13.3
Impact Factor
Theranostics 2012; 2(2):127-138. doi:10.7150/thno.3476 This issue Cite
Review
Analysis of Protease Activity Using Quantum Dots and Resonance Energy Transfer
Department of Life Science, Hanyang University, Seoul 133-791, Republic of Korea
Received 2011-9-5; Accepted 2011-10-4; Published 2012-2-1
Abstract
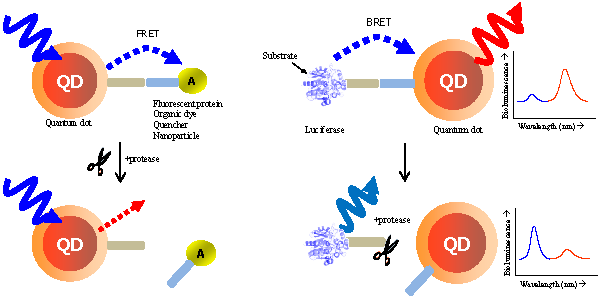
This review demonstrates the detection of protease activity based on the energy transfer of quantum dots (QDs). By incorporation of varying protease substrates into designed QD probes both in fluorescence resonance energy transfer (FRET) and bioluminescence resonance energy transfer (BRET) system, proteolytic activity led to changes in the energy transfer efficiency. Especially due to the superior properties of QDs, it can be served as an excellent probe for a multiplexed and high-throughput protease assay with high sensitivity. It is anticipated that the QD-based FRET/BRET probes will have a great potential for dissecting the fundamental roles of proteases and designing potential protease inhibitors as therapeutic drugs in biology and nanomedicine.
Keywords: protease, quantum dot, energy transfer, BRET, FRET, multiplex, nanoparticle, luciferase, quencher
1. Introduction
The ability of enzymes to target the specific substrate in complex milieu is essential for understanding the fidelity of most biological functions. Among enzymes, proteases, owing to their ability to regulate the physiological functions of many proteins through initiating irreversible hydrolysis at the post-translational level, have been an increasing interest in major human diseases such as cancer, AIDS, inflammation and neurodegenerative diseases [1, 2]. To this end, methods to assay proteases and their inhibitors have been of great interest in diagnosis of protease-relevant diseases and development of potential drugs [3-10]. Over the past decade, for probing the abundance of proteases, most of attempts have relied on affinity-based methods such as immunoassay [11, 12], rather than the activity-based assay which can provide the more specificity for the diagnostic value and drug discovery [13, 14]. A number of approaches to assay protease activity have been reported traditionally based on liquid chromatography or gel electrophoresis [15, 16], but most of them are time-consuming and laborious.
With the recent advance of evaluating the proteolytic processes through genomic and proteomic approaches, short peptide substrates for many proteases have been identified, which could allow for a route to analyze and trace the activities of their matching proteases in a rapid and facile manner [17, 18]. Therefore, assaying proteolytic activity can be easily designed by combining such peptide substrates with appropriate reporters, such as fluorescence and bioluminescence [19, 20]. The most useful system to assay the protease is based on the principle of fluorescence resonance energy transfer (FRET) where two fluorophores (energy donor and acceptor) are typically attached to the ends of the substrate with a distance of less than 10 nm. FRET is a process in which energy is transferred nonradiatively from an exited state donor to a proximal ground state acceptor via resonant dipole-dipole interactions. While a peptide substrate is adjacent to both the donor and the acceptor, the activity of proteases gives rise to the change in FRET efficiency. As a result of the cleavage of the peptide substrate by the protease, the energy transfer is disrupted and the ratio of donor to acceptor fluorescence emission changes when both chromophores are fluorescent, or the fluorescent intensity increases when the acceptor is a quencher. Although popular methods to adapt this FRET-based principle for monitoring in vivo protease activity has been made by using genetically engineered fluorescence protein (typically CFP-YFP), the use of stable, genetically engineered cell lines expressing the CFP-YFP substrate is much tedious and the real analysis of energy transfer is often hindered by cross-talk and bleed-through between two fluorescent proteins with narrow Strokes shift. Thus, this system may not be suitable for monitoring the rapid turnover rate of many proteases.
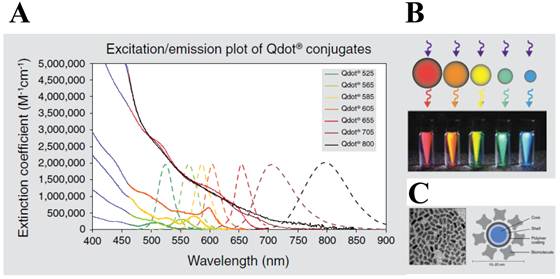
More recently, much attention has been given to the development of highly sensitive protease assay especially by merging numerous types of nanomaterials with improved detection properties. Among them, quantum dots (QDs) are the most attractable for biosensing because they have over-performed photophysical properties than those of organic dyes and fluorescent proteins. First, QDs typically have very broad absorption spectra with very large molar extinction coefficients of the order of 0.5-5×106 M−1 cm−1 [21, 22] at wavelengths extending from the ultraviolet to the visible (Figure 1). This means that QDs are able to absorb 10-50 times more photons than organic dyes at the same excitation photon flux, providing a sufficient brightness for the sensing system [23]. Second, size-tunable QDs are characterized by non-overlapping emission band profiles due to their narrow and symmetric emission bands (full width at half maximum of 25-40 nm) that can span the light spectrum from the ultraviolet to the infrared (400-1350 nm). This property enables the QDs to be useful for multiplexing assay in a single run [24-26]. For bioconjugation, QDs can be coated with organic molecules and macromolecules to provide aqueous solubility. These coatings can be broadly classified as ligand-based or polymer-based, and neutral or charged. Third, they possess an excellent photostability (thousand-fold longer than conventional organic fluorophores) [27], quantum yield (the ratio of emitted to absorbed photons) as high as 65−75% in most organometallic methods [28, 29] (but 20−25 % in the water-phase synthesis), and long fluorescence lifetimes (>10 ns) [30, 31]; thus, QD-based sensing and imaging have been successfully applied for continuous tracking studies over a long period of time. Therefore, QDs can provide a potential solution for RET system because the conventional organic dyes cause some technical problems (e.g. fast photobleaching and significant emission overlap between donor and acceptor, which complicate the development of robust and sensitive RET-based biosensors).
