, Liang G, Guo Z, Guo Z, Xu B. Intracellular hydrogelation of small molecules inhibits bacterial growth. Angew Chem Int Edit. 2007;46:8216-19
8. Spruell JM, Hawker CJ. Triggered structural and property changes in polymeric nanomaterials. Chem Sci. 2011;2:18-26
9. Liang G, Ren H, Rao J. A biocompatible condensation reaction for controlled assembly of nanostructures in living cells. Nat Chem. 2010;2:54-60
10. Razgulin A, Ma N, Rao J. Strategies for in vivo imaging of enzyme activity: an overview and recent advances. Chem Soc Rev. 2011;40:4186-216
11. Lee S, Xie J, Chen X. Peptides and peptide hormones for molecular imaging and disease diagnosis. Chem Rev. 2010;110:3087-111
12. Koo H, Huh MS, Ryu JH, Lee D-E, Sun I-C, Choi K, Kim K, Kwon IC. Nanoprobes for biomedical imaging in living systems. Nano Today. 2011;6:204-20
13. Ye Y, Chen X. Integrin targeting for tumor optical imaging. Theranostics. 2011;1:102-06
14. Jeong H, Huh M, Lee SJ, Koo H, Kwon IC, Jeong SY, Kim K. Photosensitizer-conjugated human serum albumin nanoparticles for effective photodynamic therapy. Theranostics. 2011;1:230-39
15. Bhirde A, Guo N, Chen X. Targeted nanoprobes reveal early time point kinetics in vivo by time-resolved MRI. Theranostics. 2011;1:274-76
16. Schnepp ZAC, Gonzalez-McQuire R, Mann S. Hybrid biocomposites based on calcium phosphate mineralization of self-assembled supramolecular hydrogels. Adv Mater. 2006;18:1869-72
17. Yang Z, Xu B. A simple visual assay based on small molecule hydrogels for detecting inhibitors of enzymes. Chem Commun. 2004:2424-25
18. Yang Z, Liang G, Ma M, Gao Y, Xu B. In vitro and in vivo enzymatic formation of supramolecular hydrogels based on self-assembled nanofibers of a beta-amino acid derivative. Small. 2007;3:558-62
19. Yang Z, Liang G, Wang L, Xu B. Using a kinase/phosphatase switch to regulate a supramolecular hydrogel and forming the supramoleclar hydrogel in vivo. J Am Chem Soc. 2006;128:3038-43
20. Yang Z, Ho P, Liang G, Chow K, Wang Q, Cao Y, Guo Z, Xu B. Using beta-lactamase to trigger supramolecular hydrogelation. J Am Chem Soc. 2007;129:266-67
21. Toledano S, Williams RJ, Jayawarna V, Ulijn RV. Enzyme-triggered self-assembly of peptide hydrogels via reversed hydrolysis. J Am Chem Soc. 2006;128:1070-71
22. de Loos M, Friggeri A, van Esch J, Kellogg RM, Feringa BL. Cyclohexane bis-urea compounds for the gelation of water and aqueous solutions. Org Biomol Chem. 2005;3:1631-39
23. Yang Z, Xu K, Guo Z, Guo Z, Xu B. Intracellular enzymatic formation of nanofibers results in hydrogelation and regulated cell death. Adv Mater. 2007;19:3152-56
24. Ye D, Liang G, Ma M, Rao J. Controlling intracellular macrocyclization for the imaging of protease activity. Angew Chem Int Edit. 2011;50:2275-79
25. Yang Z, Liang G, Xu B. Enzymatic hydrogelation of small molecules. Accounts Chem Res. 2008;41:315-26
26. Yang S, Zhang S. Self-assembling behavior of designer lipid-like peptides. Supramol Chem. 2006;18:389-96
27. Becker AL, Johnston APR, Caruso F. Peptide nucleic acid films and capsules: assembly and enzymatic degradation. Macromol Biosci. 2010;10:488-95
28. Cohen GM. Caspases: the executioners of apoptosis. Biochem J. 1997;326:1-16
29. Woo M, Hakem R, Soengas MS, Duncan GS, Shahinian A, Kagi D, Hakem A, McCurrach M, Khoo W, Kaufman SA, Senaldi G, Howard T, Lowe SW, Mak TW. Essential contribution of caspase 3 CPP32 to apoptosis and its associated nuclear changes. Genes Dev. 1998;12:806-19
30. Wang C, Guttridge DC, Mayo MW, Baldwin AS. NF-kappa B induces expression of the Bcl-2 homologue A1/Bfl-1 to preferentially suppress chemotherapy-induced apoptosis. Mol Cell Biol. 1999;19:5923-29
31. Talanian RV, Quinlan C, Trautz S, Hackett MC, Mankovich JA, Banach D, Ghayur T, Brady KD, Wong WW. Substrate specificities of caspase family proteases. J Biol Chem. 1997;272:9677-82
32. Cao C, Chen Y, Wu F, Deng Y, Liang G. Caspase-3 controlled assembly of nanoparticles for fluorescence turn on. Chem Commun. 2011;47:10320-22
33. Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Copper-free click chemistry for dynamic in vivo imaging. Proc Natl Acad Sci U S A. 2007;104:16793-97
34. Thomas G. Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3:753-66
35. Shapiro J, Sciaky N, Lee J, Bosshart H, Angeletti RH, Bonifacino JS. Localization of endogenous furin in cultured cell lines. J Histochem Cytochem. 1997;45:3-12
36. Dragulescu-Andrasi A, Liang G, Rao J. In vivo bioluminescence imaging of furin activity in breast cancer cells using bioluminogenic substrates. Bioconjugate Chem. 2009;20:1660-66
37. Mbikay M, Sirois F, Yao J, Seidah NG, Chretien M. Comparative analysis of expression of the proprotein convertases furin, PACE4, PC1 and PC2 in human lung tumours. Br J Cancer. 1997;75:1509-14
38. Bassi DE, Mahloogi H, Al-Saleem L, De Cicco RL, Ridge JA, Klein-Szanto AJP. Elevated furin expression in aggressive human head and neck tumors and tumors cell lines. Mol Carcinog. 2001;31:224-32
39. Bassi DE, De Cicco RL, Mahloogi H, Zucker S, Thomas G, Klein-Szanto AJP. Furin inhibition results in absent or decreased invasiveness and tumorigenicity of human cancer cells. Proc Natl Acad Sci U S A. 2001;98:10326-31
40. Sounni NE, Baramova EN, Munaut C, Maquoi E, Frankenne F, Foidart JM, Noel A. Expression of membrane type 1 matrix metalloproteinase (MT1-MMP) in A2058 melanoma cells is associated with MMP-2 activation and increased tumor growth and vascularization. Int J Cancer. 2002;98:23-28
41. Hosaka M, Nagahama M, Kim WS, Watanabe T, Hatsuzawa K, Ikemizu J, Murakami K, Nakayama K. Arg-X-Lys/Arg-Arg motif as a signal for precursor cleavage catalyzed by furin within the constitutive secretory pathway. J Biol Chem. 1991;266:12127-30
42. Saha S, Bardelli A, Buckhaults P, Velculescu VE, Rago C, St Croix B, Romans KE, Choti MA, Lengauer C, Kinzler KW, Vogelstein B. A phosphatase associated with metastasis of colorectal cancer. Science. 2001;294:1343-46
43. Hutton JC, Eisenbarth GS. A pancreatic beta-cell-specific homolog of glucose-6-phosphatase emerges as a major target of cell-mediated autoimmunity in diabetes. Proc Natl Acad Sci U S A. 2003;100:8626-28
44. Yuan J, Yankner BA. Apoptosis in the nervous system. Nature. 2000;407:802-09
45. Auch CJ, Saha RN, Sheikh FG, Liu X, Jacobs BL, Pahan K. Role of protein kinase R in double-stranded RNA-induced expression of nitric oxide synthase in human astroglia. FEBS Lett. 2004;563:223-28
46. Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc Natl Acad Sci U S A. 2002;99:10335-40
47. Rowinsky EK, Donehower RC. Drug-therapy - paclitaxel (taxol). N Engl J Med. 1995;332:1004-14
48. Gao Y, Kuang Y, Guo Z, Guo Z, Krauss IJ, Xu B. Enzyme-instructed molecular self-assembly confers nanofibers and a supramolecular hydrogel of taxol derivative. J Am Chem Soc. 2009;131:13576-77
49. Helblingleclerc A, Zhang X, Topaloglu H, Cruaud C, Tesson F, Weissenbach J, Tome FMS, Schwartz K, Fardeau M, Tryggvason K, Guicheney P. Mutations in the laminin alpha-2-chain gene (LAMA2) cause merosin-deficient congenital muscular-dystrophy. Nature Genet. 1995;11:216-18
50. Robbins PB, Sheu SM, Goodnough JB, Khavari PA. Impact of laminin 5 beta 3 gene versus protein replacement on gene expression patterns in junctional epidermolysis bullosa. Hum Gene Ther. 2001;12:1443-48
51. Igoucheva O, Kelly A, Uitto J, Alexeev V. Protein therapeutics for junctional epidermolysis bullosa: Incorporation of recombinant beta 3 chain into laminin 332 in beta 3-/- keratinocytes in vitro. J Invest Dermatol. 2008;128:1476-86
52. Rooney JE, Gurpur PB, Burkin DJ. Laminin-111 protein therapy prevents muscle disease in the mdx mouse model for Duchenne muscular dystrophy. Proc Natl Acad Sci U S A. 2009;106:7991-96
53. Williams RJ, Hall TE, Glattauer V, White J, Pasic PJ, Sorensen AB, Waddington L, McLean KM, Currie PD, Hartley PG. The in vivo performance of an enzyme-assisted self-assembled peptide/protein hydrogel. Biomaterials. 2011;32:5304-10
Author contact
![]() Corresponding author: email: gliangedu.cn
Corresponding author: email: gliangedu.cn
 Global reach, higher impact
Global reach, higher impact