Characters of quantum dot (QD). (A) Excitation (solid line) and emission spectra (dotted line) of CdSe quantum dots. (B) Size tuneability and emission color of five different QDs with the same long-wavelength UV lamp. (C) Structure of a QD nanocrystal: transmission electron microscope image of core shell QD at 200,000x magnification (left) and schematic of the overall structure (right). Figures were obtained from Invitrogen website (www.invitrogen.com).
In FRET system, QDs not only showed reliable switches from a quenched state (off) to a fluorescent state (on) [32, 33] without the crosstalk between the donor QD and the acceptor counterpart due to their large Stokes shift, but also contibuted to multiple binding of an energy acceptor, leading to the increase in overall energy transfer efficiency [34, 35]. Most significantly, the continuously tunable emissions that can be matched to any desired acceptor, makes it possible to use many fluorophores for multiplexed assay [36]. The quencher (orgarnic or metal [28, 37-40] substances) or emissive fluorescent molecules (fluorophores, proteins, or other QDs) [41] can be promising acceptors. Although QDs are primarily used as donors in FRET, they may also play a critical role as energy acceptors in bioluminescence resonance energy transfer (BRET), with a bioluminescent protein as the energy donor. BRET resembles FRET in many aspects except that it does not require external light source for the donor excitation. The broad excitation spectra and large Strokes shift of QDs allow them to be excited by nearly all the bioluminescent proteins in BRET constructs. Feasibility of QDs as the BRET acceptor for a mutant of Renilla luciferase (Luc8 with improved chemical stability and light efficiency) has been recently realized both in vitro and in vivo [42, 43]. This capability becomes even more attractive for imaging for protease activity which could not be easily detected by other organic dyes.
Extensive reviews on QD-RET can be found in recent references [44-56], we will focus on the QD and energy transfer-based protease detection to avoid redundancy in a myriad of applications of QDs. While much emphasis in recent years has been placed on “enzyme activity” in drug discovery, such QD-based RET systems offer a highly sensitive and real-time monitoring strategy. Much attention to utilize the QD system in proteolytic study for biotechnology and nanomedicine applications is still on the way. A short overview regarding QD-FRET, QD-BRET, and multiplexed assay system is followed by some highlights of broad applications for the detection of protease activity. Most importantly, promising properties of QD-based probes, alternative to organic dyes for fluorescence-based applications, is discussed in this review.
2. Analysis of protease activity using QD-FRET
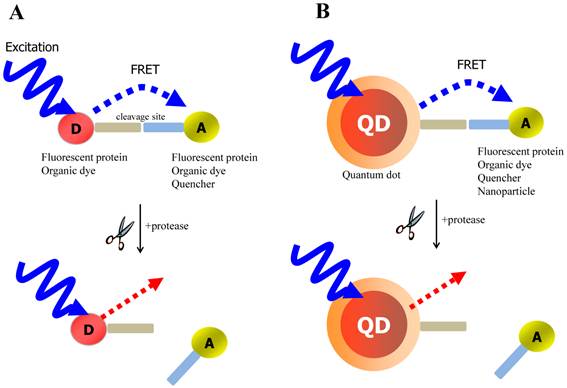
FRET is the most commonly utilized technique in these applications because of the high sensitivity, good reproducibility, and real-time monitoring capabilities. Conventional methods to detect protease activity are based on peptide-linked organic fluorophores. Examples for protease activity are auto-quenched probes [57, 58], dual chromophore probes [59], and multiphoton FRET-based probes [60], as depicted in Figure 2A. However, such organic fluorophores often have problems such as photobleaching, susceptibility to environment, difficulty in multiplexed analysis by specific paring between donor and acceptor. As aforementioned, these problems in FRET assays can be overcome when appropriate fluorophore or quencher is used in conjunction with quantum dots (QDs)[61, 62] (Figure 2B). Since QDs have broad absorption and narrow emission spectra as well as high photostability, they are favorable in long-term monitoring and multiplexed detection, where QDs are typically used as fluorescence donors while the fluorescent (or quenchable) acceptor is often an appropriately labeled peptide.
In order to detect protease activity, energy acceptor groups (organic fluorophore or quencher [40, 61-63]) were first bound to the surface of a QD through a peptide sequence. This close proximity allows for quenching of the QD emission via FRET, while subsequent cleavage of the peptide sequence by protease activity leads to a recovery of the QD fluorescence. A recent study by Shi et al. has shown a ratiometric QD-FRET probe to measure protease activity in vivo, consisting of the donor QD and the acceptor rhodamine labeled peptide [62, 64]. Local excitation of the QDs is able to induce efficient energy transfer into the adjacent rhodamine dye. Approximately 48 numbers of a rhodamine dye labeled substrate (RGDC) for collagenase were conjugated on a single QD through a sulfhydryl group of cysteine residue. When the probes were first tested for trypsin (500 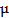 g/mL for 15 min) in solution, they gave rise to 60% increase in the photoluminescence of the QDs and a corresponding decrease in the emission of the rhodamine molecules, based on FRET signal changes [64]. Similarly, upon the cleavage by collagenase, fluorescence from QD at 545 nm was recovered up to 60% while that of rhodamine at 560 nm were diminished. Thus, ratiometric change in fluorescence emissions of QD and rhodamine allowed for the real-time detection and quantification of collagenase activity. The activity of collagenase, matrix metalloproteinases (MMPs) was monitored in normal (HTB 125) and cancerous breast cells (HTB 126) in which collagenase is negligible. This result clearly showed that QD-dye FRET sensors could be very useful to detect protease activity by measuring the ratiometric change.
g/mL for 15 min) in solution, they gave rise to 60% increase in the photoluminescence of the QDs and a corresponding decrease in the emission of the rhodamine molecules, based on FRET signal changes [64]. Similarly, upon the cleavage by collagenase, fluorescence from QD at 545 nm was recovered up to 60% while that of rhodamine at 560 nm were diminished. Thus, ratiometric change in fluorescence emissions of QD and rhodamine allowed for the real-time detection and quantification of collagenase activity. The activity of collagenase, matrix metalloproteinases (MMPs) was monitored in normal (HTB 125) and cancerous breast cells (HTB 126) in which collagenase is negligible. This result clearly showed that QD-dye FRET sensors could be very useful to detect protease activity by measuring the ratiometric change.
Schematic of FRET-based probe for the detection of protease activity. (A) conventional FRET (B) QD-based FRET. D and A indicate energy donor and energy acceptor, respectively.
Mattoussi group has reported the similar approach to measure the activities of different proteases by tuning the appropriate peptide substrate [61]. Modular peptides were rationally designed to have four different parts; i) an N-terminal hexahistidine (His6) domain for self-assembly with QD, (ii) a helix-linker spacer region, (iii) an exposed protease recognition/cleavage sequence, and (iv) a C-terminal site-specific location (cysteine thiol) for dye attachment. In particular, the artificial residue alpha-amino isobutyric acid (Aib) and alanine were incorporated into the helix linker to provide rigidity, and an organic fluorophore or a quencher was attached to the C-term via thiol chemistry. For the detection of caspase-1 activity, dihydrolipoic acid (DHLA)-capped QD538 and the Cy3 was employed as the FRET pair, and QD-FRET probes for three other proteases (thrombin, collagenase and chymotrypsin) were also designed to have the quencher (QXL-520) instead of Cy3. They described that the relative FRET efficiency within each QD nano-assembly can be controlled through the number of peptides assembled per QD. Therefore, these assays provide quantitative data for protolytic activity by measuring enzymatic velocity, Michaelis-Menten kinetic parameters, and mechanisms of enzymatic inhibition. Unlike the conventional protease assay format in which substrate concentration has to be used in the micromolar range, they demonstrated that the developed format can be consistently used with lower substrate concentrations (200nM QD and peptide concentration of 0.2-1.0 μM) over a diverse selection of enzymes that manifest different specific activities. Moreover, it is likely that the substrates incorporated on the QD complex allow easy access to the desired protease and provide the high sensitivity and low background by being easily tuned. More recently, they utilized this system to detect caspase 3 activity, a key downstream effector of apoptosis. Along with that, the presence of calcium ions with the acceptor CaRbCl can increase the FRET efficiency, enabling calcium-sensitizing sensors [65]. Also they have reported this QD-dye FRET system to serve as a papain or proteinase K sensor, based on tunable coupler, 520 QD and Cy3 [66]. In a similar way, Biswas et al. has showed that a genetically programmable protein module was designed to have a His6, a cleavage site labeled with a Alexa dye via cysteine residue, an elastin-like peptide (ELP) domain for purification, and a flanking TAT peptide [67]. This QD-dye FRET module was used for the detection of HIV-1 Pr activity in vitro and in cancer cells, which particularly takes responsibility for drug-resistance against rapidly mutating viruses such as HIV-1. Analysis of enzymatic inhibition was also performed in the presence of specific inhibitors.
QD-FRET assay system to measure protease activity has been applied to chip-based format by Kim et al [68]. While the photoluminescence (PL) of donor streptavidin-QD525 immobilized on a surface was quenched due to the presence of an energy acceptor (peptide substrates modified with TAMRA and biotin at N- and C-terminus, respectively) in close proximity, the protease activity caused modulation in the efficiency of the energy transfer between the acceptor and donor, thus enabling the highly sensitive detection of MMP-7 activity. In contrast to a solution-based analysis, the chip-based format allowed more reliable analysis, with no aggregation of QDs. Plus, this format required a much smaller reaction volume. This method is likely to have a potential to screen the activity of disease-associated proteases for the development of therapeutics and diagnostics in a high-throughput manner.
In addition to organic fluorophores, a fluorescent protein was easily designed as an energy acceptor against QD donor. Boeneman et al [69] has demonstrated that a red fluorescent protein (mCherry) expressing the caspase 3 cleavage site and a His6 sequence were self-assembled to the surface of CdSe-ZnS DHLA QDs via metal affinity coordination, leading to FRET quenching of the QD and sensitized emission from the mCherry acceptor. Caspase-3 activity caused the FRET efficiency to be reduced. Owing to the favorable spectral overlap (Förster distance R0=4.9 nm) between QD550 and mCherry, considerable loss in QD PL was observed along with an increase in sensitized mCherry emission. A FRET efficiency of approximately 50% was measured when the number of mCherry per a single QD was six. Caspase 3-induced changes in FRET efficiency were comparable to those observed in fluorescent protein sensors. However, compared to two fluorescent proteins, some advantages of QD-fluorescent protein encompass 5-10 times less substrate and ~3 orders of magnitude less enzyme in terms of quantity to be used. As a result, they were able to detect enzymatic activity for caspase 3 concentrations as low as 20 pM. This capability seems to be mainly due to multivalent effect of QDs.
In order to construct QD-FRET probe, quenching groups (organic quenchers or metal nanoparticles) [61-63, 70] can be bound to the surface of a QD through a peptide sequence. Unlike QD-dye FRET system based on the ratiometry of dual emission, this close proximity causes only quenching of the QD emission via the resonance energy transfer, while subsequent cleavage of the peptide sequence by the corresponding protease led to a recovery of the QD fluorescence. The quenching ability of the gold nanoparticle (AuNP) has been known to be much higher than that of organic quenchers as described elsewhere [38]. As such, the use of AuNPs as energy acceptors enables the energy transfer to be valid even in the excess distance of the traditional FRET. One of the feasible mechanisms might be associated with the property of metal surface, termed nanosurface energy transfer (NSET); it was reported that the metal surface extended the effective energy transfer distance up to 22 nm (an inversely fourth power distance dependence), resulting in a high energy transfer efficiency [71-75]. Although both NSET and FRET rely on dipolar coupling, a longer distance dependence of NSET might be due to the cooperative effect of more accessible acceptor dipoles yielding more dipolar interactions. NSET, therefore is expected to have diverse applications for measuring distances in excess of 10 nm. Utilizing the NSET mechanism, the initial report of protease detection with a QD-AuNP system was made by Chang et al [63]. A peptide substrate, GGLGPAGGCG, was employed to measure the activity of collagenase. When the surface of QDs was capped with a layer of DHLA, mercaptosuccinic acid, or other carboxylic ligands, the N-terminal amines of the peptides were easily coupled to the carboxylic acids on the QDs by EDC, and the cysteine was conjugated to the maleimide functionalized gold nanoparticles. When the AuNP level was six per QD, a quenching efficiency of 71% was observed. The excess quenchers on the QD possibly decrease the probe sensitivity to protease hydrolysis even if they cause the quenching efficiency to increase. By the release of the gold quencher in the presence of collagenase (0.2 mg mL−1), QD fluorescence was recovered up to 51%, meaning that a number of enzyme molecules are not enough to be fully accessible between two large nanoparticles. This observation that multivalent effect of QDs may induce a low detection sensitivity of enzyme is not consistent with the previous study [69] where the increasing number of fluorescent proteins per single QD increase the sensitivity. This may be attributed to the different coupling format. The QD-FRET between QDs and fluorescent acceptors depends on the ratio change of donor-to-acceptor in fluorescent intensity for measuring the protease activity, while the QD-FRET using quenching acceptors relies on the single fluorescent intensity of the donor QD. Moreover, the recovery yield of QD-AuNP might be more complicated than that of QD-fluorescent protein because the released metal quenchers may significantly contribute to the QD emission in solution due to the random collision effect of metal surface, resulting in low detection sensitivity of tested enzymes (that is, low recovery efficiency from the quenching state). Therefore, further step to remove the unbound acceptors (especially in metal acceptors) is desirable to increase the enzyme sensitivity as seen in the chip-based format [68].
3. Analysis of protease activity using QD-BRET
Recent studies conducted by Rao group have demonstrated the feasibility of using QDs as the acceptor in a bioluminescence resonance energy transfer (BRET) system [42, 43]. In FRET, QDs are not able to function as effective acceptors [76] because a direct excitation of donor fluorophores has to accompany by unavoidable excitation of the QD with a broad absorption. However, when QDs can serve as an energy acceptor with a light-emitting protein, this problem can be overcome; since the bioluminescence energy of a luciferase-catalyzed reaction occurs only in its active site and cannot excite the acceptor QD, it can be successfully transferred to the QDs to produce quantum dot light emission. Upon addition of the luciferase substrate (coelenterazine) to QD-luciferase conjugate, a short blue light emission at 475 nm is transferred the QDs. Main advantage of QD-luciferase system is to eliminate the need for excitation light which causes inevitable background autofluorescence. In the case of QD-FRET that has been widely employed as activity-based probes, the excited illumination can partly increase the background noise level especially in serum sample, thus diminishing the value of acquired information as a result of the false-positive signal. Moreover, a common luciferase protein serves as the BRET donor for several QDs with different colors because QDs have similar absorption spectra and absorb blue light efficiently. Especially in contrast to QD-FRET system where the FRET efficiency improves as the number of FRET acceptors per QD increases, the BRET ratio with varying numbers of luciferase on the surface of single QD were quite similar although the intensity of both luciferase and QD emissions varied by approximately100-fold. This means that the QD-BRET system is more dependent on the donor-to-acceptor distance rather than varying donor number, thus being applied to detect the distance-dependent assay with high fidelity. In Figure 3, this QD-BRET based probe can be applied to the detection of protease activity.
Schematic of QD-BRET sensor to detect protease activity. QD and luciferase protein are closely linked together through a peptide substrate. In contrast to a quiescent state in the absence of protease (A), an active state (B) of this probe shows that protease cleavage removes the donor protein from QD and disrupts BRET, leading to a loss of bioluminescence-induced QD emission (red line).
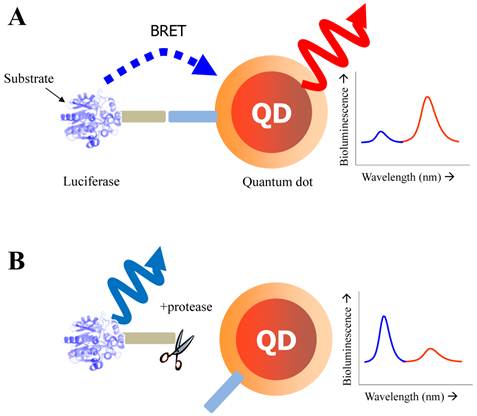
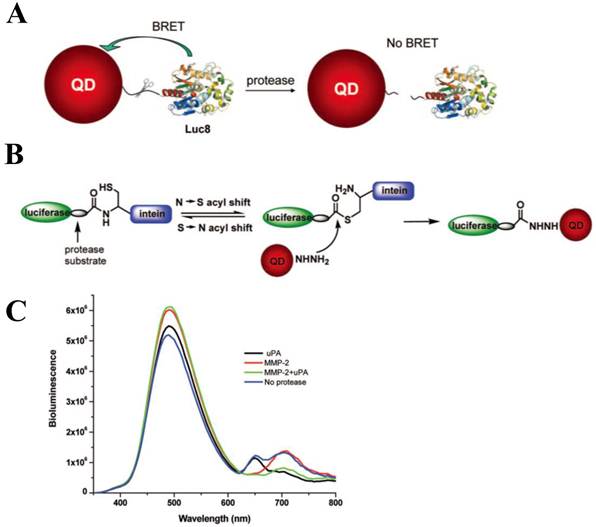
Yao et al. has focused on the detection of the activity of MMPs that is a promising cancer biomarker enzyme. Since the secretion level of MMPs in human serum is of very interest as a promising prognostic marker [77, 78] due to the up-regulation of MMPs in almost all human cancerous cells or in malignant tissues, the designed probe to measure MMP activity and related inhibitory effect is valuable for discovering drug candidates for anticancer therapeutics [79-81]. For the construction of QD-BRET energy donor, a bioluminescent protein fused to the MMP-2 substrate (GGPLGVR) and a hexahistidine tag at its C terminus was genetically expressed in E. coli. The bioluminescent protein is a Renilla reniformis luciferase mutant dubbed Luc8, which has eight mutations and shows higher stability and improved catalytic efficiency than the wild-type luciferase [82]. Since the simultaneous coordination is generated between carboxyl groups on the QD surface and the Luc8 His tag in the presence of Ni2+, a strong BRET signal was observed just immediately after luciferase-GGPLGVRGGH6 is mixed with carboxyl QD655 and Ni2+. Upon cleavage of the flanking peptide region by MMP-2, the His tag was released from the fusion Luc8 and the BRET signal decreased. In comparison to FRET-based QD sensors, BRET based QD biosensors have several attractive features. Most significantly, the large spectral separation between the BRET donor and acceptor emissions makes it easy to detect both emissions. In this case, a ratiometric measurement is far more accurate and reliable than that of QD-FRET because the integrated intensities from two separable spectra was favorable to reflect the energy-transferred efficiency [43]. In addition, considering that the BRET ratio for a concentration of MMP-2 of 2 ng mL-1 (~30 pM) decreased by about 10%, and by 15% for a concentration of MMP-2 of 5 ng mL-1 (~75 pM), the sensitivity is high probably due to a low background emission. However, since the overall sensitivity of protease assays is determined by several factors such as the peptidyl substrate, the particular protease of interest, the conditions (e.g. pH, divalent cations), the RET efficiency, the acceptor quantum yield (for ratiometric sensing), and the background signal magnitude, the sensing performance should be further evaluated by comparing other contributing factors. Main drawback of this system is that it cannot be used in complex biological media such as serum because of widespread existence of chelate complex that might affect the enzyme sensitivity as well as the electrostatic interaction of designed nanosensor. The later issue (electrostatic inhibition in serum) can be overcome with site-specific conjugation. Recently, a modified QD-BRET nanosensor with a stable covalent linkage has been developed by Xia et al., enabling the detection of protease activity in mouse sera and tumor lysates [83]. Once the QD-luciferase complex is premade with high affinity, the protease activity can be detected by dissociation of the linkage. To accomplish the site-specific conjugation, the carboxylated QDs had been functionalized with adipic dihydrazide and subsequently treated with the luciferase-protease substrate recombinant protein with an additional intein segment. Since hydrazides are excellent nucleophiles to attack the thioester intermediate of inteins that are natural protein ligation mediators. The reaction proceeded rapidly when the two components were mixed together, and resulted in cleavage of the intein and ligation of the C terminus of the recombinant protein to the QDs.
Notwithstanding the crucial role of proteases in cellular function, the prognostic attempts have much been interrupted in serum level mainly because many interferences for activity-based detection exist. By employing this strategy, a high level of sensitivity can be attained in a concentration range that would enable protease activity to be detected in complex biological media. Most significantly, this approach by using QD-BRET can provide a new family of activity-based assays for enzymes and their inhibition in a high-throughput manner.
4. Multiplexed analysis of protease activity
Highly rapid and sensitive detection in a multiplexed manner has been of great interest to study clinical diagnostics, drug screening, and bio-imaging applications. The ability to identify multiple analytes in a single assay format has many advantages over single-target systems; high-throughput screening, low consumption of reagents and samples, and decrease in errors between inter-sampling. Although many of powerful analytical methods are now available, fluorescent probes have long been used to characterize molecular interactions for the multiplexed purpose, due to their chemical stability and sensitivity [84]. However, the narrow absorption and large cross-talk of organic dyes and fluorescent proteins is a major hurdle for simultaneous detection of multiple signals, because of the requirement of an elaborate excitation and the complicated analysis of collected data. Moreover, low resistance of fluorescent probes to chemical and photodegradation can significantly limit their use in multiplexed monitoring. Since QD nanocrystals can be simultaneously excited at a single excitation wavelength, multicolored QDs make it possible to achieve simultaneous monitoring of biomolecules with high level of sensitivity and excellent photostability.
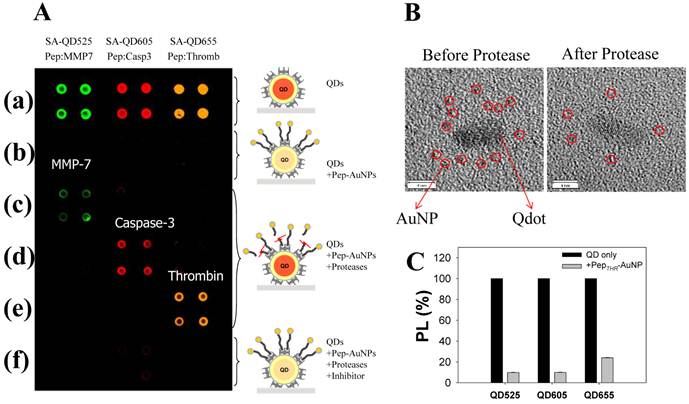
A multiplexed system to detect the activity and inhibitory effects of several proteases (MMPs, thrombin, and caspase-3) has been proposed by Kim et al, which is utilized by the principle of energy transfer between the AuNP and respective QDs on a glass slide [40]. For construction of nanoprobes, The AuNP acceptors conjugated with a peptide substrate including cysteine and biotin were associated with streptavidin (SA)-conjugated QDs (SA-QDs, energy donor) deposited on a glass slide, thus quenching the PL of the QD by the energy transfer. Upon addition of a protease to cleave the peptide substrate on the AuNP-QD conjugates, there was a significant regeneration of the photoluminescence emission of the QDs. Protease inhibitors also prevented any recovery of the photoluminescence of QDs by inhibiting the protease activity. When three types of SA-QD (SA-QD525, SAQD605, and SA-QD655) were independently complexed with the AuNPs with different peptide substrates, a specific reaction of the protease induced a strong photoluminescence intensity from each spot, at a specified wavelength (Figure 4A and 4B). Marginal cross-reactional images of the protease against other peptide substrates were observed, thus confirming the multiplexed capability of this assay system. Since the AuNPs can be employed as a common energy acceptor, a variety of QDs with different colors could be used as the energy donor (Figure 4C), thus enabling a multiplexed assay. Moreover, high quenching efficiency of AuNPs allows application of the assay system to an extended separation distance between a donor and an acceptor. This developed system also overcome some drawbacks resulting from a solution-based format, including the aggregation of nanoparticles and the fluctuation in photoluminescence, and the consumption of large amounts of reagents.
Development of multiplexed system to detect protease activity with QDs. (A) Multiplexed assay of proteases by using QDs with different colors. SA-QD525, SA-QD605, and SA-QD655 were used (from left to right). Biotinylated peptide substrates for MMP-7, caspase-3, and thrombin were conjugated to the AuNPs, and then the resulting Pep-AuNPs were associated with SA-QD525, SA-QD605, and SA-QD655, respectively. (a) SA-QDs only. (b) SA-QDs + respective Pep-AuNPs. (c) SA-QDs + Pep-AuNPs + MMP-7. (d) SA-QDs + Pep-AuNPs + caspase-3, (e) SA-QDs + Pep-AuNPs + thrombin. (f) QDs + Pep-AuNPs + mixture of the respective protease and its inhibitor. (B) High-resolution TEM images of QD-PepMMP7-AuNP assembly (left) before and (right) after incubation with MMP-7. The released AuNPs in enzyme-treated samples were removed by filtration. (C) Changes in the PL intensities of QDs having different colors (SA-QD525, SA-QD605, and SA-QD655) in the presences of biotinylated PepTHR-AuNPs in solution. The molar ratios of Pep-AuNPs to respective QDs were equally maintained at 50. Adapted with permission from [40].
This multiplexed assay using a QD-FRET nanosensor has also been applied to different types of enzymes other than proteases. Suzuki et al [84]. designed QD-based nanoprobes on a FRET with QD as donor and an appended fluorophore as acceptor in order to detect multiple cellular signaling events, including the activities of protease (trypsin), deoxyribonuclease, and DNA polymerase, as well as changes in the pH. Notably, this mixture of modified QDs showed distinct changes in emission peaks before and after enzyme treatment by simultaneous-wavelength excitation in the same tube.
Using a QD-BRET nanosensor which provides great sensitivity due to its extremely low background, the simultaneous detection of MMP-2, MMP-7, and urokinase-type plasminogen activator (uPA) has been demonstrated [83] (Figure 5A). Hydrazide and intein-mediated conjugation makes the donor (luciferase) and acceptor (QD) to be assembled (Figure 5B), and the conjugated nanosensors were capable of detecting these proteases in complex biological media with a sensitivity down to ~1 ng mL−1. In addition, they could also detect multiple proteases present in one sample (Figure 5C). The most significant point here is that the BRET ratio (i.e. the ratio of donor and acceptor emissions) between the QD and the luciferase was modulated only by the degree of the protease activity, thereby making it more reliable and sensitive in comparison with other assays. As mentioned earlier, elimination of physical light excitation resulted in high sensitivity of this QD-BRET system and makes it work in complex biological media including serum and tumor samples in a multiplexed way.
Development of QD-BRET nanosensor to detect protease activity. (A) Schematic of the nanosensor comprising of a QD and luciferase proteins (Luc8) that are linked to the QD through an MMP peptide substrate and (B) intein-mediated site-specific conjugation of Luc8 fusion proteins to QDs. (C) Simultaneous detection of MMP-2 and uPA: A mixture of QD655-MMP-2-Luc8 (0.15 μM) and QD705-uPA-Luc8 (0.2 μM) were incubated with MMP-2 (1 μg/mL, red); uPA (10 μg/mL, black); MMP-2 (1 μg/mL) + uPA (10 μg/mL) (green); or no enzyme (blue) at room temperature for 1 h in 20 mM Tris buffer (pH 7.5). Adapted with permission from [83].
5. Conclusion
In this review we demonstrate that there have been several technological advances that are likely to make important contributions to sensing protease activity. Improved analysis of protease activity is now paving the way of understanding the physiological roles of many proteases in complex living system. Also it is noteworthy that probing the protease activity is implicated in early detection of both primary tumors and some diseases that remains a major challenge. Hence, newly developed sensing and imaging system using QDs will be tremendously applied to the areas that require the rapid analysis and high sensitivity. However, some limitations regarding specificity and imaging resolution in vivo still remain formidable. For example, temporal and spatial localization of protease activity with its specific substrate should be required in future studies to dissect the roles of many proteases. It is anticipated that the integration of biological information with improved nanosensors and toolkits will allow us to track protease-specific activity over the long period. Moreover, by combining the QD-RET with protease-modulated sensors, therapeutic approaches (e.g. siRNA delivery and photodynamic therapy) will have a great potential in biomedicine and nanomedicine.
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2011-0022757).
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Puente XS, Sanchez LM, Overall CM, Lopez-Otin C. Human and mouse proteases: a comparative genomic approach. Nat Rev Genet. 2003;4:544-58
2. Overall CM, Tam EM, Kappelhoff R, Connor A, Ewart T, Morrison CJ. et al. Protease degradomics: mass spectrometry discovery of protease substrates and the CLIP-CHIP, a dedicated DNA microarray of all human proteases and inhibitors. Biol Chem. 2004;385:493-504
3. Neurath H. The versatility of proteolytic enzymes. J Cell Biochem. 1986;32:35-49
4. Lopez-Otin C, Overall CM. Protease degradomics: a new challenge for proteomics. Nat Rev Mol Cell Biol. 2002;3:509-19
5. Turk B. Targeting proteases: successes, failures and future prospects. Nat Rev Drug Discov. 2006;5:785-99
6. Mason SD, Joyce JA. Proteolytic networks in cancer. Trends Cell Biol. 2011;21:228-37
7. Overall CM, Kleifeld O. Tumour microenvironment - opinion: validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat Rev Cancer. 2006;6:227-39
8. Concha NO, Abdel-Meguid SS. Controlling apoptosis by inhibition of caspases. Curr Med Chem. 2002;9:713-26
9. Schwienhorst A. Direct thrombin inhibitors - a survey of recent developments. Cell Mol Life Sci. 2006;63:2773-91
10. White CM. Thrombin-directed inhibitors: pharmacology and clinical use. Am Heart J. 2005;149:S54-60
11. Laack E, Kohler A, Kugler C, Dierlamm T, Knuffmann C, Vohwinkel G. et al. Pretreatment serum levels of matrix metalloproteinase-9 and vascular endothelial growth factor in non-small-cell lung cancer. Ann Oncol. 2002;13:1550-7
12. Ohuchi E, Azumano I, Yoshida S, Iwata K, Okada Y. A one-step sandwich enzyme immunoassay for human matrix metalloproteinase 7 (matrilysin) using monoclonal antibodies. Clin Chim Acta. 1996;244:181-98
13. Srinivasan R, Huang X, Ng SL, Yao SQ. Activity-based fingerprinting of proteases. Chembiochem. 2006;7:32-6
14. Sun H, Chattopadhaya S, Wang J, Yao SQ. Recent developments in microarray-based enzyme assays: from functional annotation to substrate/inhibitor fingerprinting. Anal Bioanal Chem. 2006;386:416-26
15. Bjurlin MA, Bloomer S, Nelson CJ. Characterization of proteolytic activity of protease. Biotechnol Lett. 2002;24:191-5
16. Zhao Z, Raftery MJ, Niu XM, Daja MM, Russell PJ. Application of in-gel protease assay in a biological sample: characterization and identification of urokinase-type plasminogen activator (uPA) in secreted proteins from a prostate cancer cell line PC-3. Electrophoresis. 2004;25:1142-8
17. Schilling O, Overall CM. Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat Biotechnol. 2008;26:685-94
18. Thomas DA, Francis P, Smith C, Ratcliffe S, Ede NJ, Kay C. et al. A broad-spectrum fluorescence-based peptide library for the rapid identification of protease substrates. Proteomics. 2006;6:2112-20
19. Nagai T, Miyawaki A. A high-throughput method for development of FRET-based indicators for proteolysis. Biochem Bioph Res Co. 2004;319:72-7
20. Mahajan NP, Harrison-Shostak DC, Michaux J, Herman B. Novel mutant green fluorescent protein protease substrates reveal the activation of specific caspases during apoptosis. Chem Biol. 1999;6:401-9
21. Hawrylak P, Narvaez GA, Bayer M, Forchel A. Excitonic absorption in a quantum Dot. Phys Rev Lett. 2000;85:389-92
22. Moreels I, Hens Z. On the interpretation of colloidal quantum-dot absorption spectra. Small. 2008;4:1866-8
23. Gao X, Cui Y, Levenson RM, Chung LW, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol. 2004;22:969-76
24. Chan WCW, Maxwell DJ, Gao XH, Bailey RE, Han MY, Nie SM. Luminescent quantum dots for multiplexed biological detection and imaging. Curr Opin Biotech. 2002;13:40-6
25. Wu XY, Liu HJ, Liu JQ, Haley KN, Treadway JA, Larson JP. et al. Immunofluorescent labeling of cancer marker Her2 and other cellular targets with semiconductor quantum dots. Nat Biotechnol. 2002;21:41-6
26. Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nat Biotechnol. 2003;21:47-51
27. Bruchez M Jr, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013-6
28. Hines MA, Guyot-Sionnest P. Synthesis and characterization of strongly luminescing ZnS-Capped CdSe nanocrystals. J Phys Chem. 1996;100:468-71
29. Yu JS, Liu YF. Selective synthesis of CdTe and high luminescence CdTe/CdS quantum dots: The effect of ligands. J Colloid Interf Sci. 2009;333:690-8
30. Michalet X, Pinaud F, Lacoste TD, Dahan M, Bruchez MP, Alivisatos AP. et al. Properties of fluorescent semiconductor nanocrystals and their application to biological labeling. Single Mol. 2001;2:261-76
31. Lounis B, Bechtel HA, Gerion D, Alivisatos P, Moerner WE. Photon antibunching in single CdSe/ZnS quantum dot fluorescence. Chem Phys Lett. 2000;329:399-404
32. Tran PT, Goldman ER, Anderson GP, Mauro JM, Mattoussi H. Use of luminescent CdSe-ZnS nanocrystal bioconjugates in quantum dot-based nanosensors. Phys Status Solidi B. 2002;229:427-32
33. Willard DM, Van Orden A. Quantum dots - Resonant energy-transfer sensor. Nat Mater. 2003;2:575-6
34. Clapp AR, Medintz IL, Mauro JM, Fisher BR, Bawendi MG, Mattoussi H. Fluorescence resonance energy transfer between quantum dot donors and dye-labeled protein acceptors. J Am Chem Soc. 2004;126:301-10
35. Zhang CY, Yeh HC, Kuroki MT, Wang TH. Single-quantum-dot-based DNA nanosensor. Nat Mater. 2005;4:826-31
36. Mauro JM, Medintz IL, Clapp AR, Mattoussi H, Goldman ER, Fisher B. Self-assembled nanoscale biosensors based on quantum dot FRET donors. Nat Mater. 2003;2:630-8
37. Oh E, Hong MY, Lee D, Nam SH, Yoon HC, Kim HS. Inhibition assay of biomolecules based on fluorescence resonance energy transfer (FRET) between quantum dots and gold nanoparticles. J Am Chem Soc. 2005;127:3270-1
38. Oh E, Lee D, Kim YP, Cha SY, Oh DB, Kang HA. et al. Nanoparticle-based energy transfer for rapid and simple detection of protein glycosylation. Angew Chem Int Ed. 2006;45:7959-63
39. Wargnier R, Baranov AV, Maslov VG, Stsiapura V, Artemyev M, Pluot M. et al. Energy transfer in aqueous solutions of oppositely charged CdSe/ZnS core/shell quantum dots and in quantum dot-nanogold assemblies. Nano Lett. 2004;4:451-7
40. Kim YP, Oh YH, Oh E, Ko S, Han MK, Kim HS. Energy transfer-based multiplexed assay of proteases by using gold nanoparticle and quantum dot conjugates on a surface. Anal Chem. 2008;80:4634-41
41. Wang SP, Mamedova N, Kotov NA, Chen W, Studer J. Antigen/antibody immunocomplex from CdTe nanoparticle bioconjugates. Nano Lett. 2002;2:817-22
42. So MK, Xu C, Loening AM, Gambhir SS, Rao J. Self-illuminating quantum dot conjugates for in vivo imaging. Nat Biotechnol. 2006;24:339-43
43. Rao JH, So MK, Loening AM, Gambhir SS. Creating self-illuminating quantum dot conjugates. Nat Protoc. 2006;1:1160-4
44. Chaniotakis N, Frasco MF. Bioconjugated quantum dots as fluorescent probes for bioanalytical applications. Anal Bioanal Chem. 2010;396:229-40
45. Biju V, Mundayoor S, Omkumar RV, Anas A, Ishikawa M. Bioconjugated quantum dots for cancer research: Present status, prospects and remaining issues. Biotechnol Adv. 2010;28:199-213
46. Krull UJ, Algar WR, Tavares AJ. Beyond labels: A review of the application of quantum dots as integrated components of assays, bioprobes, and biosensors utilizing optical transduction. Anal Chim Acta. 2010;673:1-25
47. Rao JH, Xia ZY. Biosensing and imaging based on bioluminescence resonance energy transfer. Curr Opin Biotech. 2009;20:37-44
48. Mattoussi H, Sapsford KE, Pons T, Medintz IL. Biosensing with luminescent semiconductor quantum dots. Sensors-Basel. 2006;6:925-53
49. Rogach AL, Klar TA, Lupton JM, Meijerink A, Feldmann J. Energy transfer with semiconductor nanocrystals. J Mater Chem. 2009;19:1208-21
50. Rogach AL. Fluorescence energy transfer in hybrid structures of semiconductor nanocrystals. Nano Today. 2011;6:355-65
51. Mattoussi H, Clapp AR, Medintz IL. Forster resonance energy transfer investigations using quantum-dot fluorophores. Chemphyschem. 2006;7:47-57
52. Medintz IL, Sapsford KE, Berti L. Materials for fluorescence resonance energy transfer analysis: Beyond traditional donor-acceptor combinations. Angew Chem Int Ed. 2006;45:4562-88
53. Rotello VM, Agasti SS, Rana S, Park MH, Kim CK, You CC. Nanoparticles for detection and diagnosis. Adv Drug Deliver Rev. 2010;62:316-28
54. Krull UJ, Algar WR. New opportunities in multiplexed optical bioanalyses using quantum dots and donor-acceptor interactions. Anal Bioanal Chem. 2010;398:2439-49
55. Mattoussi H, Medintz IL. Quantum dot-based resonance energy transfer and its growing application in biology. Phys Chem Chem Phys. 2009;11:17-45
56. Chaniotakis N, Frasco MF. Semiconductor Quantum Dots in Chemical Sensors and Biosensors. Sensors. 2009;9:7266-86
57. Weissleder R, Tung CH, Mahmood U, Bogdanov A. In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nat Biotechnol. 1999;17:375-8
58. Kim K, Lee S, Ryu JH, Park K, Lee A, Lee SY. et al. Polymeric Nanoparticle-Based Activatable Near-Infrared Nanosensor for Protease Determination In Vivo. Nano Lett. 2009;9:4412-6
59. Kircher MF, Weissleder R, Josephson L. A dual fluorochrome probe for imaging proteases. Bioconjugate Chem. 2004;15:242-8
60. Stockholm D, Bartoli M, Sillon G, Bourg N, Davoust J, Richard I. Imaging calpain protease activity by multiphoton FRET in living mice. J Mol Biol. 2005;346:215-22
61. Medintz IL, Clapp AR, Brunel FM, Tiefenbrunn T, Uyeda HT, Chang EL. et al. Proteolytic activity monitored by fluorescence resonance energy transfer through quantum-dot-peptide conjugates. Nat Mater. 2006;5:581-9
62. Shi L, De Paoli V, Rosenzweig N, Rosenzweig Z. Synthesis and application of quantum dots FRET-based protease sensors. J Am Chem Soc. 2006;128:10378-9
63. Chang E, Miller JS, Sun J, Yu WW, Colvin VL, Drezek R. et al. Protease-activated quantum dot probes. Biochem Bioph Res Co. 2005;334:1317-21
64. Rosenzweig Z, Shi LF, Rosenzweig N. Luminescent quantum dots fluorescence resonance energy transfer-based probes for enzymatic activity and enzyme inhibitors. Anal Chem. 2007;79:208-14
65. Medintz IL, Prasuhn DE, Feltz A, Blanco-Canosa JB, Susumu K, Stewart MH. et al. Quantum Dot Peptide Biosensors for Monitoring Caspase 3 Proteolysis and Calcium Ions. Acs Nano. 2010;4:5487-97
66. Clapp AR, Goldman ER, Uyeda HT, Chang EL, Whitley JL, Medintz IL. Monitoring of Enzymatic Proteolysis Using Self-Assembled Quantum Dot-Protein Substrate Sensors. J Sensors. 2008;2008:1-10
67. Chen W, Biswas P, Cella LN, Kang SH, Mulchandani A, Yates MV. A quantum-dot based protein module for in vivo monitoring of protease activity through fluorescence resonance energy transfer. Chem Commun. 2011;47:5259-61
68. Kim HS, Kim YP, Oh YH, Oh E. Chip-based protease assay using fluorescence resonance energy transfer between quantum dots and fluorophores. Biochip J. 2007;1:228-33
69. Medintz IL, Boeneman K, Mei BC, Dennis AM, Bao G, Deschamps JR. et al. Sensing Caspase 3 Activity with Quantum Dot-Fluorescent Protein Assemblies. J Am Chem Soc. 2009;131:3828-9
70. Yun CS, Javier A, Jennings T, Fisher M, Hira S, Peterson S. et al. Nanometal surface energy transfer in optical rulers, breaking the FRET barrier. J Am Chem Soc. 2005;127:3115-9
71. Jennings TL, Singh MP, Strouse GF. Fluorescent lifetime quenching near d = 1.5 nm gold nanoparticles: probing NSET validity. J Am Chem Soc. 2006;128:5462-7
72. Jennings TL, Schlatterer JC, Singh MP, Greenbaum NL, Strouse GF. NSET molecular beacon analysis of hammerhead RNA substrate binding and catalysis. Nano Lett. 2006;6:1318-24
73. Dulkeith E, Morteani AC, Niedereichholz T, Klar TA, Feldmann J, Levi SA. et al. Fluorescence quenching of dye molecules near gold nanoparticles: radiative and nonradiative effects. Phys Rev Lett. 2002;89:203002
74. Pons T, Medintz IL, Sapsford KE, Higashiya S, Grimes AF, English DS. et al. On the quenching of semiconductor quantum dot photoluminescence by proximal gold nanoparticles. Nano Lett. 2007;7:3157-64
75. Nabiev I, Sukhanova A, Devy M, Venteo L, Kaplan H, Artemyev M. et al. Biocompatible fluorescent nanocrystals for immunolabeling of membrane proteins and cells. Anal Biochem. 2004;324:60-7
76. Lein M, Nowak L, Jung K, Koenig F, Lichtinghagen R, Schnorr D. et al. Analytical aspects regarding the measurement of metalloproteinases and their inhibitors in blood. Clin Biochem. 1997;30:491-6
77. Nikkola J, Vihinen P, Vuoristo MS, Kellokumpu-Lehtinen P, Kahari VM, Pyrhonen S. High serum levels of matrix metalloproteinase-9 and matrix metalloproteinase-1 are associated with rapid progression in patients with metastatic melanoma. Clin Cancer Res. 2005;11:5158-66
78. Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387-92
79. Denis LJ, Verweij J. Matrix metalloproteinase inhibitors: present achievements and future prospects. Invest New Drugs. 1997;15:175-85
80. Hidalgo M, Eckhardt SG. Development of matrix metalloproteinase inhibitors in cancer therapy. J Natl Cancer Inst. 2001;93:178-93
81. Loening AM, Fenn TD, Wu AM, Gambhir SS. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng Des Sel. 2006;19:391-400
82. Rao JH, Xia ZY, Xing Y, So MK, Koh AL, Sinclair R. Multiplex Detection of Protease Activity with Quantum Dot Nanosensors Prepared by Intein-Mediated Specific Bioconjugation. Anal Chem. 2008;80:8649-55
83. Tong AK, Li Z, Jones GS, Russo JJ, Ju J. Combinatorial fluorescence energy transfer tags for multiplex biological assays. Nat Biotechnol. 2001;19:756-9
84. Suzuki M, Husimi Y, Komatsu H, Suzuki K, Douglas KT. Quantum dot FRET Biosensors that respond to pH, to proteolytic or nucleolytic cleavage, to DNA synthesis, or to a multiplexing combination. J Am Chem Soc. 2008;130:5720-5
Author contact
![]() Corresponding author: Young-Pil Kim, PhD, Department of Life Science, Hanyang University, Seoul 133-791, Republic of Korea. Tel: +82-2-2220-2560; Fax: +82-2-2299-3495. E-mail: ypilkimac.kr
Corresponding author: Young-Pil Kim, PhD, Department of Life Science, Hanyang University, Seoul 133-791, Republic of Korea. Tel: +82-2-2220-2560; Fax: +82-2-2299-3495. E-mail: ypilkimac.kr
 Global reach, higher impact
Global reach, higher impact